Introduction
Breast cancer is among the leading causes of
cancer-related death in women, with an estimated 1.7 million cases
and 521,900 mortalities registered in 2012 worldwide, according to
the global cancer statistics published in 2015 (1). Despite improvements in therapeutic
strategies for the treatment of breast cancer, such as surgery,
radiotherapy and chemotherapy, the survival rate of breast cancer
is only ~20% for metastatic disease, compared with 90% for
localized disease (2). Thus, breast
cancer continues to be a major medical issue among women worldwide,
and the development of novel therapies for the treatment of breast
cancer is urgently required.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs of ~22 nucleotides in length, which affects the expression of
target genes via binding to their 3′-untranslated regions
(3′-UTRs). The interactions between miRNAs and their target genes
can affect various biological behaviors, such as cell
proliferation, differentiation, apoptosis, invasion and metastasis.
In recent years, certain miRNAs have been confirmed as oncogenes or
tumor suppressors in the tumorigenesis and metastasis of breast
cancer. For example, Zhang et al (3) demonstrated that miR-409-3p suppressed
the growth and invasive ability of breast cancer cells by targeting
Akt1, and Chen et al (4)
found that miR-340 inhibited cell migration and invasion through
the targeting of MYO10 in breast cancer. In addition, Zhang et
al (5) showed that miR-33a
suppressed breast cancer cell proliferation and metastasis by
targeting ADAM9 and ROS1, and Song et al (6) suggested that miR-200c inhibited breast
cancer proliferation by targeting KRAS. Furthermore, a recent study
indicated that miR-520d serves a key role in the progression and
metastasis of colorectal cancer by targeting CTHRC1 (7). However, the role of miR-520c-3p in
breast cancer cells remains unclear.
Interleukin-8 (IL-8) is a chemokine that serves key
roles in numerous biological processes, including cell
proliferation, migration, angiogenesis and metastasis (8–12).
Studies have identified high levels of serum IL-8 in patients with
various types of cancer (13),
including pancreatic (14),
prostate (15) and breast cancer
(16) Furthermore, this was
correlated with the survival rate of patients with breast cancer,
and was associated with increased rates of metastasis (16). IL-8 is also secreted by cancer cells
undergoing epithelial-mesenchymal transition (EMT) and promotes
adjacent epithelial-like cells to enter EMT, thus promoting the
development and metastasis of carcinoma (17). These findings indicate that
interruption of the IL-8 signaling pathway may be a potential
treatment target in mesenchymal cells or aggressive malignant
cancers. In our previous study, it was predicted that miR-520c-3p
directly targeted the 3′-UTR of IL-8, and to the best of our
knowledge, no published studies have investigated the relationship
between miR-520c-3p and IL-8 in breast cancer. Therefore, the
present study was conducted to further investigate whether
miR-520c-3p interacts with IL-8 and regulate EMT in breast cancer
cells.
Materials and methods
Cell lines and culture
The human breast cancer cell lines MCF-7 and T47D,
and 293T cells were purchased from the Chinese Academy of Sciences
Cell Bank (Shanghai, China). 293T and MCF-7 cells were grown in
Dulbeccos modified Eagles medium (DMEM), and T47D cells were
cultured in RPMI-1640 medium (both from Gibco, Carlsbad, CA, USA).
All culture medium was supplemented with 10% fetal bovine serum
(FBS; HyClone, Logan, UT, USA) and 1% penicillin-streptomycin, and
cells were cultured in 100% humidity in a cell culture incubator at
37°C with 5% CO2, and passaged every 2–3 days.
Bioinformatic analysis and target
prediction
The mature mRNA sequence of IL-8 was acquired from
the NCBI GenBank (reference sequence: NM_000584.3). The miRNAs
predicted to target IL-8 were obtained from the online miRNA target
prediction programs TargetScan (human) (http://www.targetscan.org), miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
and miRanda (http://www.microRNA.org). Only miRNAs
common to at least two online searches were considered for further
investigation in the present study. The miRNA sequences used for
analysis were retrieved from miRbase (http://www.mirbase.org).
Plasmid constructs
The wild-type (wt) 3′-UTR of human IL-8 containing
the predicted target site of miR-520c-3p was cloned from MCF-7
human genomic DNA, and amplified by PCR with the following primers:
5′-ATCGCCGTGTAAAAGGAAGAGGGCTGAGGAATTCAT-3′ (forward) and
5′-CACTGGACTAGTGGAGGAGAGCACATAAAAACATC-3′ (reverse). The fragments
were inserted into the HindIII and BamHI sites of a
pcDNA3.1-luciferase vector to generate
pcDNA3.1-luciferase-IL-8-3′-UTR-wt, according to a previously
reported method (18). A mutant
(mut)-type pcDNA3.1-luciferase-IL-8-3′-UTR-mut, containing 10
mutations in the 3′-UTR target site of the miR-520c-3p seed
sequence, was constructed using the following primers:
5′-GTGATGTTGTGAGGACATGTGGCGTTACAATAAGTTTTTTCATCA-3′ (forward) and
5′-TGTTATGATGAAAAAACTTATTGTAACGCCACATGTCCTCACAAC-3′ (reverse),
using a previously reported mutation method (19). hsa-pri-miR520c-3p microRNA
overexpression plasmids was constructed into pdsAAV-CB-EGFP using
the following primers: 5′-AATTTCAGGTCCCGGAGGAGGATTGCCCGTTGATGA-3′
(forward) and 5′-CACCACCACCGGATCCTACATACTAGTGCTTGGGC-3′ (reverse).
