Introduction
The aryl hydrocarbon receptor (AHR) is a
ligand-activated transcription factor that is best known to mediate
the toxicities of dioxins and dioxin-like compounds (1). After ligand binding, AHR translocates
to the nucleus and dimerizes with the aryl hydrocarbon nuclear
translocator (ARNT) and activates the expression of a battery of
genes containing specific DNA-enhancer sequences, which are known
as xenobiotic-responsive elements (XREs) (2). These genes encode
xenobiotic-metabolizing enzymes, which are involved in
biotransformation and cellular detoxification process (3,4).
Phenotype of AHR-null mice showed developmental defects in the
hepatic, hematopoietic, cardiovascular and immune systems (5–9). As
such, AHR pathway is believed to contribute significantly to sense
the environmental chemicals. In addition to cellular
detoxification, AHR also regulates signal transduction pathways
involved in cellular melabolism, proliferation, differentiation,
and apoptosis (10). Several
studies have shown that AHR, which is expressed at aberrantly high
levels in a wide panel of tumors, plays a critical role in tumor
progression by enhancing tumor invasion and migration (11,12).
Increasing expression of AHR and its binding partner, ARNT, has
been noted in hepatocellular carcinoma (HCC) (13). Furthermore, recent evidence suggests
that the possibility that AHR contributes to cell invasion and
migration through upregulation of stem cell (e.g., Notch1,2,
Sox2 and Pou5F1/Oct4) and invasion/migration-associated
genes (e.g., Snail1, Snail2, Twist1,2, NDRG1 and p53)
(14–16).
It has been reported that ligand-induced expression
of AHR occurs at the transcriptional level. So far, no report
exists on the regulation of AHR expression at the translational
level (17,18). Translation initiation of mRNAs in
eukaryotic cells is a complex process and affects the overall rate
of protein synthesis (19). For
most mRNAs, the global translation initiation is mediated by
ribosome scanning whereby the translation initiation complexes
recognizes and binds to the cap structure at the 5′-end of mRNA and
scans the mRNA in 5′-3′ direction until the first AUG codon within
an optimal context is encountered. The global translation
initiation was inhibited by a variety of conditions including
nutrient deprivation, heat shock, apoptosis and viral infection.
Yet, despite this occurrence, a minority of cellular mRNA can still
be translated by internal ribosome entry sites (IRES)-dependent
translation initiation (20,21).
IRES elements were first characterized in picornaviruses that
contain long 5′-UTRs that naturally lack 5′-terminal cap structures
(22,23). IRES elements were also discovered in
several mammalian mRNAs encoding regulatory proteins induced in
growth control and programmed cell death. Fibroblast growth factor
2 (FGF2), insulin-like growth factor II (IGF2), the protooncogene
c-myc, NF-κB repressing factor (NRF), X-linked inhibitor of
apoptosis protein (XIAP), heat shock protein 70 (Hsp70) and Aurora
A kinase, have all been shown to contain IRES (24–30).
Since many transcription factors contain IRES, it was hypothesized
the AHR mRNA would also contain an IRES. In the present study,
evidence is provided demonstrating that the 5′-UTR of AHR mRNA
contains an IRES. Importantly, overexpression of AHR through
IRES-dependent translation initiation contributes to cancer cells
survival under drug stress. Therefore, these results imply that
translation initiation by internal ribosome entry may represent an
important mechanism for the regulation of AHR expression in cancer
cells.
Materials and methods
Construction of dicistronic reporter
plasmids
The initial dicistronic vector pRF containing the
reporter genes for Renilla luciferase (first cistron, RL)
and firefly luciferase (second cistron, FL) was described
previously (31,32). To obtain plasmids pR-NRF-F and
pR-AHR-F, we inserted the 5′-UTRs of AHR and NRF which were
synthesized by Sangon Biotech. Co. (Shanghai, China) into pRF
respectively. pBR-AHR-F was constructed by deleting SV40 promoter
from pR-AHR-F and used to analyze the cryptic promoter of AHR
5′-UTR. To detect the core region for IRES activity, AHR 5′-UTR was
deleted to different length sequences by PCR. The primers with
EcoRI and NdeI restriction sites are listed in
Table I. PCRs were performed using
PrimerSTAR Max DNA polymerase (Takara, Dalian, China) according to
the manufacturer's instructions and the resulting products were
inserted into pRF. All PCRs were conducted at 98°C for 3 min
followed by 32 cycles of 98°C for 10 sec, 68°C for 20 sec and 72°C
for 1 min, and 72°C for 5 min.
 | Table 1.Oligonucleotide primers used in PCR
for plasmids construct. |
Table 1.
Oligonucleotide primers used in PCR
for plasmids construct.
| No. | Sequences of
oligonucleotide primers 5′-3′ |
|---|
| –518 to −1 | F:
GGAATTCCATATGGCTCAGAACAGGGGCAGCCGTG |
|
| R:
CGGAATTCGGTGCCCAGCCGACGGCG |
| –493 to −1 | F:
GGAATTCCATATGCCCGAGCTCCGCAGGCGG |
|
| R:
CGGAATTCGGTGCCCAGCCGACGGCG |
| –404 to −1 | F:
GGAATTCCATATGCGGTCACGGGGCGCGGC |
|
| R:
CGGAATTCGGTGCCCAGCCGACGGCG |
| –313 to −1 | F:
GGAATTCCATATGGCGGGCATTGCCGCGCCG |
|
| R:
CGGAATTCGGTGCCCAGCCGACGGCG |
| –212 to −1 | F:
GGAATTCCATATGAGCTCACCTGTACTGGCGCGGG |
|
| R:
CGGAATTCGGTGCCCAGCCGACGGCG |
| –613 to −212 | F:
GGAATTCCATATGAGTGGCTGGGGAGTCCCGTCGA |
|
| R:
CGGAATTCTGCCTGGGCCTGGCGCAGTGA |
| –613 to −313 | F:
GGAATTCCATATGAGTGGCTGGGGAGTCCCGTCGA |
|
| R:
CGGAATTCCAGGTGAGGCGGCCCGGG |
| –613 to −493 | F:
GGAATTCCATATGAGTGGCTGGGGAGTCCCGTCGA |
|
| R:
CGGAATTCGCGCGGGCCACCAGTCCC |
| –613 to −518 | F:
GGAATTCCATATGAGTGGCTGGGGAGTCCCGTCGA |
|
| R:
CGGAATTCCCCAGCTTCCGTTCGGCTACACG |
Cell culture and treatment
All cell lines were obtained from American Type
Culture Collection. Cell lines HCT-8, Bel7402 and A2780/PTX were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific),
where other cell lines were cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific). A2780/PTX supplemented
with 0.3 mg/ml PTX and all cell lines were maintained in medium
supplemented with 10% fetal bovine serum (FBS) at 37°C, 5%
CO2. For drug treatment, A2780, MB231, Bel7302 and SW620
cell lines were cultured in medium supplemented with clinically
relevant concentration of PTX (0.2 and 0.4 µg/ml) and without PTX
for 24 h. PTX was purchased from Sigma.
Luciferase assays and transient
transfection
Cells were grown to 60–70% confluency in 24-well
plate and transfected with 1 µg of plasmid DNA using 2 µl
Lipofectamine™ 2000 (Invitrogen, Thermo Fisher Scientific, Inc.)
per well, according to the manufacturer's instructions. Cells were
harvested after incubation for 24 h at 37°C. Renilla and
firefly luciferase activities were quantitated using the
Dual-luciferase reporter system (Promega Corp.) and a GloMAX™ 2020
luminometer. Cell lysate (20 µl) was combined sequentially with
firefly then Renilla luciferase substrates (100 µl)
according to the manufacturer's instructions. Light emission was
measured 3 sec after addition of each of the substrates and
integrated over a 10-sec interval. These experiments were repeated
more than three times.
