Introduction
Oesophageal carcinoma (EC) is one of the most common
and aggressive cancers, and the sixth leading cause of
cancer-related death worldwide, particularly in Africa and Asia
(1,2). Oesophageal squamous cell carcinoma
(ESCC) is the predominant histological subtype in China. Even with
the development of treatment strategies, surgical techniques and
perioperative nursing, the overall survival (OS) of EC patients
remains at 15–25% (3). Therefore,
identification of key prognostic biomarkers and effective
therapeutic targets is important for the current clinical
management of ESCC. In the present study, we explored the clinical
significance and the function of STMN1 in tumour cells of ESCC.
Stathmin 1 (STMN1), also known as p17, p18, p19,
19K, metablastin, oncoprotein 18 and LAP 18, is a
microtubule-regulated protein that plays an important role in the
disassembly of the mitotic spindle and cell cycle progression.
STMN1 is activated by dephosphorylation and binds two α- and
β-tubulin heterodimers during the process of microtubule
disassembly (4). Furthermore, STMN1
also has been found to affect cell proliferation, differentiation,
migration and signal transduction. The dysfunction of STMN1,
resulting in consistent microtubule assembly, cell cycle disorder,
microtubule dynamic destabilization and abnormal signal
transduction, is closely related to tumour metastasis (4,5). STMN1
is highly expressed in many malignancies including acute leukaemia
(6), lymphoma (7), ovarian carcinoma (8), prostate (9), breast (10) and head and neck cancer (11), hepatocellular carcinoma (12), osteosarcoma (13), lung cancer (14,15)
and mesothelioma (7). These
findings indicate that STMN1 is closely related to human cancers
and an important biomarker for diagnosis and prognosis.
Overexpressed STMN1 was found to be associated with lymph node
metastasis, poor prognosis and recurrence in ESCC (16,17).
However, the correlation between STMN1 expression and
clinicopathological characteristics, prognosis and biological
function of STMN1 in ESCC remains largely unclear. The aim of the
present study was to identify the expression and investigate the
biological function of STMN1 in ESCC.
Materials and methods
Patients and specimens
Tissue specimens from 276 patients who underwent
surgical resection were collected between November 2007 and January
2010 at The Shantou Central Hospital. None of them had distant
metastasis. All tumours were confirmed by pathologists as ESCC
staged according to the 7th Edition of the American Joint Committee
on Cancer (AJCC) Tumor-Node-Metastasis (TNM) Staging System for
ESCC (18). Tumour grade was
defined as well differentiated, moderately differentiated or poorly
differentiated, according to the World Health Organization (WHO)
classification of oesophageal tumours (19). The main clinicopathological
characteristics of the patients are summarised in Table I. OS was defined as the interval
between surgery and death from the tumour or between surgery and
the last observation taken for surviving patients. Disease-free
survival (DFS) was defined as the interval between surgery and the
diagnosis of relapse or death. Ethical approval was obtained from
the Ethics Committees of the Central Hospital of Shantou City, the
Medical College of Shantou University and the West China Hospital.
Only resected samples from surgical patients giving written
informed consent were included for use in the research.
 | Table I.Correlation between STMN1 expression
and clinicopathological characteristics in ESCC. |
Table I.
Correlation between STMN1 expression
and clinicopathological characteristics in ESCC.
|
|
| STMN1
expression | Correlation
analysis |
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | No. of cases
(%) | Low, n (%) | High, n (%) | Corr.