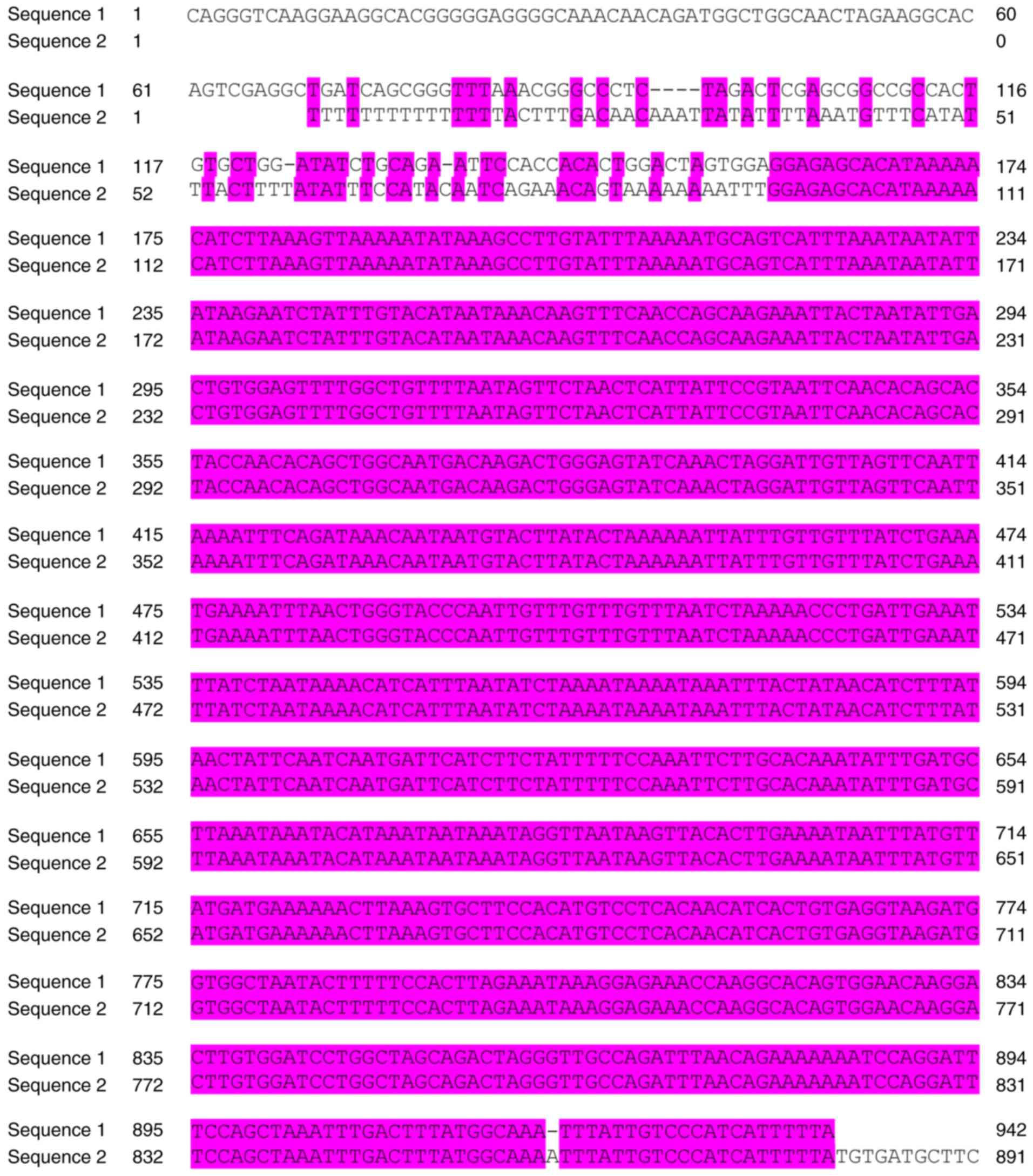
The sequence alignments of human miR-520c-3p, the predicted binding
site in the wt IL-8 3′-UTR, and the point-mutated sequence are
shown in Fig. 1. The
pcDNA3.1-luciferase plasmid was amplified from a pGL3-basic vector
(Promega, Madison, WI, USA) into pcDNA3.1(+) (Invitrogen, Carlsbad,
CA, USA), and recombined with insertion of a firefly luciferase
gene (20). pRL-SV40, containing
wild-type Renilla luciferase, was purchased from Promega for
fluorescence labeling.
Dual-luciferase reporter assays
The constructs containing the wt or mut 3′-UTR
sequences of IL-8, the miR-520c-3p overexpression or control
vector, and pRL-SV40 were transfected using a calcium phosphate
cell transfection method for the dual-luciferase reporter assays.
Cells were cultured in 24-well plates (0.1–0.2 million cells/well)
in triplicate for 18–24 h prior to transfection, then 500 ng of
pcDNA3.1-luciferase-IL-8-3′-UTR-wt or
pcDNA3.1-luciferase-IL-8-3′-UTR-mut and 500 ng of
pdsAAV-CB-EGFP-miR-520c-3p or pdsAAV-CB-EGFP were co-transfected
into cells, along with 10 ng pRL-SV40 as an internal control.
Luciferase activity was detected 24 h after transfection with a
Dual-Luciferase Reporter Assay System (Promega), according to the
manufacturer's instructions. To control for variations in vector
transfection, the activity of the reporter vector was normalized to
Renilla luciferase activity; thus, the ratio of firefly
luciferase/Renilla luciferase was assumed to represent the
effects of the pdsAAV-CB-EGFP-miR-520c-3p or pdsAAV-CB-EGFP vector
on IL-8.
Transfection
miR-520c-3p RNA mimics (AAAGUGCUUCC UUUUAGAGGGU)
were synthesized by RiboBio Co., Ltd. (Guangzhou, China) and stored
at −80°C. The miR-520c-3p mimics (50 nM) or scrambled miRNA
controls were transfected into MCF-7 and T47D cells using
Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's
protocol.
RNA extraction and reverse
transcription (RT)-quantitative PCR (qPCR)
At 48 h after transfection with miR-520c-3p mimics
or scrambled miRNA controls, total RNA was harvested from MCF-7 and
T47D cells with TRIzol reagent (Invitrogen), following the
supplier's protocol. The primers used for IL-8 and EMT-related
marker genes are shown in Table I
and were purchased from Invitrogen. All PCR amplifications were
performed using a SYBR-Green RT-qPCR kit (GenePharma, Shanghai,
China), and relative gene expression was analyzed based on the
comparative quantification cycle (Cq) method (21).
 | Table I.The primers of IL-8 and EMT-related
marker genes. |
Table I.