SiRNA transfection
RNAi duplexes for AHR and siNC (negative control)
duplexes were purchased from Santa Cruz Biotechnology (sc-29654;
sc-37007). Cells were seeded in RPMI-1640 or DMEM containing 10%
FBS and maintained for 24 h. The cells were transfected with siRNA
or siNC using Lipofectamine 2000 (Invitrogen, Thermo Fisher
Scientific) according to the manufacturer's protocol.
MTT assay
A2780, A549, MB231, Bel7402, HCT-8, SW620 and HEK293
cells were seeded at a density of 3,000 cells per well containing
100 µl of medium in a 96-well plate for 24 h, then transfected with
10 pmol siNC or siAHR using 1 µl Lipofectamine 2000 (Invitrogen,
Thermo Fisher Scientific) per well. After 72 h, the surviving cell
number was determined as described previously (33).
Reverse transcriptase (RT)-PCR and
quantitative (q) PCR
Total RNA was isolated from treated cells using
TRIzol reagent (Invitrogen) and was reverse transcribed using a
reserves transcription system (Takara) to generate a cDNA template
according to the manufacturer's instructions. RT-PCR was performed
with PrimerSTAR Max DNA polymerase (Takara) according to the
manufacturer's instructions. The PCR was conducted at 98°C for 3
min, followed by 32 cycles of 98°C for 10 sec, 68°C for 50 sec, and
72°C for 5 min. Amplification products were visualized on a 1%
agarose gel staining by ethidium bromide under UV light and
quantified by ImageJ software (NIH, Bethesda, MD, USA). qRCR was
performed with SYBR® Premix Ex Taq™ (Takara) according
to the manufacturer's instructions and quantified with a Roach
Lightcycler 480. The PCR was conducted at 95°C for 10 min, followed
by 40 cycles of 95°C for 10 sec, 60°C for 30 sec, and 95°C for 10
sec, 60°C for 60 sec and 95°C for 15 sec. The primers were: for R1
(41–210), forward, 5′-TGATAACTGGTCCGCAGTGGT-3′ and reverse,
5′-TACTGGCTCAATATGTGGCACAA-3′; for R2 (701–865), forward,
5′-TGCTTATCTACGTGCAAGTGATG-3′ and reverse,
5′-CCCATTTCATCAGGTGCATCTT-3′; for F1 (69–340) forward,
5′-CGAGCAGCTGCACAAAGCCAT-3′ and reverse,
5′-GCTCGCGCTCGTTGTAGATG-3′; for AHR 5′-UTR, forward,
5′-AGTGGCTGGGGAGTCCCGTC-3′ and reverse, 5′-GGTGCCCAGCCGACGGCG-3′;
for AHR, forward, 5′-CCCATATCCGAATGATTAAGACTG-3′ and reverse,
5′-CGTAAATGCTCTGTTCCTTCC-3; for β-actin, forward,
5′-TGAAGTGTGACGTGGACATC-3′ and reverse,
5′-GGAGGAGCAATGATCTTGAT-3′.
Protein analysis
Western blot analyses were performed using standard
procedures. Cells were harvested in cell lysis buffer RIPA (Roche)
and phosphatase inhibitors (Pierce). Cell extracts were separated
by 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)
membranes. The membranes were incubated with 5% milk in
Tris-buffered saline (TBS) overnight at 4°C. The blots were washed
three times in TBS, incubated with polyclonal antibodies directed
against AHR (1:1,000; sc-74571; Santa Cruz Biotechnology),
caspase-3 (1:500; ab32042; Abcam), survivin (1:5,000; ab76424;
Abcam) and β-actin (1:1,000; AA128; Beyotime Institute of
Biotechnology) for 4 h at room temperature, washed, incubated with
secondary antibodies respectively (1:1,000; A0216; Beyotime
Institute of Biotechnology) for 2 h, washed. Then proteins were
detected by a chemiluminescent detection system (Tanton, Shanghai,
China) and quantified by ImageJ software (NIH).
Statistical analysis
All experiments were performed at least three times.
The data were analyzed using Graphpad Prism 5.0 (La Jolla, CA, USA)
and presented as mean values ± SEM. Statistical analysis was
performed using a t-test. p<0.05 was set as the statistically
significant difference.
Results
AHR knockdown inhibits the growth of
cancer cells
AHR is highly expressed in a wide panel of tumors
and previous studies indicated that overexpression of AHR could
help cancer cells survive under cellular stress (34,35).
To address whether AHR imparts survival to cancer cells, AHR was
knocked down in a variety of cell lines including human ovarian
cancer cell line (A2780), human lung cancer cell line (A549), human
breast cancer cell line (MDA-MB231), human hepatocellular carcinoma
cell line (Bel7402), human colon cancer cell line (HCT-8 and SW620)
and human embryonic kidney 293 cells (HEK293). After transfection
with siNC or siAHR for 72 h, the protein expression of AHR was
examined in each cell line via western blotting (Fig. 1A). The results showed that the
levels of AHR from each cell line in the siAHR group were
significantly reduced compared to the siNC group. In addition,
knockdown of AHR expression by siRNA specific for human AHR
resulted in an increased expression level of caspase-3 and a
decreased expression level of survivin (a member of the inhibitor
of apoptosis family), especially in A549 and HCT-8 cell lines,
indicating that the critical role of AHR for the anti-apoptotic
effect. Next, a dimethylthiazolyl diphenyl tetrazolium (MTT) assay
was performed and showed that the reduction of AHR decreased the
growth of HEK293 and multiple cancer cell lines, especially in
A2780 and A549 cell lines (2.5- and 2.4-fold). These data reveal
that knocking down AHR could suppress the proliferation of various
cancer cells. Therefore, overexpression of AHR may facilitate the
growth of cancer cells.