coefficient | P-value |
|---|
| Sex |
|
|
| −0.31 | 0.601 |
|
Female | 58 (21.0) | 37 (63.8) | 21 (36.2) |
|
|
|
Male | 218 (79.0) | 147 (67.4) | 71 (32.6) |
|
|
| Age (years) |
|
|
| 0.000 | 1.000 |
|
≤55 | 108 (39.1) | 72 (66.7) | 36 (33.3) |
|
|
|
>55 | 168 (60.9) | 112 (66.7) | 56 (33.3) |
|
|
| Location |
|
|
| −0.042 | 0.483 |
|
Upper | 17 (6.2) | 10 (58.8) | 7 (41.2) |
|
|
|
Middle | 117 (42.4) | 77 (65.8) | 40 (34.2) |
|
|
|
Lower | 142 (51.4) | 97 (68.3) | 45 (31.7) |
|
|
| Tumour
sizea (cm) |
|
|
| −0.054 | 0.347 |
| ≤3 | 62 (22.6) | 38 (61.3) | 24 (38.7) |
|
|
|
3–5 | 131 (47.8) | 88 (67.2) | 43 (32.8) |
|
|
| ≥5 | 81 (29.6) | 56 (69.1) | 25 (30.9) |
|
|
|
Differentiation |
|
|
| 0.127 | 0.037 |
| G1 | 43 (15.6) | 31 (72.1) | 12 (27.9) |
|
|
| G2 | 212 (76.8) | 145 (68.4) | 67 (31.6) |
|
|
| G3 | 21 (7.6) | 8 (38.1) | 13 (61.9) |
|
|
| pT |
|
|
| 0.033 | 0.561 |
| T1 | 11 (4.0) | 8 (72.7) | 3 (27.3) |
|
|
| T2 | 43 (15.6) | 30 (69.8) | 13 (30.2) |
|
|
| T3 | 221 (80.1) | 145 (65.6) | 76 (34.4) |
|
|
|
T4a | 1 (0.4) | 1 (100) | 0 |
|
|
| pN |
|
|
| 0.087 | 0.124 |
| N0 | 137 (49.6) | 97 (70.8) | 40 (29.2) |
|
|
| N1 | 72 (26.1) | 46 (63.9) | 26 (36.1) |
|
|
| N2 | 50 (18.1) | 32 (64.0) | 18 (36.0) |
|
|
| N3 | 17 (6.2) | 9 (52.9) | 8 (47.1) |
|
|
| TNM stage |
|
|
| 0.110 | 0.053 |
| I | 22 (8.0) | 18 (81.8) | 4 (18.2) |
|
|
| II | 129 (46.7) | 89 (69.0) | 40 (31.0) |
|
|
|
III | 125 (45.3) | 77 (61.6) | 48 (38.4) |
|
|
| Adjuvant
therapy |
|
|
| 0.070 | 0.223 |
| No | 148 (53.6) | 102 (68.9) | 46 (31.1) |
|
|
|
Radiotherapy | 41 (14.9) | 27 (65.9) | 14 (34.1) |
|
|
|
Chemotherapy | 59 (21.4) | 42 (71.2) | 17 (28.8) |
|
|
|
Radiochemotherapy | 28 (10.1) | 13 (46.4) | 15 (53.6) |
|
|
Cell culture
ESCC cell lines (KYSE150 and KYSE450) were generous
gifts from Professor Ming-Zhou Guo, Department of Gastroenterology
and Hepatology, Chinese PLA General Hospital. KYSE150 cells were
cultured in RPMI-1640 medium (Thermo Fisher Scientific Inc.,
Waltham, MA, USA) with 10% foetal bovine serum (Gibco, Grand
Island, NY, USA) and KYSE450 cells were cultured in Dulbecco's
modified Eagle's medium (Thermo Fisher Scientific Inc.) with 10%
foetal bovine serum. All cell lines were incubated at 37°C in a
humidified atmosphere containing 5% CO2.
Tissue microarrays (TMAs) and
immunohistochemistry (IHC)
TMAs were constructed based on standard techniques
and IHC was carried out using a two-step protocol (PV-9000 Polymer
Detection System; ZSGB-BIO, Beijing, China) according to the
manufacturer's instructions. These techniques were previously
described (20,21).
All sections were analysed blindly by two
experienced pathologists. Each separate tissue core was scored on
the basis of the intensity and positive staining proportion
according to the literature (22).
The intensity of positive staining was scored as follows: 0,
negative; 1, weak; 2, moderate; and 3, strong. The proportion of
positive cells was scored on a 0–4 scale as follows: 0, 0–5%; 1,
6–25%; 2, 26–50%; 3, 51–75% and 4, >75%. If the positive
staining was homogeneous, a final score was achieved by
multiplication of the two scores, producing a total range of 0–12.
When the staining was heterogeneous, each component was
independently scored and summed for the results. The mean value of
the two scores was considered representative of one tumour. For
statistical analysis, X-Tile software was used to separate the
STMN1 expression score into two subgroups: high expression and low
expression.
siRNA transfection
Double-stranded small interfering RNAs (siRNAs) were
synthesised in duplex and purified forms by GenePharma Co.