The primers of IL-8 and EMT-related
marker genes.
|
| Forward primer
5′-3′ |
|---|
| Gene names | Reverse primer
5′-3′ |
|---|
| IL-8 | F
5′-GGTGCAGTTTTGCCAAGGAG-3′ |
|
| R
5′-TTCCTTGGGGTCCAGACAGA-3′ |
| E-cadherin | F
5′-TCCAGTGAACAACGATGGCA-3′ |
|
| R
5′-CCTGGGCAGTGTAGGATGTG-3′ |
| Vimentin | F
5′-GGACCAGCTAACCAACGACA-3′ |
|
| R
5′-AAGGTCAAGACGTGCCAGAG-3′ |
| Fibronectin | F
5′-AGCCTGGGAGCTCTATTCCA-3′ |
|
| R
5′-CTTGGTCGTACACCCAGCTT-3′ |
| GAPDH | F
5′-CAGCGACACCCACTCCTC-3′ |
|
| R
5′-TGAGGTCCACCACCCTGT-3′ |
Western blotting
The breast cancer cell lines MCF-7 and T47D were
transfected with miR-520c-3p mimics or scrambled miRNA controls
using Lipofectamine 2000, following the manufacturer's
instructions. Cells were collected 48 h post-transfection in RIPA
lysis buffer (Boster, Wuhan, China), and were boiled at 100°C for
10 min to extract proteins with 5X SDS loading buffer. A BCA
protein assay kit (KeyGen Biotech Co., Ltd., Nanjing, China) was
used to measure the protein concentration. The extracted proteins
were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
and electro-transferred onto polyvinylidene fluoride (PVDF)
membranes (Millipore, Billerica, MA, USA). After blocking in 5%
fat-free milk in Tris-buffered saline with Tween-20 (TBST) at room
temperature, the membranes were incubated at 4°C overnight (>12
h) with the following primary antibodies: anti-IL-8 (1:2,000;
Abcam, Shanghai, China), anti-E-cadherin (1:1,000), anti-vimentin
(1:1,000), anti-fibronectin (1:1,000; all from Bioworld, Shanghai,
China), and anti-GAPDH (1:1,000; KangChen Bio-tech, Shanghai,
China). Horseradish peroxidase-conjugated secondary antibodies were
subsequently incubated with the membranes for 2 h at room
temperature. The protein bands were visualized using a Pierce ECL
Plus Western Blotting Substrate (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and GAPDH was used as an internal control to
normalize the expression levels of the aforementioned proteins.
Invasion and migration assays
The invasive abilities of MCF-7 and T47D cells were
assessed with Transwell assays. At 24 h post-transfection,
4×104 cells suspended in serum-free culture medium were
plated in the upper chambers of 24-well plates containing Transwell
inserts pre-coated with Matrigel (BD Biosciences, San Jose, CA,
USA), following the manufacturer's protocol, and the lower chambers
were filled with culture medium containing 10% FBS. After 48 h, the
cells that had invaded to the bottom of the chambers were fixed
with 4% paraformaldehyde and stained with 0.1% crystal violet. All
the assays were performed in triplicate.
The post-transfection migratory ability of cells was
assessed with a wound-healing assay. MCF-7 and T47D cells were
seeded into 6-well plates. When cells reached 80% confluency 24 h
after transfection, a 200-µl plastic pipette tip was used to draw
across the center of the cultured cells to generate a 1-mm ‘+ wound
area. The migration of MCF-7 and T47D cells into the wound area was
examined by microscopy.
Statistical analysis
All assays were performed in triplicate, and all
steps were executed independently at least three times. Data are
presented as the mean ± standard error of the mean (SEM).
Statistical analyses were performed using GraphPad Prism 5.0
(GraphPad Software, San Diego, CA, USA). Differences were
considered statistically significant at P<0.05.
Results
Bioinformatic analysis of IL-8
3′-UTR
The whole 3′-UTR of IL-8 is 1,192 bp and is
conserved in Homo sapiens, as shown in Fig. 2. To identify the potential miRNAs
that interact with the 3′-UTR of IL-8, three publically available
online databases (miRWalk, TargetScan and miRanda) were used. As
illustrated in Table II, the
miR-520 family (miR-520a, b, c, d and e) was commonly identified in
the three databases, and miR-520c-3p was predicted by all three
databases to regulate the 3′-UTR of IL-8.
 | Table II.The potential miRNAs occupy the
putative site on the 3′-UTR of IL-8. |
Table II.
The potential miRNAs occupy the
putative site on the 3′-UTR of IL-8.
| miRWalk | microRNA.org | TargetScan |
|---|
|
|
|
|---|
| Potential miRNAs | Seed sequences | Located sites | Potential miRNAs | Seed sequences | Located sites | Potential miRNAs | Seed sequences | Located sites |