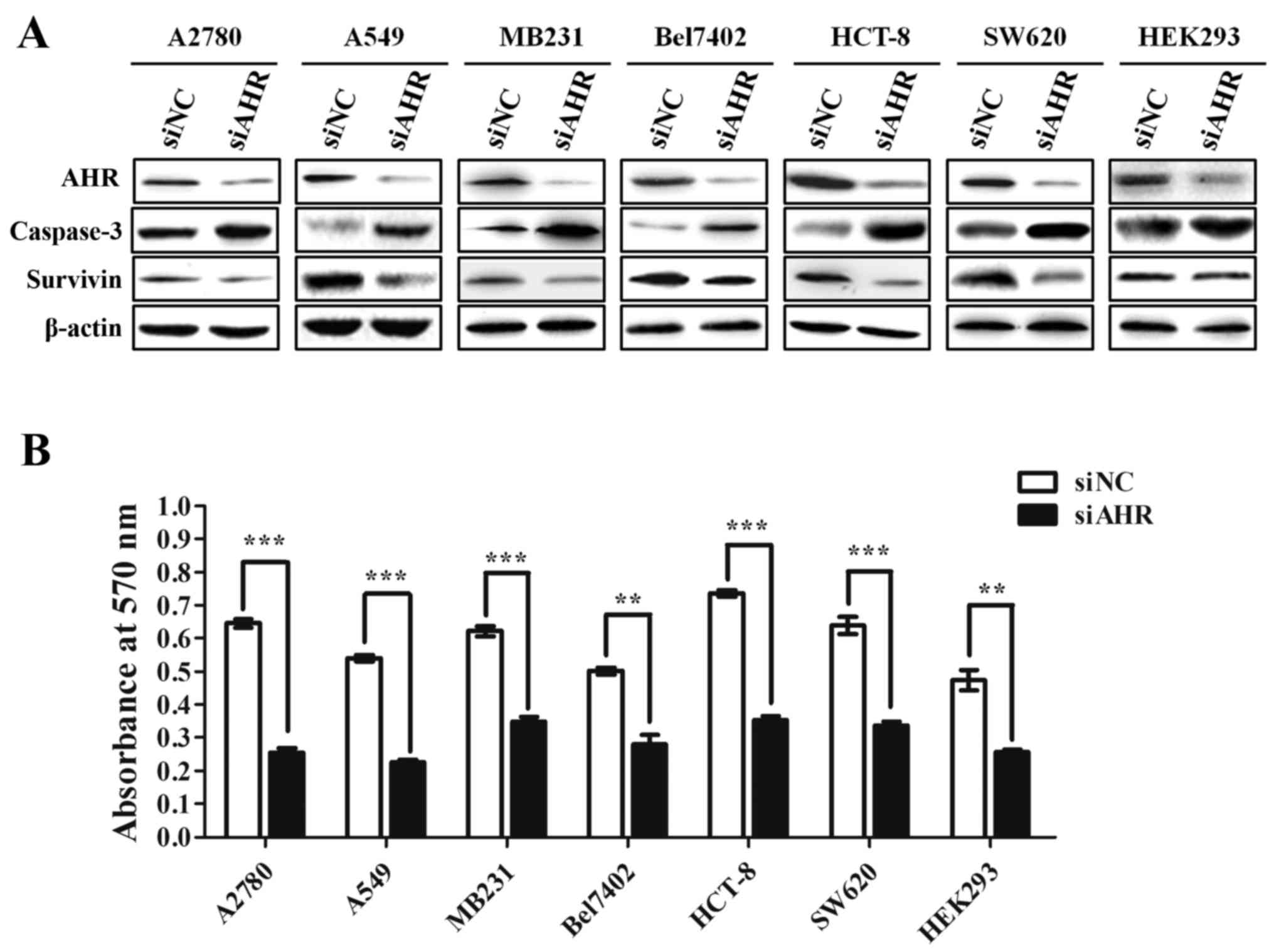 | Figure 1.AHR knockdown inhibits the growth of
cancer cells. (A) A2780, A549, MB231, Bel7402, HCT-8, SW620 and
HEK293 cell lines were transfected with siNC (control) or siAHR,
respectively. The AHR, caspase-3 and survivin protein levels from
each cell line were analyzed 72 h post-transfection by western
blotting. β-actin served as a loading control. (B) MTT assays show
the proliferation of A2780, A549, MB231, Bel7402, HCT-8, SW620 and
HEK293 cell lines after transfection with siRNA against AHR or
control siRNA. Data represent the mean ± SEM of three independent
experiments. **p<0.001; ***p<0.0001. |
Characterization of the 5′-UTR of AHR
mRNA
This study aimed to identify elements that regulate
AHR translation. After searching the NCBI database, we found that
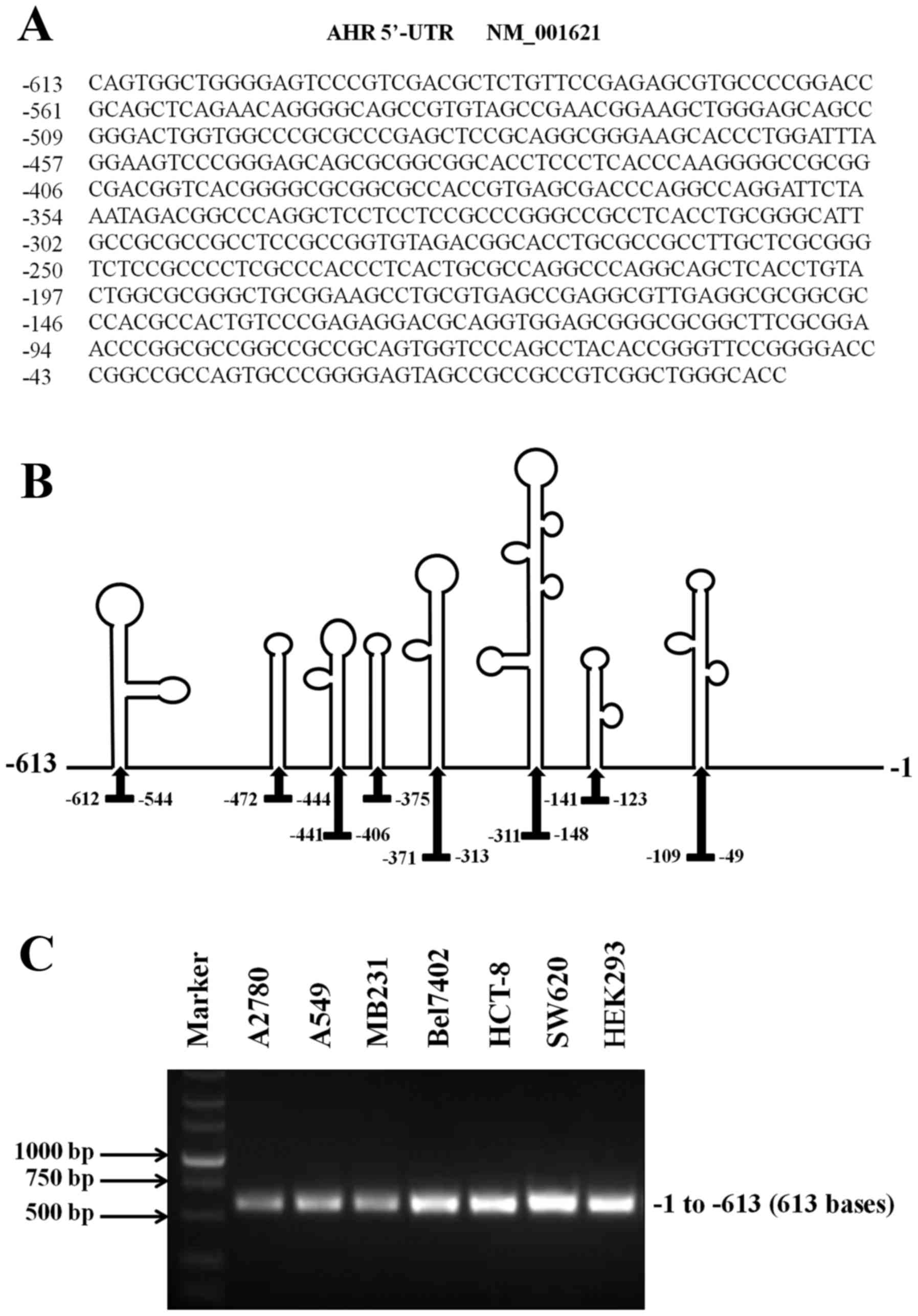
AHR mRNA has an usually long 5′-UTR (613 bases), which may present
the activity of the IRES (Fig. 2A).
In addition, this sequence had an average predicted folding energy
(∆G = −321.48 kcal/mole) and contained eight distinct stem-loop
structures, which are often detected in RNAs known to initiate
translation via IRES (Fig. 2B).
Then, RT-PCR assays were carried out to analyze the expression of
AHR 5′-UTR in HEK293 and various cancer cell lines (A2780, A549,
MB231, Bel7402, HCT-8 and SW620). The results demonstrated and
confirmed the presence of the 5′-UTR of AHR mRNA in HEK293 and all
cancer cells studied (Fig. 2C).