(Shanghai, China). Four sequences were designed and filtered out to
select the one that was most interference efficient. The sequences
were as follows: siRNA 1 F, 5′-AGAAGAAGGAUCUUUCCCU-3 and R,
3-AGGGAAA GAUCCUUCUUCU-5; siRNA 2 F, 5′-AAUGGCAGAAGAGAAACUG-3′ and
R, 3-CAGUUUCUCUUCUGCCAUU-5′; siRNA 3 F, 5-AAGAGUAUGUAGUGGCUUC-3 and
R, 3-GAAGCCACUACAUACUCUU-5′; siRNA 4 F, 5-AAG CACAAGCGUGUUUCUA-3
and R, 3′-UAGAAACACG CUUGUGCUU-5. STMN1 was silenced using
Lipofectamine® RNAiMAX Transfection reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol. The
controls were treated with nonsense siRNA. After 48 h, proteins
were extracted and the interference efficiency was confirmed.
Plasmid transfection
The plasmids Flag-STMN1 were purchased from Sino
Biological Inc. (Beijing, China). Flag-STMN1 was transiently
transfected into KYSE150 and KYSE450 cells, using
Lipofectamine® 3000 transfection reagent (Invitrogen)
according to the manufacturer's instructions.
Western blot analysis
Standard western blot analysis was performed as
previously described (23).
Briefly, proteins were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride (PVDF) membranes (Millipore,
Billerica, MA, USA). The membranes were blocked in 5% non-fat milk
for 1 h, followed by the addition of anti-STMN1 (Santa Cruz
Biotechnology, Santa Cruz, CA, USA) overnight at 4°C. The membranes
were then washed and incubated with secondary antibody coupled to
horseradish peroxidase for 1 h. Antigen-antibody complexes were
detected by Western Blotting Luminol reagent (Santa Cruz
Biotechnology). Photography and quantitative analyses of
related-immunoreactive bands were performed using a FluorChem™
IS-8900 (Alpha Innotech, San Leandro, CA, USA).
Wound healing assay
At 48 h after transfection, cells were spread in a
layer and serum-starved for 12 h before wounding. A 1,000-µl
pipette tip was used to create a wound across the diameter of the
well. Cell migration across the wound surface was then assessed by
microscopy after every 24 h. Images were captured under a
magnification of ×40 (DMI3000B; Leica, Wetzlar, Germany). Adobe
Illustrator CS5 was used to measure the distance between the edges
of the wound.
Cell migration and invasion
assays
Cell invasion and migration assays were performed as
described below. At 48 h after transfection with siSTMN1 or
Flag-STMN1 or their corresponding negative control, cells were
starved for 12 h and 6×105 starved cells were seeded
into Transwells (BD Biosciences, San Jose, CA, USA) with 8-µm pore
size membranes coated with or without Matrigel (for invasion and
migration assays, respectively). After 48 h, cells within the
Transwells were removed and migrated/invaded cells on the bottom of
the Transwell were stained with crystal violet. Images of
migrated/invaded cells on the Transwell membrane were captured
under a magnification of ×200, and the numbers of migratory/invaded
cells were counted from at least 10 different fields.
Colony formation assay
Transfected cells were trypsinised, counted with a
cell counter (Bio-Rad, Hercules, CA, USA), and then 1,000 cells (or
500 cells) were inoculated in each well of 6-well (or 12-well)
plates. Cultures were maintained for two weeks, and cells were then
fixed, stained and photographed.
Cell proliferation assay
The CellTiter 96® AQueous One Solution
Cell Proliferation Assay (MTS; Promega, Madison, WI, USA) was used
to measure cell proliferation. Cells that had been transfected for
48 h were seeded in a 96-well plate. Every 24 h, 20 µl of MTS
reagent was added to the plate. After incubation at 37°C for 2 h,
the absorbance was measured at 492 nm.
Cell cycle analysis
For cell cycle analysis by flow cytometry,
transfected cells were fixed with 70% ethanol overnight at 4°C.