|---|
| miR-302a | UGGAAGCACUU | 588–598 | miR-124 | GUGCCUU | 508–514 | miR-302c-3p | AAGCACU | 591–597 |
| miR-302b | UGGAAGCACUU | 588–598 | miR-506 | GUGCCUU | 508–514 |
miR-520f-3p | AAGCACU | 591–597 |
| miR-302c | UGGAAGCACUU | 588–598 | miR-128 | CACUGUG | 504–510 | miR-302d-3p | AGCACUU | 592–598 |
| miR-302d | UGGAAGCACUU | 588–598 | miR-93 | AGCACUUU | 592–599 | miR-302a-3p | AGCACUU | 592–598 |
|
miR-520e | GGAAGCACUUU | 589–599 | miR-519d | GCACUUU | 593–599 | miR-302b-3p | AGCACUU | 592–598 |
| miR-526b | GGAAGCACUUU | 589–599 | miR-20b | AGCACUUU | 592–599 | miR-302c-3p | AGCACUU | 592–598 |
|
miR-520b | GGAAGCACUUU | 589–599 | miR-20a | AAGCACUUU | 591–599 | miR-302e-3p | AGCACUU | 592–598 |
| miR-302e | UGGAAGCACUU | 588–598 | miR-17 | AAGCACUUU | 591–599 | miR-372-3p | AGCACUU | 592–598 |
| miR-520f | GGAAGCACUU | 589–598 | miR-106b | AGCACUUU | 592–599 | miR-373-3p | AGCACUU | 592–598 |
|
miR-520a-3p | GGAAGCACUU | 589–598 | miR-106a | AAGCACUUU | 591–600 |
miR-520e | AGCACUU | 592–598 |
|
miR-520c-3p | GGAAGCACUU | 589–598 | miR-372 | AGCACUU | 592–598 |
miR-520d-3p | AGCACUU | 592–598 |
|
miR-520d-3p | GAAGCACUUU | 590–599 |
miR-520e | GGAAGCACUU | 589–598 |
miR-520a-3p | AGCACUU | 592–598 |
| miR-567 | AGAACAUACU | 991–1000 |
miR-520d-3p | GAAGCACUU | 590–598 |
miR-520b | AGCACUU | 592–598 |
| miR-17 | AAGCACUUU | 588–596 |
miR-520a-3p | GGAAGCACUU | 589–598 |
miR-520c-3p | AGCACUU | 592–598 |
| miR-20a | AAGCACUUU | 588–596 | miR-373 | GAAGCACUU | 590–598 | miR-20a-5p | GCACUUUA | 593–600 |
| miR-106a | AAGCACUUU | 588–596 | miR-302d | UGGAAGCACUU | 588–598 | miR-93-5p | GCACUUUA | 593–600 |
| miR-106b | AGCACUUUA | 587–595 | miR-302c | UGGAAGCACUU | 588–598 | miR-526b-3p | GCACUUUA | 593–600 |
| miR-373 | GAAGCACUU | 589–596 | miR-302b | UGGAAGCACUU | 588–598 | miR-106a-5p | GCACUUUA | 593–600 |
| miR-656 | UAUAAUAUU | 87–95 | miR-302a | UGGAAGCACUU | 588–598 | miR-106b-5p | GCACUUUA | 593–600 |
| miR-23a | AAUGUGAU | 817–824 | miR-302e | UGGAAGCACUU | 588–598 | miR-519d-3p | GCACUUUA | 593–600 |
| miR-93 | AGCACUUU | 588–595 |
miR-520c-3p | GGAAGCACUU | 589–598 | miR-20b-5p | GCACUUUA | 593–600 |
| miR-23b | AAUGUGAU | 817–824 |
miR-520b | GGAAGCACUU | 589–598 | miR-17-5p | GCACUUUA | 593–600 |
| miR-106b | AGCACUUU | 588–595 | miR-154 | UAACCU | 644–649 |
| miR-372 | AGCACUUU | 588–595 |
|
|
|
|
|
|
| miR-340 | GCUUUAUA | 50–57 |
|
|
|
|
|
|
| miR-20b | AGCACUUU | 588–595 |
|
|
|
|
|
|
| miR-621 | UGCUAGCC | 705–712 |
|
|
|
|
|
|
| miR-634 | UGCUGGUU | 127–134 |
|
|
|
|
|
|
| miR-653 | UUCAACAC | 1113–1120 |
|
|
|
|
|
|
| miR-656 | UAUAAUAU | 88–95 |
|
|
|
|
|
|
| miR-889 | GAUAUUAA | 412–419 |
|
|
|
|
|
|
| miR-944 | AAUAAUUU | 941–948 |
|
|
|
|
|
|
| miR-1294 | ACCUCACA | 619–626 |
|
|
|
|
|
|
miR-520c-3p regulates IL-8 expression
by directly targeting its 3′-UTR
The plasmids pcDNA3.1(+)-luciferase-IL-8-3′-UTR-wt
and pcDNA3.1(+)-luciferase-IL-8-3′-UTR-mut were cloned for use in
dual-luciferase reporter assays to validate the 3′-UTR of IL-8 as a
direct target of miR-520c-3p. The relative effect of miR-520c-3p
was determined as the ratio of firefly/Renilla luciferase
activities in the 293T cells in reference to the researches of Wang
et al and Shi et al (22,23).
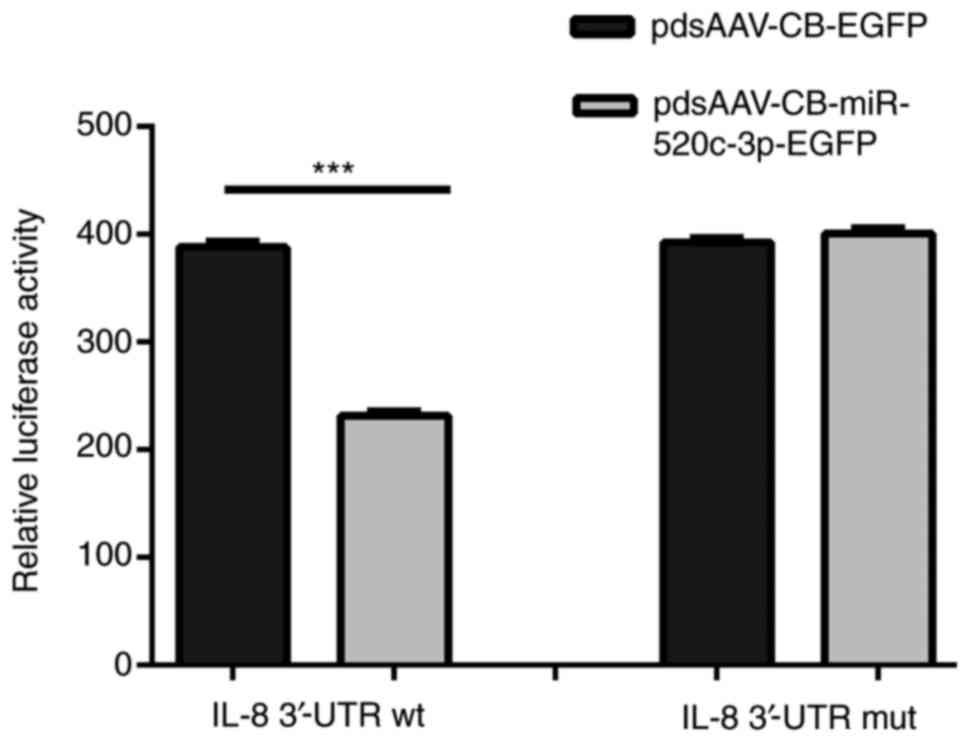
As shown in Fig. 3, the results
verified that the relative luciferase activity of
pcDNA3.1(+)-luciferase-IL-8-3′-UTR-wt in the 293T cells
co-transfected with miR-520c-3p was markedly decreased compared
with that in cells transfected with scrambled miRNA control
(P<0.001). However, no significant inhibitory effect of
miR-520c-3p was detected when the 3′-UTR of IL-8 was mutated,
indicating that miR-520c-3p inhibited luciferase reporter activity
by direct interaction with the wt 3′-UTR of IL-8. These results
were consistent with bioinformatic predictions, and indicated that
IL-8 may be directly targeted and functionally regulated by
miR-520c-3p in MCF-7 and T47D cells. RT-qPCR, western blotting,
Transwell and wound-healing assays were subsequently performed to
further verify the effects of miR-520c-3p on IL-8 expression in
MCF-7 and T47D cells.