Analysis of IRES activity in the
5′-UTR of AHR mRNA
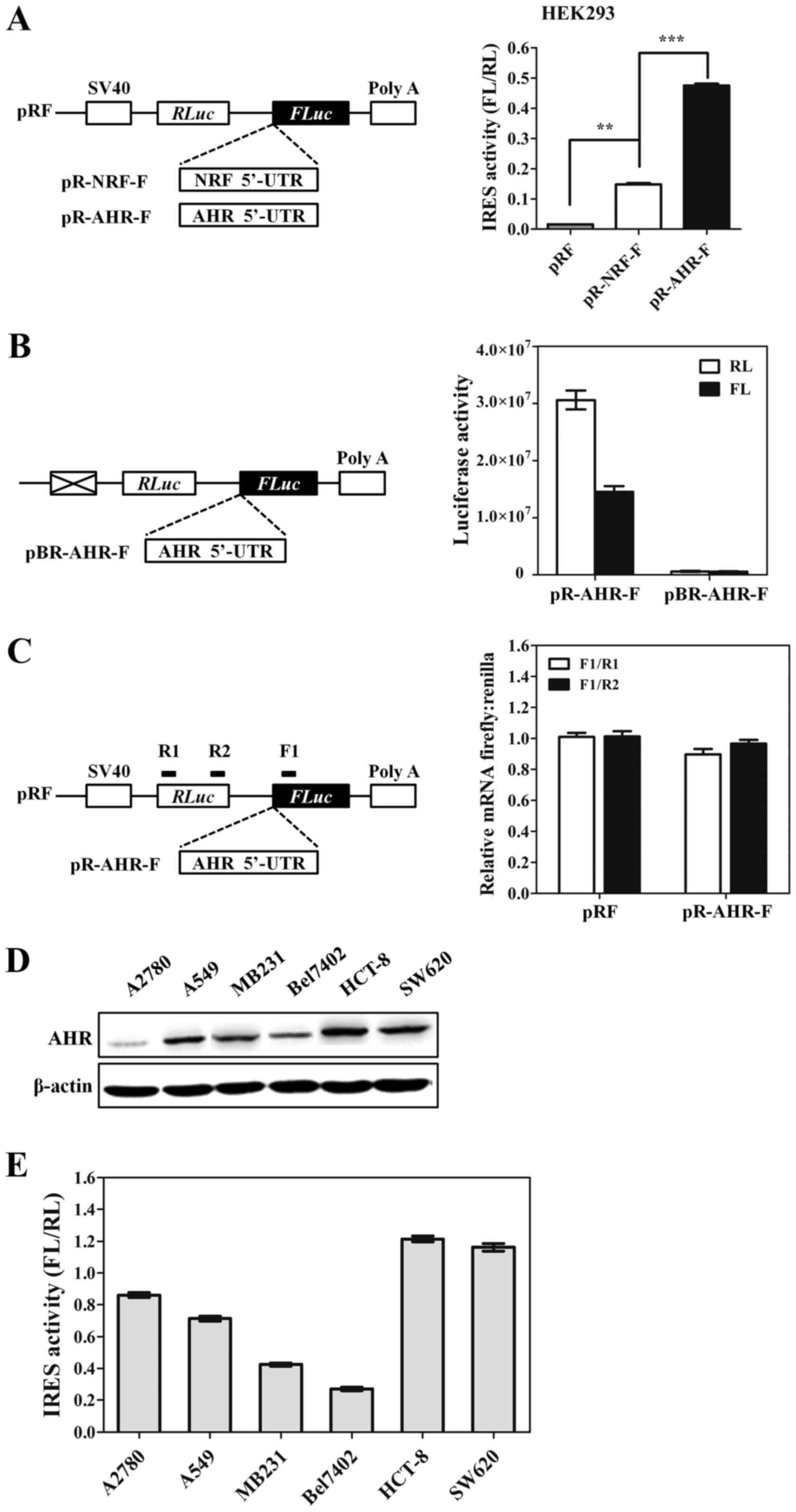
To determine if the AHR 5′-UTR contains an IRES, we
employed the dicistronic reporter plasmid pRF (31). The dicistronic construct contains a
Renilla luciferase (RL) open reading frame (ORF) and a
firefly luciferase (FL) ORF that are separated by an intercistronic
region. Translation of the Renilla cistron occurs by a
cap-dependent mechanism, while translation of the second cistron
will not occur unless there is an IRES present to initiate
translation. Accordingly, the AHR 5′-UTR was inserted into the
intercistronic region (Fig. 3A).
The 5′-UTR of the NRF mRNA, which is known to harbor a strong IRES
element, was utilized as a positive control (30). HEK293 was transfected with the
dicistronic constructs and both luciferase activities were
measured. As predicted, the firefly luciferase activity was
markedly increased by the insertion of the AHR 5′-UTR. An
approximately 30-fold increase was observed in the pR-AHR-F
construct. AHR 5′-UTR was also 3.2 times better at promoting
IRES-mediated translation than the well-characterized IRES in NRF
mRNA, the positive control. To ensure that the increase in firefly
luciferase activity was a result of enhanced translation rather
than the presence of cryptic promoter activity in AHR 5′-UTR
sequence, a promoterless plasmid pBR-AHR-F was constructed. HEK293
cells were transfected with pR-AHR-F and pBR-AHR-F plasmids,
respectively, and luciferase activity was measured. No activity of
Renilla and firefly luciferase was detected in cells
transfected with pBR-AHR-F (Fig.
3B). The production of functional monocistronic firefly
luciferase mRNAs may be caused by alternative splicing events that
removed the Renilla ORF. Therefore, reporter mRNA of firefly
and Renilla was analyzed by qRT-PCR to obtain the ratio of
FL and RL mRNA. The ratio from intact dicistronic message is 1:1
such as pRF in Fig. 3C and the
existence of splicing site results a ratio higher than 1. After
analyzing the detected sequence AHR and control, the ratio of each
was lower than 1, indicating that there was no evidence for a
cryptic splicing event that result in loss of the Renilla
cistron. Taken together, these results suggested that an effective
IRES is present in AHR 5′-UTR sequence. To determine whether the
relative activity of the AHR IRES is dependent upon the cell type,
we tested AHR expression levels and IRES activities in 6 cancer
cell lines. The results shown in Fig.
3D and E demonstrate that AHR IRES activity parallels AHR
protein expression in all cancer cell lines except for A2780 cells,
indicating that IRES-dependent translation may be contributing to
the overexpression of the AHR protein observed in the subset of
cancer cell lines.