Cell pellets were incubated in phosphate-buffered saline (PBS)
containing 0.1% Triton X-100 for 10 min at room temperature, and
then were treated with RNase (50 mg/ml) for 10 min and propidium
iodide (PI; 5 mg/ml) for 30 min, respectively. FlowJo 7.6 software
was used to determine the cell cycle phases.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
software (SPSS, Inc., Chicago, IL, USA). Categorical data were
compared using the Chi-square test or Fisher's exact probability
test. Correlation analysis was performed using Kendall's tau
coefficient. Kaplan-Meier survival analyses were used to estimate
5-year OS and DFS in patients with ESCC. The log-rank test was used
to assess differences in survival between groups. The Cox
proportional hazard regression model was used to perform a
multivariate analysis, and the risk ratio and its 95% confidence
interval were recorded. A two-sided P-value <0.05 was considered
to indicate a statistically significant result.
Results
Overexpression of STMN1 predicts a
poorer prognosis in ESCC
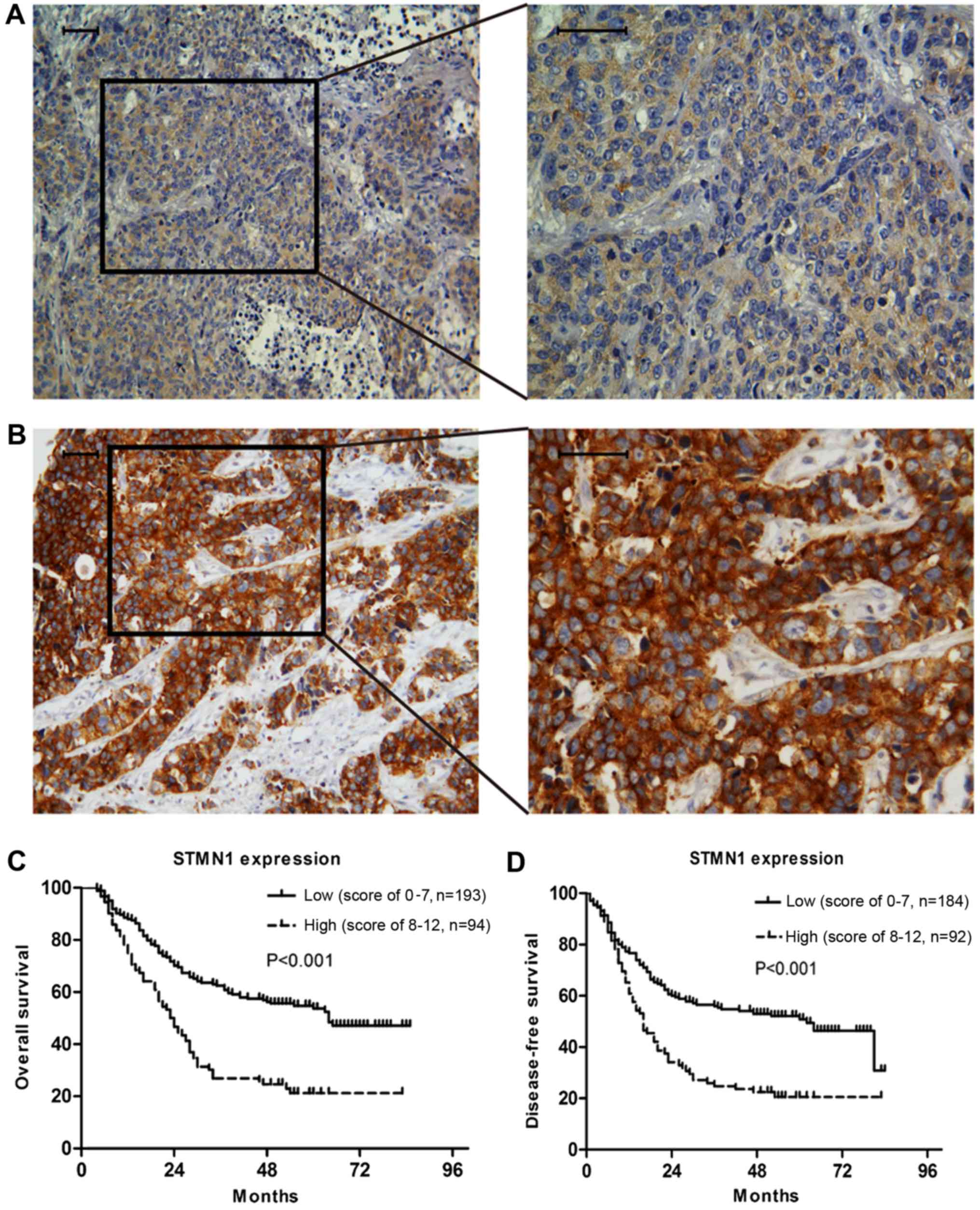
The IHC staining showed that the expression of STMN1
in ESCC was mainly located in the cytoplasm (Fig. 1). The impact of STMN1 expression on
the OS and DFS of ESCC patients was evaluated by Kaplan-Meier
survival analysis. The median length of OS and DFS was 34.0 (range,
4–85) and 24 (range, 0–84) months, respectively. The IHC results of
276 patients showed that patients with high STMN1 expression (IHC
score, 8–12) had a poorer prognosis compared with the patients with
low STMN1 expression (IHC score, 0–7). The 5-year OS rates of ESCC
patients with high and low STMN1 expression were 21.2 and 53.7%
(P<0.001) (Fig. 1C), and the DFS
rates were 20.6 and 50.9% (P<0.001), respectively (Fig. 1D).