miR-520c downregulates the expression
of IL-8 and alters the expression of EMT-related factors in breast
cancer cell lines
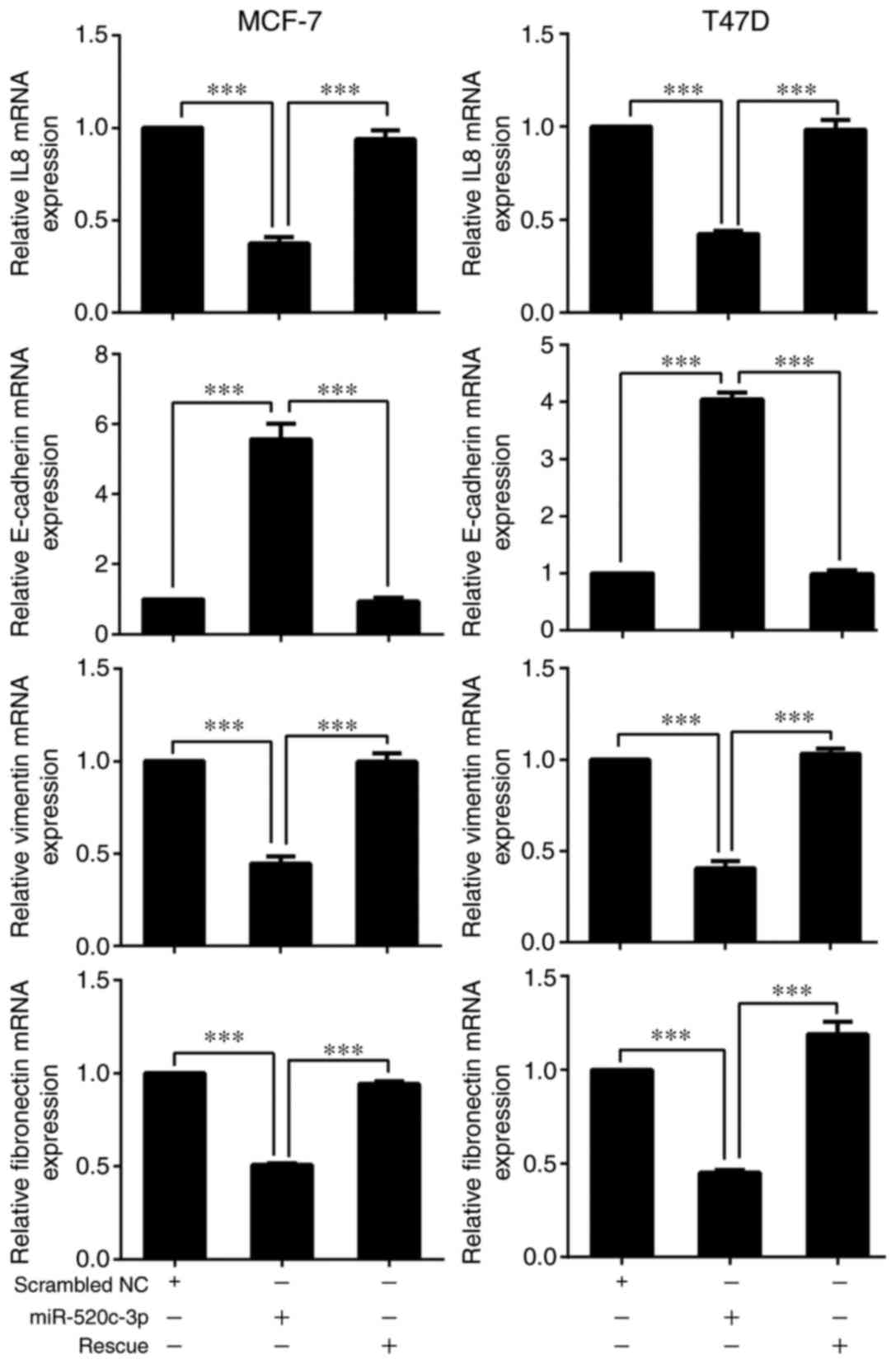
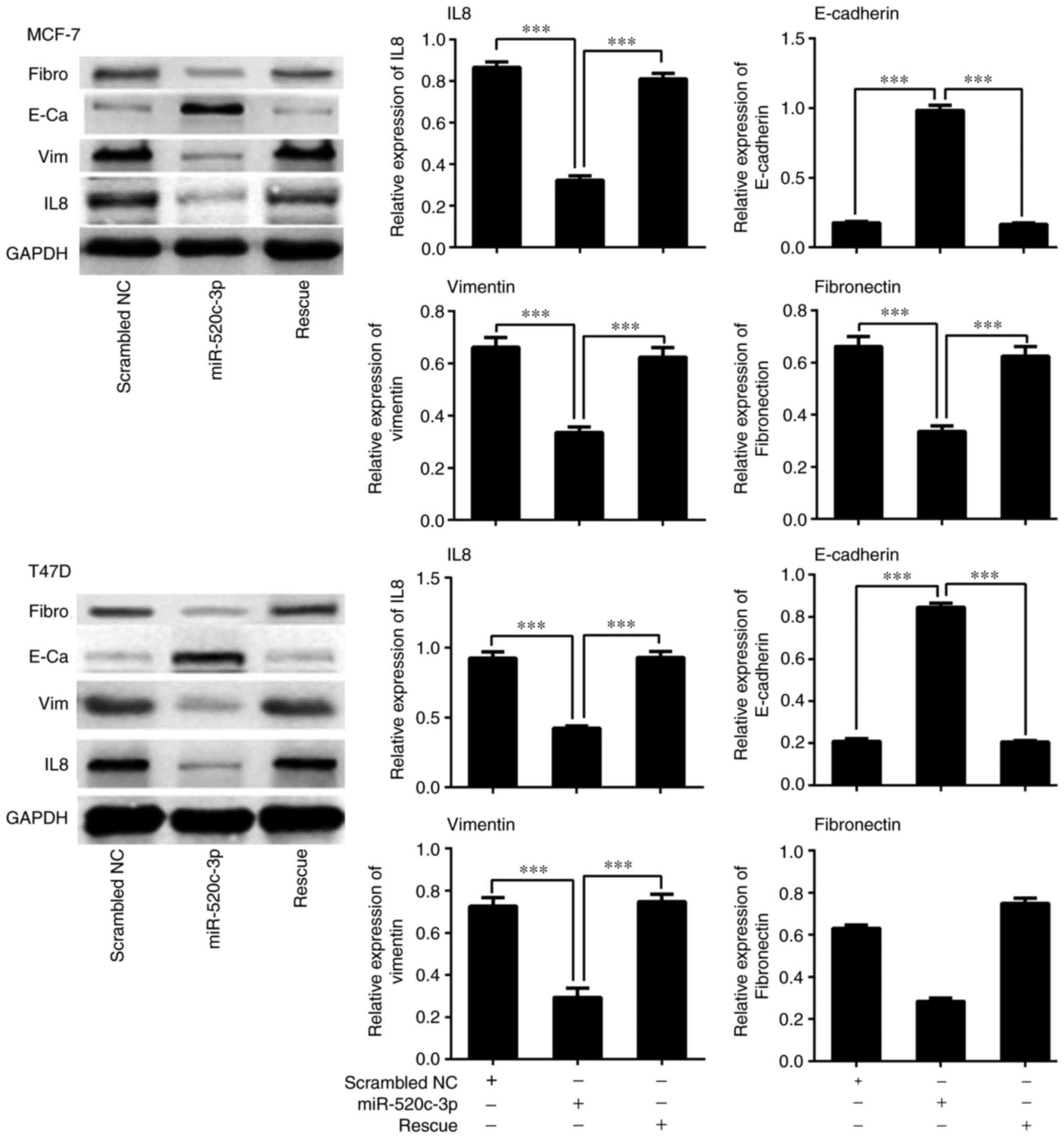
To further investigate whether miR-520c-3p
effectively regulates the expression of IL-8 and EMT-related genes,
including E-cadherin, vimentin and fibronectin, in MCF-7 and T47D
cells, RT-qPCR and western blot analyses were performed. The
results confirmed that overexpression of miR-520c-3p significantly
reduced the expression of IL-8 and resulted in increased E-cadherin
and decreased vimentin and fibronectin levels in MCF-7 and T47D
cells, as compared with scrambled miRNA control transfectants