Mapping the AHR IRES
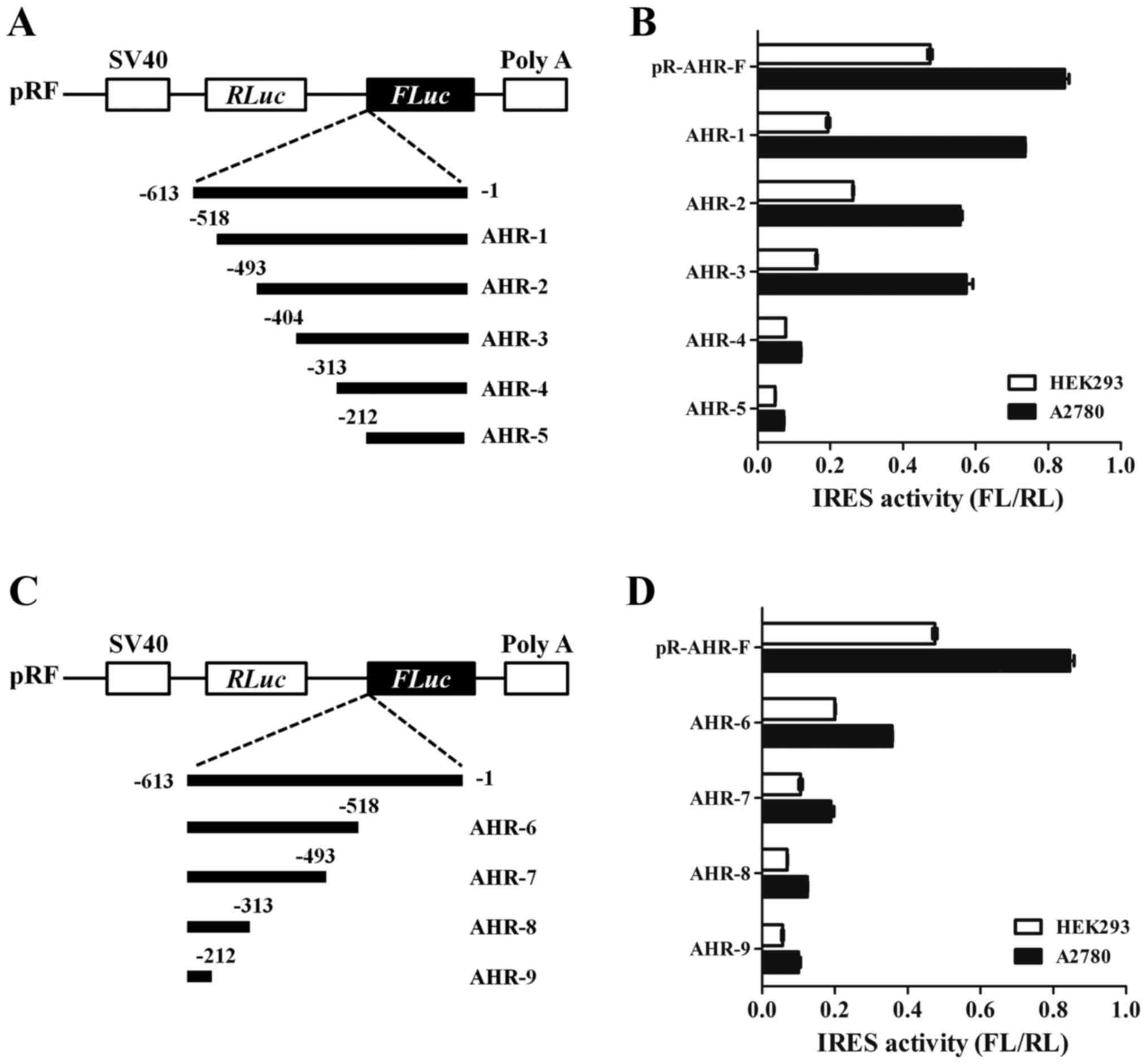
To further determine the position of the IRES within
the AHR 5′-UTR, a series of deletions from the 5′-end of the UTR to
−518 (−518 to −1), −493 (−493 to −1), −404 (−404 to −1), −313 (−313
to −1) and −212 (−212 to −1) was cloned into the dicistronic vector
(Fig. 4A). The plasmids were
transfected into HEK293 and A2780 cells and the relative luciferase
activity was determined for each of these truncations. As seen in
Fig. 4B, deletion of the 5′-end 209
nt resulted in little change in IRES activity (cf. −518 to −1, −493
to −1 and −404 to −1) in A2780 cells. Conversely, truncation of the
IRES element from −404 to −313 or −212 nucleotides diminished IRES
activity to <20%, suggesting that the region between −404 and
−313 is required for maximal IRES activity. To establish a 3′
border, deletions from the 3′-end were tested in the dicistronic
vector: −613 to −212, −613 to −313. −613 to −493 and −613 to −518
(Fig. 4C). At the 3′-end, deletion
of 211 nt (−613 to −212) resulted in 2.3-fold decrease in
downstream cistron activity in HEK293 and A2780 cells (Fig. 4D). Further three mutations
approximately abolish their translation initiation activity (cf.
−613 to −313, −613 to −493 and −613 to −518). The deletion analysis
indicated that sequences at both the 5′ and 3′ ends of the AHR are
essential for the integrity of the IRES.
AHR is overexpressed by IRES mechanism
after PTX treatment
Cellular IRES is generally used under
patho-physiological conditions when cap-dependent scanning is
compromised, such as cellular stress and apoptosis. In order to
determine whether AHR 5′-UTR regulates its protein expression in
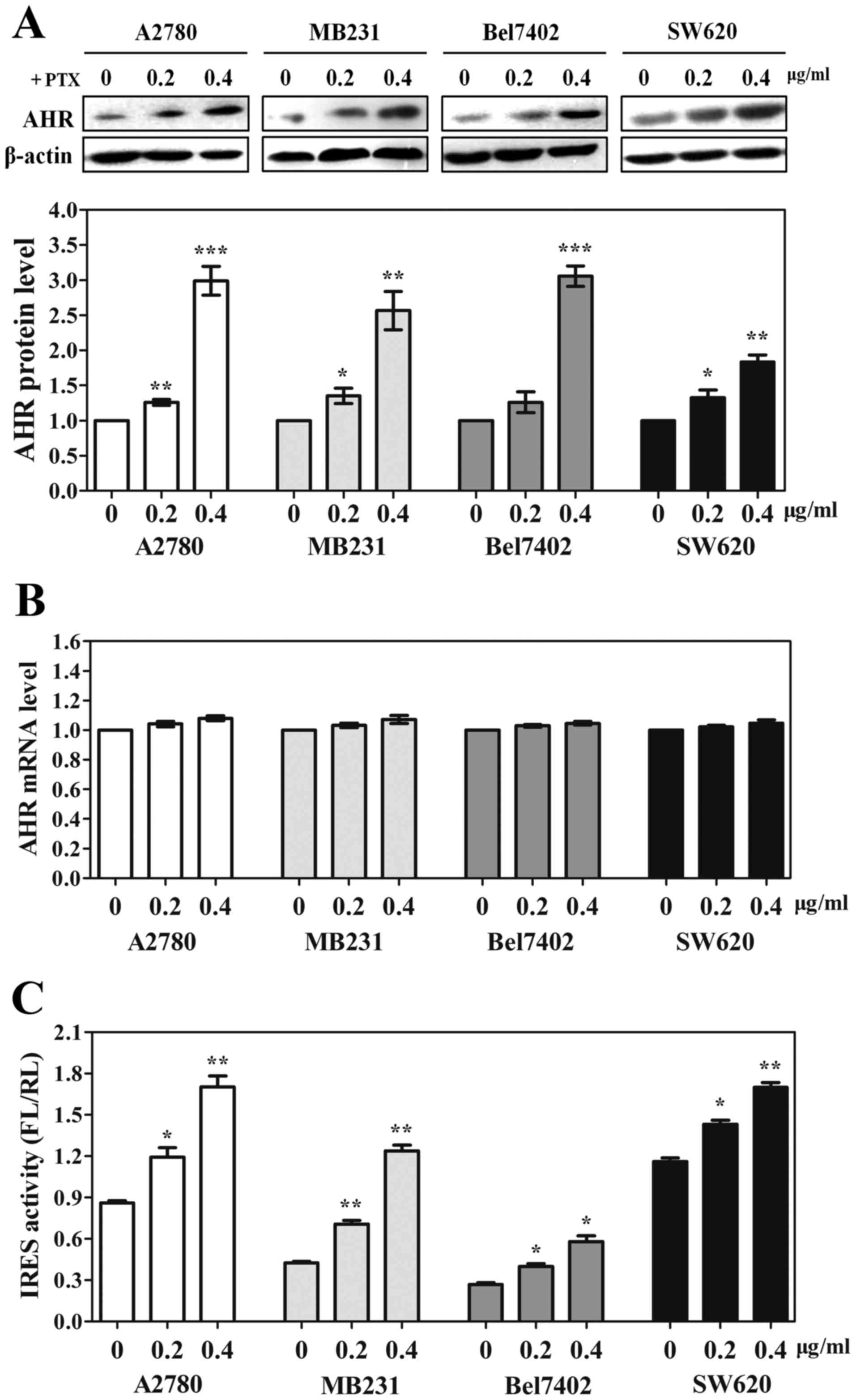
cancer cells after PTX treatment, the endogenous expression of AHR
was first measured by western blotting. A2780, MB231, Bel7402 and
SW620 cells were treated with increasing concentrations (0.2 and
0.4 µg/ml) of PTX for 24 h and AHR protein expression was examined.