In addition, we performed univariate and
multivariate analyses to investigate the prognostic factors for
ESCC. We found that age (P=0.002; 95% CI, 1.228–2.434), tumour
length (P=0.046; 95% CI, 1.004–1.560), pN stage (P=0.005; 95% CI,
1.117–1.840) and STMN1 expression (P<0.001; 95% CI, 1.559–2.970)
were independent prognostic factors for OS (Table II). Tumour length (P=0.010; 95% CI,
1.072–1.678), pN stage (P=0.010; 95% CI, 1.081–1.775) and STMN1
expression (P=0.001; 95% CI, 1.278–2.444) were independent
prognostic factors for DFS (Table
III).
 | Table II.Univariate and multivariate analyses
of overall survival (OS) on clinicopathological
characteristics. |
Table II.
Univariate and multivariate analyses
of overall survival (OS) on clinicopathological
characteristics.
|
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|
|---|
| Patient
characteristics | No. of
patients | MST (month) | P-value | HR | 95% CI | P-value |
|---|
| Gender |
|
| 0.599 |
|
|
|
|
Male | 218 | 37.0 |
|
|
|
|
|
Female | 58 | 41.0 |
|
|
|
|
| Age (years) |
|
| 0.019 | 1.729 | 1.228–2.434 | 0.002 |
|
≤55 | 108 | 63.0 |
|
|
|
|
|
>55 | 168 | 30.0 |
|
|
|
|
| Location |
|
| 0.492 |
|
|
|
| Ut | 17 | 24.0 |
|
|
|
|
| Mt | 117 | 48.0 |
|
|
|
|
| Lt | 142 | 37.0 |
|
|
|
|
|
Differentiation |
|
| 0.018 | 1.201 | 0.856–1.685 | 0.290 |
| G1 | 43 | 64.0 |
|
|
|
|
| G2 | 212 | 35.0 |
|
|
|
|
| G3 | 21 | 22.0 |
|
|
|
|
| Length (cm) |
|
| 0.043 | 1.252 | 1.004–1.560 | 0.046 |
| ≤3 | 62 | 65.0 |
|
|
|
|
|
3–5 | 131 | 34.0 |
|
|
|
|
| ≥5 | 81 | 30.0 |
|
|
|
|
| pT |
|
| 0.169 |
|
|
|
| T1 | 11 | 55.3a |
|
|
|
|
| T2 | 43 | 46.0 |
|
|
|
|
| T3 | 221 | 34.0 |
|
|
|
|
| T4 | 1 | 20.0 |
|
|
|
|
| pN |
|
| <0.001 | 1.434 | 1.117–1.840 | 0.005 |
| N0 | 137 | 57.3a |
|
|
|
|
| N1 | 72 | 31.0 |
|
|
|
|
| N2 | 50 | 24.0 |
|
|
|
|
| N3 | 17 | 14.0 |
|
|
|
|
| TNM stage |
|
| <0.001 | 1.201 | 0.856–1.685 | 0.290 |
| I | 22 | 64.1a |
|
|
|
|
| II | 129 | 64.0 |
|
|
|
|
|
III | 125 | 24.0 |
|
|
|
|
| Adjuvant
therapy |
|
| 0.147 |
|
|
|
| No | 148 | 30.0 |
|
|
|
|
| RT | 41 | 63.0 |
|
|
|
|
| CT | 59 | 54.0 |
|
|
|
|
|
CRT | 28 | 30.0 |
|
|
|
|
| STMN1 |
|
| <0.001 | 2.149 | 1.555–2.970 | <0.001 |
|
Low | 184 | 64.0 |
|
|
|
|
|
High | 92 | 23.0 |
|
|
|
|
 | Table III.Univariate and multivariate analyses
of disease-free survival (DFS) on clinicopathological
characteristics. |
Table III.