(Figs. 4 and 5; P<0.001). Conversely, overexpression
of IL-8 led to a decrease in E-cadherin expression and increases in
vimentin and fibronectin levels (P<0.001) (Figs. 4 and 5).
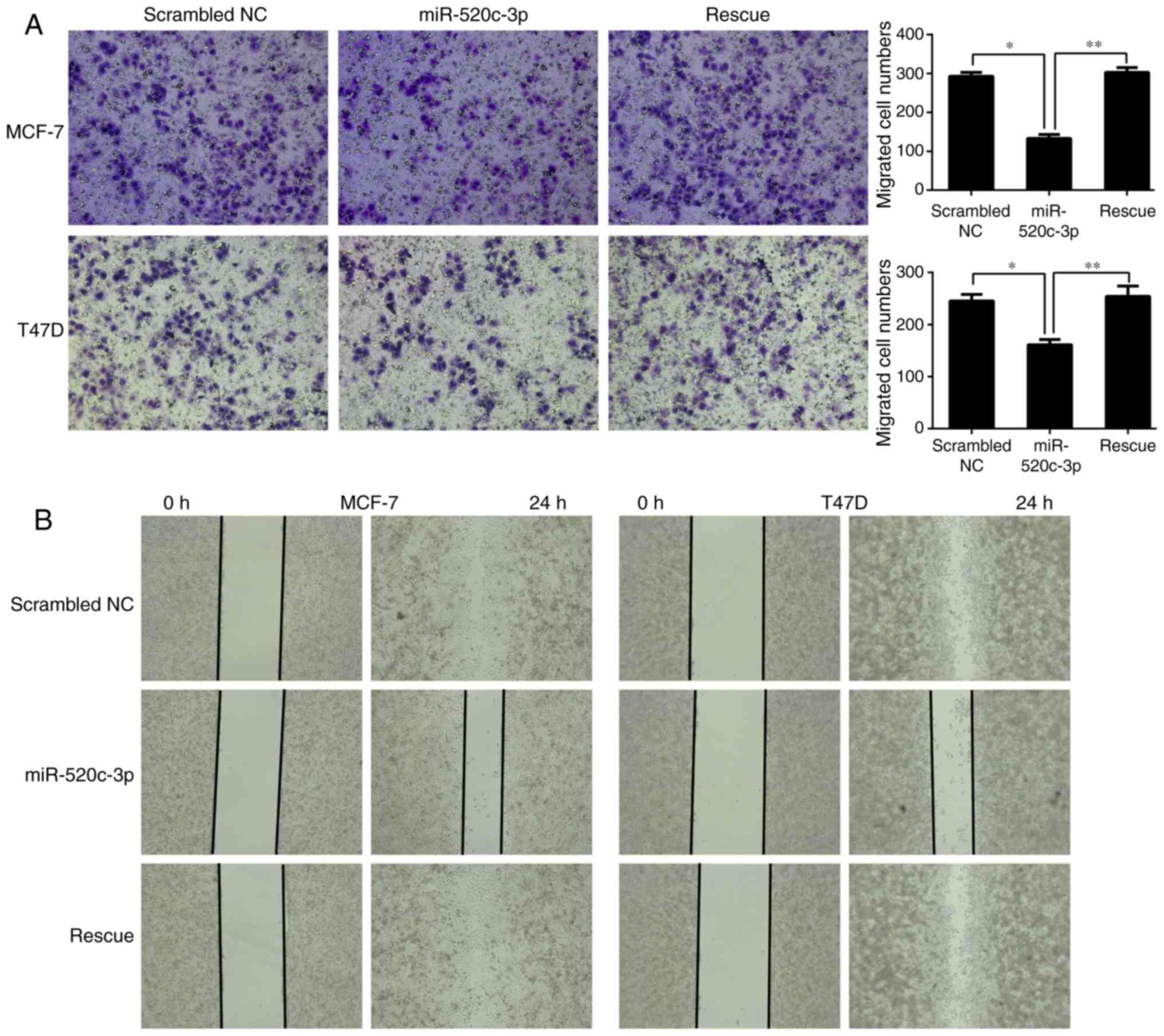
miR-520c-3p inhibits the invasion and
migration of MCF-7 and T47D cells
IL-8 has been reported to regulate tumor invasion
and metastasis (9–16), and the present study verified that
miR-520c-3p regulates the expression of IL-8 by directly targeting
its 3′-UTR. Transwell and wound-healing assays were subsequently
performed to evaluate whether miR-520c-3p co-transfected with
pcDNA3.1(+)-luciferase-IL-8-3′-UTR affected the invasive and
migratory abilities of the MCF-7 and T47D breast cancer cell lines.