As shown in Fig. 5A, AHR expression
was significantly increased in each cell line treated with PTX.
Moreover, we observed that the PTX-induced AHR expression was
dose-dependent. To evaluate whether the induction of AHR occurred
at transcriptional level, we performed qRT-PCR to examine AHR mRNA
expression in each cell line. We found that PTX had no effect on
AHR transcription in any of the cancer cell lines (Fig. 5B). These results indicated the
elevated AHR may be translated by IRES-dependent mechanism. Next,
we performed gene transfections and reporter assays to measure the
IRES activity of AHR 5′-UTR in each cell line with PTX treatment.
As expected, following with the elevation of the dose, the
inducible IRES activity was obviously increased in all cell lines,
especially in MB231 cells (Fig.
5C). Taken together, these data suggest that IRES-dependent
translation mechanism plays a critical role in tumor cell
adaptation to PTX treatment.
IRES activity is increased in
PTX-resistant ovarian cancer cells
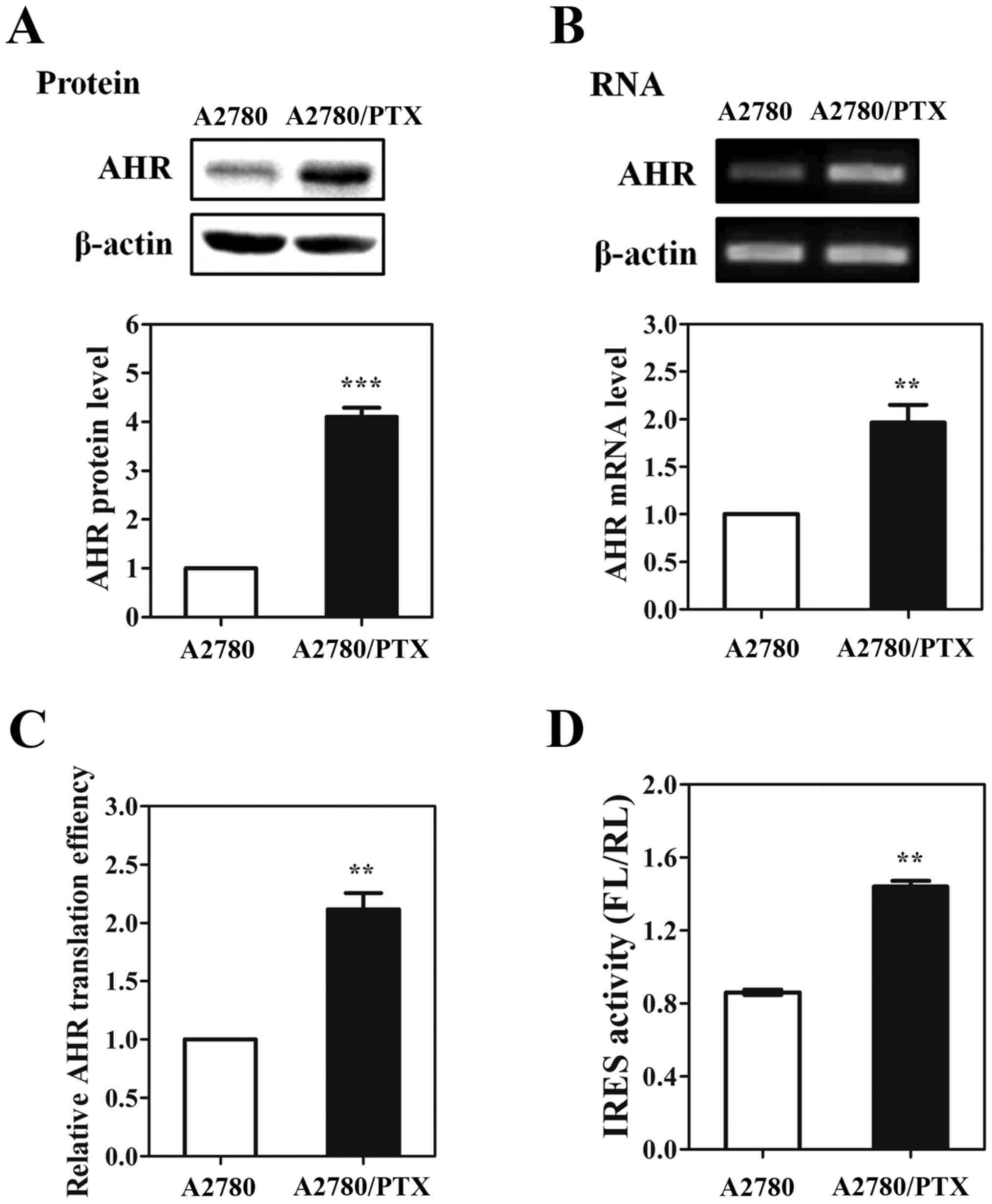
In this study, the AHR protein levels were enhanced
in various wild-type cancer cells after treating with PTX. To study
whether AHR was overexpressed in drug-resistant cancer cells, we
measured the endogenous protein and mRNA levels of AHR in
PTX-resistant and wild-type ovarian cancer cell lines,
respectively. We found that AHR protein expression level was
significantly increased in A2780/PTX cells (Fig. 6A). In addition, there was also an
increase in AHR mRNA level in the drug-resistant cells compared to
the wild-type cells (Fig. 6B). To
test whether the translational efficacy was increased in A2780/PTX
cells, MDR1 protein level was normalized to the mRNA level in each
cell line (Fig. 6C). Interestingly,
the protein-to-mRNA level was 2.1-fold higher in A2780/PTX cells,
indicating that the elevated protein synthesis contributed to the
enhanced AHR protein in A2780/PTX cells. To detect AHR IRES
activity in drug-resistant cancer cells, we performed the
dual-luciferase assay in PTX-resistant A2780 cells. Results, as
shown in Fig. 6D, demonstrated that
the AHR 5′-UTR had more efficiently IRES activity (~1.67-fold
higher) in PTX-resistant ovarian cancer cells. Therefore, we
proposed that the increased IRES activity may promote the AHR
protein production in drug-resistant ovarian cancer cells.