Univariate and multivariate analyses
of disease-free survival (DFS) on clinicopathological
characteristics.
|
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|
|---|
| Patient
characteristics | No. of
patientsa | MST (month) | P-value | HR | 95% CI | P-value |
|---|
| Gender |
|
| 0.736 |
|
|
|
|
Male | 213 | 27.0 |
|
|
|
|
|
Female | 56 | 25.0 |
|
|
|
|
| Age (years) |
|
| 0.124 |
|
|
|
|
≤55 | 106 | 47.0 |
|
|
|
|
|
>55 | 163 | 23.0 |
|
|
|
|
| Location |
|
| 0.242 |
|
|
|
| Ut | 17 | 20.0 |
|
|
|
|
| Mt | 117 | 47.0 |
|
|
|
|
| Lt | 135 | 23.0 |
|
|
|
|
|
Differentiation |
|
| 0.004 | 1.304 | 0.921–1.847 | 0.135 |
| G1 | 41 | 64.0 |
|
|
|
|
| G2 | 208 | 28.0 |
|
|
|
|
| G3 | 20 | 14.0 |
|
|
|
|
| Length (cm) |
|
| 0.017 | 1.341 | 1.072–1.678 | 0.010 |
| ≤3 | 59 | 62.0 |
|
|
|
|
|
3–5 | 129 | 23.0 |
|
|
|
|
| ≥5 | 79 | 17.0 |
|
|
|
|
| pTb |
|
| 0.103 |
|
|
|
| T1 | 10 |
|
|
|
|
|
| T2 | 43 |
|
|
|
|
|
| T3 | 221 |
|
|
|
|
|
| T4 | 1 |
|
|
|
|
|
| pN |
|
| <0.001 | 1.385 | 1.081–1.775 | 0.010 |
| N0 | 133 | 81.0 |
|
|
|
|
| N1 | 70 | 21.0 |
|
|
|
|
| N2 | 49 | 16.0 |
|
|
|
|
| N3 | 17 | 10.0 |
|
|
|
|
| TNM stage |
|
| <0.001 | 1.149 | 0.743–1.778 | 0.532 |
| I | 20 | 63.4c |
|
|
|
|
| II | 125 | 64.0 |
|
|
|
|
|
III | 124 | 17.0 |
|
|
|
|
| Adjuvant
therapy |
|
| 0.174 |
|
|
|
| No | 143 | 29.0 |
|
|
|
|
| RT | 41 | 62.0 |
|
|
|
|
| CT | 58 | 23.0 |
|
|
|
|
|
CRT | 27 | 22.0 |
|
|
|
|
| STMN1 |
|
| <0.001 | 1.767 | 1.278–2.444 | 0.001 |
|
Low | 177 | 62.0 |
|
|
|
|
|
High | 92 | 16.0 |
|
|
|
|
Correlation between STMN1 expression
and clinicopathological characteristics in ESCC
We investigated the relationship between STMN1
expression and the clinicopathological characteristics of the ESCC
patients. The results showed that STMN1 expression was found to be
correlated with tumour grade (Table
I). There were 12 patients (27.9%) with high STMN1 expression
out of 43 patients with tumours that were well differentiated, 67
patients (31.6%) with high STMN1 expression out of 212 patients
with tumours that were moderately differentiated, and 13 patients
(61.9%) with high STMN1 expression out of 21 patients with tumours
that were poorly differentiated (correlation coefficient, 0.127,
P=0.037).