As shown in Fig. 6, the results
demonstrated that overexpression of miR-520c-3p significantly
inhibited the invasion and migration of MCF-7 and T47D cells, while
rescue experiments attenuated this inhibition, restoring the
migratory and invasive abilities. This indicated that the
metastatic capacities of cells in the rescue group were promoted by
IL-8 expression.
Discussion
Breast cancer is a malignant cancer that is highly
prevalent in females, and has a particularly high incidence rate
and mortality rate in China (24).
Over the past several decades, significant advances have been made
in our understanding of breast cancer-related mechanisms and in the
treatment of breast cancer. Tumor grade, stage and lymph node
status are known to be important factors that affect the prognosis
of patients with breast cancer, and metastasis is a leading
contributor to poor prognosis and a high rate of mortality. Many
patients with breast cancer develop metastases prior to diagnosis,
which markedly influences therapeutic efficacy and prognosis. It is
well established that EMT in tumor cells promotes migration and
invasion (25,26). Therefore, EMT is closely associated
with the metastasis of solid cancers, and suppressing EMT in tumor
cells is an important therapeutic strategy.
Numerous studies have confirmed that inflammation
serves crucial roles in the development and progression of cancer,
and many types of cancer originate from a background of infection
and inflammation. Inflammatory cells in the tumor microenvironment,
along with their secreted chemokines and cytokines, can affect the
progression of various types of tumors by regulating the
development, transformation and differentiation of tumor cells
(27,28). For example, growth factors and
inflammatory factors in the microenvironment of pancreatic cancer
can sustain and promote the growth of cancer cells, and have the
ability to inhibit immune responses (29). Therefore, cytokines and their
functional regulation have become topics of great interest in the
context of cancer research and treatment.
IL-8 is a cytokine that induces the chemotaxis of
lymphocytes and neutrophils, and may promote tumor angiogenesis;
the antisense nucleotide of IL-8 suppresses macrophage-induced
angiogenesis, which suggests a function of IL-8 in mediating
angiogenesis in inflammation and tumor progression of various types
of cancer (11,30,31). A
large number of clinical studies and experimental research have
demonstrated that IL-8 serves critical roles in the development of
breast cancer (32). Research has
shown that EMT in cancer depends on comprehensive stimulation by
soluble growth factors, cytokines and extracellular matrix
components in the tumor microenvironment; TGF-β, FGF, EGF, IL-8 and
IL-6 promotes the occurrence of EMT in various kinds of tumors. The
metastasis of breast cancer is closely associated with the
occurrence of EMT; however, the relationship between IL-8 and EMT
in breast cancer and its molecular mechanisms remain unclear. Thus,
the present study aimed to investigate the effect of miR-520c-3p
overexpression on IL-8 and EMT in the breast cancer cell lines
MCF-7 and T47D, to explore its potential molecular mechanisms and
provide an experimental basis for the treatment of breast
cancer.
Accumulating studies have demonstrated that many
different miRNAs serve critical roles in the diagnosis and
treatment of breast cancer by influencing tumorigenesis, cell cycle
regulation and differentiation, or by regulating the expression of
target genes involved in the development and prognosis of breast
cancer (3–6). Previous research has indicated that
IL-8 regulates EMT and contributes to cell proliferation, migration
and the regulation of angiogenesis in different types of tumor,
including breast cancer. However, to the best of our knowledge, no
study has confirmed the association between miR-520c-3p and IL-8 in
breast cancer. The data from the present study demonstrated that
overexpression of miR-520c-3p negatively regulated the expression
of IL-8 in the MCF-7 and T47D cells. Therefore, we hypothesized
that miR-520c-3p may inhibit EMT in breast cancer cells in
vitro by directly targeting IL-8.
Dual-luciferase reporter assays, western blotting
and RT-qPCR verified that overexpression of miR-520c-3p could
directly suppress the expression of endogenous IL-8 in MCF-7 and
T47D cells at a post-transcriptional level, confirming IL-8 to be a
target gene of miR-520c-3p, as predicted by the bioinformatic
analysis.
The effects of miR-520c-3p expression on MCF-7 and
T47D cells were also investigated in vitro. Transwell and
wound-healing assays showed that miR-520c-3p mimics effectively
suppressed the invasion and migration of breast cancer cells at a
post-transcriptional level, whereas the rescue of IL-8 expression
attenuated the effects of the miR-520c-3p mimics, increasing the
invasion and migration of MCF-7 and T47D cells. Thus, miR-520c-3p
appears to act as a suppressor of invasion and migration in breast
cancer cells. In addition, western blot analysis indicated a
regulatory effect of miR-520c-3p on the expression of the
EMT-related proteins E-cadherin, vimentin and fibronectin,
indicating that EMT may be significantly suppressed by miR-520c-3p
in breast cancer cells. Therefore, we concluded that overexpression
of miR-520c-3p inhibits IL-8 protein expression and suppresses EMT
in the breast cancer cell lines MCF-7 and T47D.
In conclusion, to the best of our knowledge, the
present study was the first to identify IL-8 as a target gene of
miR-520c-3p. The invasion, migration, and EMT of MCF-7 and T47D
cells was negatively regulated by miR-520c-3p overexpression, and
this result was likely associated with the expression of IL-8. The
mechanism regarding the regulation of IL-8 expression by
miR-520c-3p remains to be established.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang X, Lu H, Urvalek AM, Li T, Yu L,
Lamar J, DiPersio CM, Feustel PJ and Zhao J: KLF8 promotes human
breast cancer cell invasion and metastasis by transcriptional
activation of MMP9. Oncogene. 30:1901–1911. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang G, Liu Z, Xu H and Yang Q:
miR-409-3p suppresses breast cancer cell growth and invasion by
targeting Akt1. Biochem Biophys Res Commun. 469:189–195. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen CP, Sun ZL, Lu X, Wu WX, Guo WL, Lu
JJ, Han C, Huang JQ and Fang Y: MiR-340 suppresses cell migration
and invasion by targeting MYO10 in breast cancer. Oncol Rep.
35:709–716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang C, Zhang Y, Ding W, Lin Y, Huang Z
and Luo Q: MiR-33a suppresses breast cancer cell proliferation and
metastasis by targeting ADAM9 and ROS1. Protein Cell. 6:881–889.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye
F and Xie X, Chen J, Tang H and Xie X: miR-200c inhibits breast
cancer proliferation by targeting KRAS. Oncotarget. 6:34968–34978.