Discussion
AHR is present in a variety of organisms and has
important function in both physiological and toxicological
processes. At the cellular level, AHR involves in governing cell
proliferation and cell cycle, cell morphology, cell adhesion and
cell migration (36). Moreover, in
ovarian cancer, it has been demonstrated that ITE regulates cancer
cell proliferation and migration via AHR (37). In human pluripotent embryonic
teratocarcinoma NTERA2 cells, it has been proven that Alu
retrotransposons promote cell differentiation through AHR (36). Also, in breast cancer cells,
overexpression of AHR facilitated cell proliferation and migration
via upregulation of NDRG1 under hypoxia (15). Similarly, our study indicated that
knocking down AHR was able to decrease cell proliferation in human
ovarian, lung, breast, hepatocellular and colon cancer cells
(Fig. 1).
As AHR is highly expressed in a wide panel of
tumors, molecules with antagonistic AHR activity could be
considered potential candidates for the treatment of such diseases.
The results of the present study provide evidence that AHR
expression can also be regulated at the initiation stage of
translation through an IRES located within the 5′-UTR of its mRNA.
An IRES is experimentally defined as a sequence element that can
facilitate the initiation of translation of a downstream cistron in
a dicistronic mRNA. By this assay, we demonstrated that AHR 5′-UTR
has significant IRES activity with the luminescence ratio 3.2-fold
higher than the activity of positive control (NRF) in HEK293 cells
(Fig. 3). To investigate the
function of AHR IRES element in cancer cells, we performed gene
transfections and reporter assays in seven cancer cell types which
were categorized as high or low AHR protein expressing cells
(Fig. 1A). We found that AHR IRES
activity correlated with AHR protein expression in most cancer cell
lines, suggesting that IRES-dependent translation initiation
mediated by the AHR 5′-UTR is used to a greater extent in cells
overexpressing the AHR protein.
Based on our deletion analysis, it appears that the
full-length 5′-UTR of AHR mRNA exhibited the maximal IRES activity.
However, deletion of sequence between −404 and −303 or between −212
and −1 produces a remarkable negative effect on the IRES activity
of AHR mRNA, indicating that these regions are important parts of a
larger sequence forming the IRES element of 5′-UTR of AHR mRNA
(Fig. 4). The IRES-dependent
translation requires the presence of an additional complex set of
trans-acting factors (ITAFs) for translational activity to occur
(38–41). We propose that the important regions
of AHR 5′-UTR may bind to the trans-acting factors, inducing the
conformational changes that facilitate recruitment of the ribosome
to the AHR IRES.
Generally, IRES becomes activated under conditions
in which cap-dependent protein synthesis is greatly reduced,
whereupon the activated IRES initiates translation of only those
specific proteins that are able to protect cells from stress. For
instance, the IRES activity of TNF receptor-associated factor 1
(TRAF1) is simulated in leukemic cells treated with vincristine,
the 5′-UTR of human sensitivity to nitrogen mustard (hSNM1)
upregulates protein expression in HT-1080 cells during mitosis, and
several apoptosis-inducing agents induce the IRES activity of
cellular inhibitor of apoptosis protein 1 (c-IAP1) in 293T cells
(42–44). Furthermore, previous studies have
demonstrated that AHR nuclear translocator (ARNT isoform 1) impairs
chemo-resistance and survival to cancer cells (45). Thus, the increasing concentration of
chemotherapeutic drug PTX was used to treat A2780, MB231, Bel7402
and SW620 cell lines. Here, we observed that PTX induced AHR IRES
activity, which plays a key regulatory role in controlling
overexpressed AHR protein in each cell line (Fig. 5). Moreover, we found IRES-mediated
translation mechanism contributing to enhanced AHR protein
expression in PTX-resistant ovarian cancer cells. Indeed, our
results demonstrated that the AHR IRES-dependent mechanism provides
the cancer cell with the plasticity needed to respond to rapid
changes in the environment, such as chemotherapeutic drug pressure
or apoptosis.
In conclusion, we demonstrated that the human AHR
5′-UTR contains a strong IRES regulating its translation. In an
examination of various cell lines, IRES activity was a pivotal
mechanism that correlated with AHR protein expression. Moreover,
this mechanism seems to be upregulated during cellular stress,
indicating that targeting this mechanism may be beneficial for
treating multiple tumors.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81101667) and the Natural Science
Foundation of Jiangsu Province (BK2009071).
References
|
1
|
Tian J, Feng Y, Fu H, Xie HQ, Jiang JX and
Zhao B: The aryl hydrocarbon receptor: A key bridging molecule of
external and internal chemical signals. Environ Sci Technol.
49:9518–9531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beischlag TV, Morales J Luis, Hollingshead
BD and Perdew GH: The aryl hydrocarbon receptor complex and the
control of gene expression. Crit Rev Eukaryot Gene Expr.
18:207–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Safe S and Krishnan V: Cellular and
molecular biology of aryl hydrocarbon (Ah) receptor-mediated gene
expression. Arch Toxicol (Suppl). 17:99–115. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanieh H: Toward understanding the role of
aryl hydrocarbon receptor in the immune system: Current progress
and future trends. BioMed Res Int. 2014:5207632014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gonzalez FJ and Fernandez-Salguero P: The
aryl hydrocarbon receptor: Studies using the AHR-null mice. Drug
Metab Dispos. 26:1194–1198. 1998.PubMed/NCBI
|
|
6
|
Schmidt JV, Su GH, Reddy JK, Simon MC and
Bradfield CA: Characterization of a murine Ahr null allele:
Involvement of the Ah receptor in hepatic growth and development.
Proc Natl Acad Sci USA. 93:pp. 6731–6736. 1996; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lahvis GP, Lindell SL, Thomas RS, McCuskey
RS, Murphy C, Glover E, Bentz M, Southard J and Bradfield CA:
Portosystemic shunting and persistent fetal vascular structures in
aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci USA.
97:pp. 10442–10447. 2000; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lahvis GP, Pyzalski RW, Glover E, Pitot
HC, McElwee MK and Bradfield CA: The aryl hydrocarbon receptor is
required for developmental closure of the ductus venosus in the
neonatal mouse. Mol Pharmacol. 67:714–720. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gasiewicz TA, Singh KP and Casado FL: The
aryl hydrocarbon receptor has an important role in the regulation
of hematopoiesis: Implications for benzene-induced hematopoietic
toxicity. Chem Biol Interact. 184:246–251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mulero-Navarro S and Fernandez-Salguero
PM: New trends in aryl hydrocarbon receptor biology. Front Cell Dev
Biol. 4:452016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salisbury TB and Tomblin JK:
Insulin/insulin-like growth factors in cancer: New roles for the
aryl hydrocarbon receptor, tumor resistance mechanisms, and new
blocking strategies. Front Endocrinol (Lausanne).
6:122015.PubMed/NCBI
|
|
12
|
Feng S, Cao Z and Wang X: Role of aryl
hydrocarbon receptor in cancer. Biochim Biophys Acta. 1836:197–210.
2013.PubMed/NCBI
|
|
13
|
Wang LT, Chiou SS, Chai CY, Hsi E, Wang
SN, Huang SK and Hsu SH: Aryl hydrocarbon receptor regulates
histone deacetylase 8 expression to repress tumor suppressive
activity in hepatocellular carcinoma. Oncotarget. 8:7489–7501.