2015.PubMed/NCBI
|
|
7
|
Yan L, Yu J, Tan F, Ye GT, Shen ZY, Liu H,
Zhang Y, Wang JF, Zhu XJ and Li GX: SP1-mediated microRNA-520d-5p
suppresses tumor growth and metastasis in colorectal cancer by
targeting CTHRC1. Am J Cancer Res. 5:1447–1459. 2015.PubMed/NCBI
|
|
8
|
Freund A, Chauveau C, Brouillet JP, Lucas
A, Lacroix M, Licznar A, Vignon F and Lazennec G: IL-8 expression
and its possible relationship with estrogen-receptor-negative
status of breast cancer cells. Oncogene. 22:256–265. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu YM, Webster SJ, Flower D and Woll PJ:
Interleukin-8/CXCL8 is a growth factor for human lung cancer cells.
Br J Cancer. 91:1970–1976. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seaton A, Scullin P, Maxwell PJ, Wilson C,
Pettigrew J, Gallagher R, O'Sullivan JM, Johnston PG and Waugh DJ:
Interleukin-8 signaling promotes androgen-independent proliferation
of prostate cancer cells via induction of androgen receptor
expression and activation. Carcinogenesis. 29:1148–1156. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Xu RC, Zhang XL, Niu XL, Qu Y, Li
LZ and Meng XY: Interleukin-8 secretion by ovarian cancer cells
increases anchorage-independent growth, proliferation, angiogenic
potential, adhesion and invasion. Cytokine. 59:145–155. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim JH, Frantz AM, Anderson KL, Graef AJ,
Scott MC, Robinson S, Sharkey LC, O'Brien TD, Dickerson EB and
Modiano JF: Interleukin-8 promotes canine hemangiosarcoma growth by
regulating the tumor microenvironment. Exp Cell Res. 323:155–164.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sanmamed MF, Carranza-Rua O, Alfaro C,
Oñate C, Martín-Algarra S, Perez G, Landazuri SF, Gonzalez A, Gross
S, Rodriguez I, et al: Serum interleukin-8 reflects tumor burden
and treatment response across malignancies of multiple tissue
origins. Clin Cancer Res. 20:5697–5707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Shi M, Yu GZ, Qin XR, Jin G, Chen
P and Zhu MH: Interleukin-8, a promising predictor for prognosis of
pancreatic cancer. World J Gastroenterol. 18:1123–1129. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chi N, Tan Z, Ma K, Bao L and Yun Z:
Increased circulating myeloid-derived suppressor cells correlate
with cancer stages, interleukin-8 and −6 in prostate cancer. Int J
Clin Exp Med. 7:3181–3192. 2014.PubMed/NCBI
|
|
16
|
Todorović-Raković N and Milovanović J:
Interleukin-8 in breast cancer progression. J Interferon Cytokine
Res. 33:563–570. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fernando RI, Castillo MD, Litzinger M,
Hamilton DH and Palena C: IL-8 signaling plays a critical role in
the epithelial-mesenchymal transition of human carcinoma cells.
Cancer Res. 71:5296–5306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Balhana R, Stoker NG, Sikder MH, Chauviac
FX and Kendall SL: Rapid construction of mycobacterial mutagenesis
vectors using ligation-independent cloning. J Microbiol Methods.
83:34–41. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bryksin AV and Matsumura I: Overlap
extension PCR cloning: A simple and reliable way to create
recombinant plasmids. Biotechniques. 48:463–465. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun H, Li QW, Lv XY, Ai JZ, Yang QT, Duan
JJ, Bian GH, Xiao Y, Wang YD, Zhang Z, et al: MicroRNA-17
post-transcriptionally regulates polycystic kidney disease-2 gene
and promotes cell proliferation. Mol Biol Rep. 37:2951–2958. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang N, Wei L, Huang Y, Wu Y, Su M, Pang
X, Wang N, Ji F, Zhong C, Chen T, et al: miR520c blocks EMT
progression of human breast cancer cells by repressing STAT3. Oncol
Rep. 37:1537–1544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
lyu Z, Mao Z, Wang H, Fang Y, Chen T, Wan
Q, Wang M, Wang N, Xiao J, Wei H, et al: MiR-181b targets Six2 and
inhibits the proliferation of metanephric mesenchymal cells in
vitro. Biochem Biophys Res Commun. 440:495–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kucuksayan H, Ozes ON and Akca H:
Downregulation of SATB2 is critical for induction of
epithelial-to-mesenchymal transition and invasion of NSCLC cells.
Lung Cancer. 98:122–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
da Cunha IW, Souza MJ, da Costa WH,
Amâncio AM, Fonseca FP, Zequi SC, Lopes A, Guimarães GC and Soares
F: Epithelial-mesenchymal transition (EMT) phenotype at invasion
front of squamous cell carcinoma of the penis influences
oncological outcomes. Urol Oncol. 34:433.e19–433.e26. 2016.
View Article : Google Scholar
|
|
27
|
Musolino C, Allegra A, Pioggia G and
Gangemi S: Immature myeloid-derived suppressor cells: A bridge
between inflammation and cancer (Review). Oncol Rep. 37:671–683.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Coffelt SB and de Visser KE: Cancer:
Inflammation lights the way to metastasis. Nature. 507:48–49. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roshani R, McCarthy F and Hagemann T:
Inflammatory cytokines in human pancreatic cancer. Cancer Lett.
345:157–163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee YS, Choi I, Ning Y, Kim NY,
Khatchadourian V, Yang D, Chung HK, Choi D, LaBonte MJ, Ladner RD,
et al: Interleukin-8 and its receptor CXCR2 in the tumour
microenvironment promote colon cancer growth, progression and
metastasis. Br J Cancer. 106:1833–1841. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reis ST, Leite KR, Piovesan LF,
Pontes-Junior J, Viana NI, Abe DK, Crippa A, Moura CM, Adonias SP,
Srougi M, et al: Increased expression of MMP-9 and IL-8 are
correlated with poor prognosis of bladder cancer. BMC Urol.
12:182012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Rong G and Kang H: Taxotere-induced
elevated expression of IL8 in carcinoma-associated fibroblasts of
breast invasive ductal cancer. Oncol Lett. 13:1856–1860.
2017.PubMed/NCBI
|