2017.PubMed/NCBI
|
|
14
|
Stanford EA, Wang Z, Novikov O, Mulas F,
Landesman-Bollag E, Monti S, Smith BW, Seldin DC, Murphy GJ and
Sherr DH: The role of the aryl hydrocarbon receptor in the
development of cells with the molecular and functional
characteristics of cancer stem-like cells. BMC Biol. 14:202016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li EY, Huang WY, Chang YC, Tsai MH, Chuang
EY, Kuok QY, Bai ST, Chao LY, Sher YP and Lai LC: Aryl hydrocarbon
receptor activates NDRG1 transcription under hypoxia in breast
cancer cells. Sci Rep. 6:208082016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li ZD, Wang K, Yang XW, Zhuang ZG, Wang JJ
and Tong XW: Expression of aryl hydrocarbon receptor in relation to
p53 status and clinicopathological parameters in breast cancer. Int
J Clin Exp Pathol. 7:7931–7937. 2014.PubMed/NCBI
|
|
17
|
Chang CY and Puga A: Constitutive
activation of the aromatic hydrocarbon receptor. Mol Cell Biol.
18:525–535. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Opitz CA, Litzenburger UM, Sahm F, Ott M,
Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller
M, et al: An endogenous tumour-promoting ligand of the human aryl
hydrocarbon receptor. Nature. 478:197–203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hinnebusch AG and Lorsch JR: The mechanism
of eukaryotic translation initiation: new insights and challenges.
Cold Spring Harb Perspect Biol. July 18–2012.(Epub ahead of print).
doi: 10.1101/cshperspect.a011544. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shatsky IN, Dmitriev SE, Terenin IM and
Andreev DE: Cap- and IRES-independent scanning mechanism of
translation initiation as an alternative to the concept of cellular
IRESs. Mol Cells. 30:285–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Komar AA and Hatzoglou M: Cellular
IRES-mediated translation: The war of ITAFs in pathophysiological
states. Cell Cycle. 10:229–240. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jang SK, Kräusslich HG, Nicklin MJ, Duke
GM, Palmenberg AC and Wimmer E: A segment of the 5′ nontranslated
region of encephalomyocarditis virus RNA directs internal entry of
ribosomes during in vitro translation. J Virol. 62:2636–2643.
1988.PubMed/NCBI
|
|
23
|
Pelletier J and Sonenberg N: Internal
initiation of translation of eukaryotic mRNA directed by a sequence
derived from poliovirus RNA. Nature. 334:320–325. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Holcik M, Lefebvre C, Yeh C, Chow T and
Korneluk RG: A new internal-ribosome-entry-site motif potentiates
XIAP-mediated cytoprotection. Nat Cell Biol. 1:190–192. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Subkhankulova T, Mitchell SA and Willis
AE: Internal ribosome entry segment-mediated initiation of c-Myc
protein synthesis following genotoxic stress. Biochem J.
359:183–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dobson T, Chen J and Krushel LA:
Dysregulating IRES-dependent translation contributes to
overexpression of oncogenic Aurora A kinase. Mol Cancer Res.
11:887–900. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gonzalez-Herrera IG, Prado-Lourenco L,
Pileur F, Conte C, Morin A, Cabon F, Prats H, Vagner S, Bayard F,
Audigier S, et al: Testosterone regulates FGF-2 expression during
testis maturation by an IRES-dependent translational mechanism.
FASEB J. 20:476–478. 2006.PubMed/NCBI
|
|
28
|
Pedersen SK, Christiansen J, Hansen T,
Larsen MR and Nielsen FC: Human insulin-like growth factor II
leader 2 mediates internal initiation of translation. Biochem J.
363:37–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rubtsova MP, Sizova DV, Dmitriev SE,
Ivanov DS, Prassolov VS and Shatsky IN: Distinctive properties of
the 5′-untranslated region of human hsp70 mRNA. J Biol Chem.
278:22350–22356. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oumard A, Hennecke M, Hauser H and
Nourbakhsh M: Translation of NRF mRNA is mediated by highly
efficient internal ribosome entry. Mol Cell Biol. 20:2755–2759.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao W, Li Q, Zhu R and Jin J: La
autoantigen induces ribosome binding protein 1 (RRBP1) expression
through internal ribosome entry site (IRES)-mediated translation
during cellular stress condition. Int J Mol Sci. 17:11742016.
View Article : Google Scholar :
|
|
32
|
Dai W, Ma W, Li Q, Tao Y, Ding P, Zhu R
and Jin J: The 5′-UTR of DDB2 harbors an IRES element and
upregulates translation during stress conditions. Gene. 573:57–63.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan
X, Gong Y, Yin B, Liu W, Qiang B, et al: MicroRNA-128 inhibits
glioma cells proliferation by targeting transcription factor E2F3a.
J Mol Med (Berl). 87:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fujisawa Y, Li W, Wu D, Wong P, Vogel C,
Dong B, Kung HJ and Matsumura F: Ligand-independent activation of
the arylhydrocarbon receptor by ETK (Bmx) tyrosine kinase helps
MCF10AT1 breast cancer cells to survive in an apoptosis-inducing
environment. Biol Chem. 392:897–908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin J, Sheng B, Qiu Y, Yang K, Xiao W and
Yang H: Role of AhR in positive regulation of cell proliferation
and survival. Cell Prolif. 49:554–560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morales-Hernández A, González-Rico FJ,
Román AC, Rico-Leo E, Alvarez-Barrientos A, Sánchez L, Macia Á,
Heras SR, García-Pérez JL, Merino JM, et al: Alu retrotransposons
promote differentiation of human carcinoma cells through the aryl
hydrocarbon receptor. Nucleic Acids Res. 44:4665–4683. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Li Y, Jiang YZ, Dai CF, Patankar
MS, Song JS and Zheng J: An endogenous aryl hydrocarbon receptor
ligand inhibits proliferation and migration of human ovarian cancer
cells. Cancer Lett. 340:63–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martínez-Salas E, Lozano G,
Fernandez-Chamorro J, Francisco-Velilla R, Galan A and Diaz R:
RNA-binding proteins impacting on internal initiation of
translation. Int J Mol Sci. 14:21705–21726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Costa-Mattioli M, Svitkin Y and Sonenberg
N: La autoantigen is necessary for optimal function of the
poliovirus and hepatitis C virus internal ribosome entry site in
vivo and in vitro. Mol Cell Biol. 24:6861–6870. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cobbold LC, Wilson LA, Sawicka K, King HA,
Kondrashov AV, Spriggs KA, Bushell M and Willis AE: Upregulated
c-myc expression in multiple myeloma by internal ribosome entry
results from increased interactions with and expression of PTB-1
and YB-1. Oncogene. 29:2884–2891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cobbold LC, Spriggs KA, Haines SJ, Dobbyn
HC, Hayes C, de Moor CH, Lilley KS, Bushell M and Willis AE:
Identification of internal ribosome entry segment
(IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell
Biol. 28:40–49. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang L, Gu L, Li Z and Zhou M: Translation
of TRAF1 is regulated by IRES-dependent mechanism and stimulated by
vincristine. Nucleic Acids Res. 38:4503–4513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang X, Richie C and Legerski RJ:
Translation of hSNM1 is mediated by an internal ribosome entry site
that upregulates expression during mitosis. DNA Repair (Amst).
1:379–390. 2002.PubMed/NCBI
|
|
44
|
Van Eden ME, Byrd MP, Sherrill KW and
Lloyd RE: Translation of cellular inhibitor of apoptosis protein 1
(c-IAP1) mRNA is IRES mediated and regulated during cell stress.
RNA. 10:469–481. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gardella KA, Muro I, Fang G, Sarkar K,
Mendez O and Wright CW: Aryl hydrocarbon receptor nuclear
translocator (ARNT) isoforms control lymphoid cancer cell
proliferation through differentially regulating tumor suppressor
p53 activity. Oncotarget. 7:10710–10722. 2016. View Article : Google Scholar : PubMed/NCBI
|