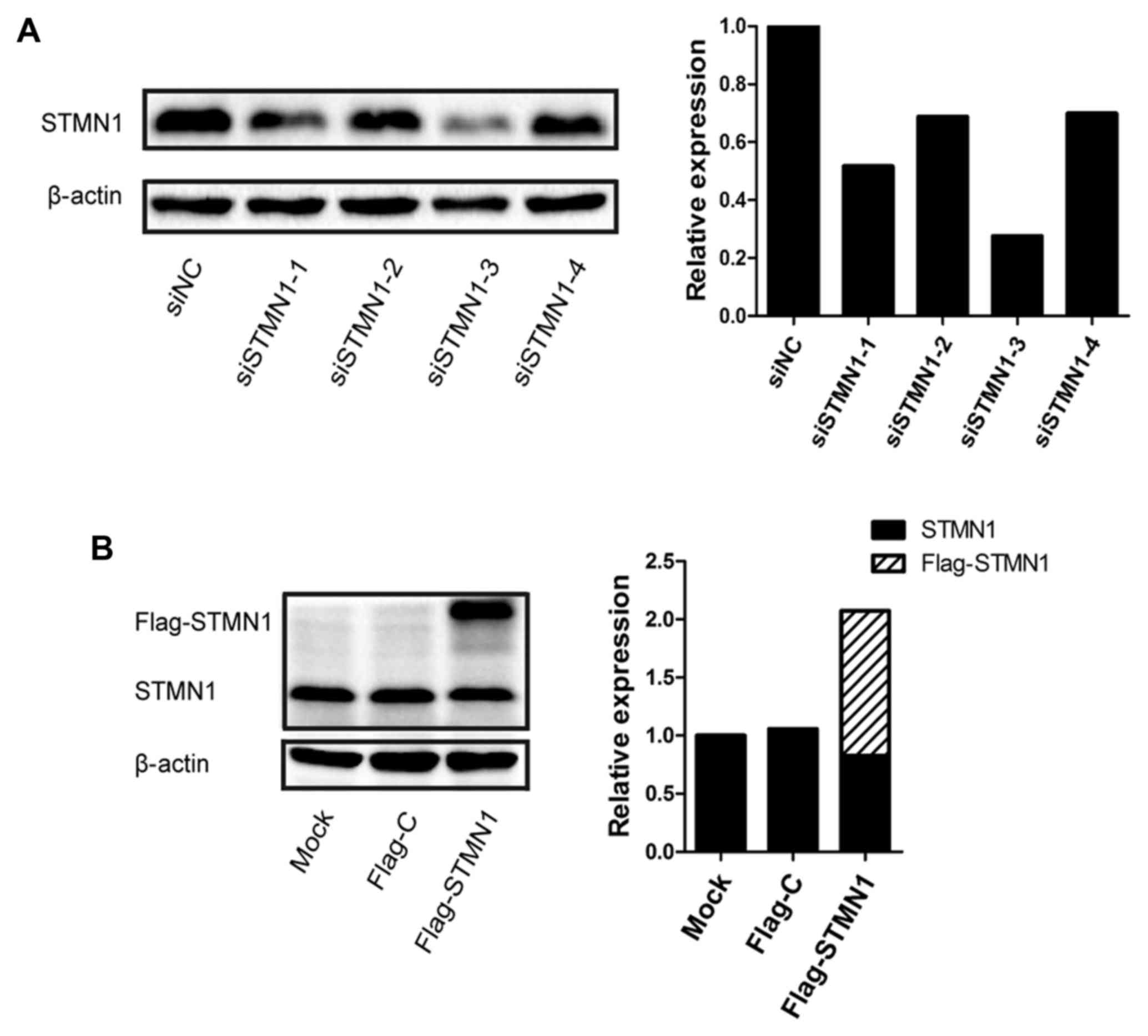
STMN1 expression following STMN1 siRNA
and plasmid transfection
To explore the effect of STMN1 on ESCC, four siRNAs
were designed to knock down STMN1 expression in KYSE150. The STMN1
expression was inhibited at the protein level up to 72.3% in the
KYSE150 cells (siRNA-3) (Fig. 2A).
Flag-STMN1 was also transfected to upregulate the STMN1 expression
in KYSE150. The STMN1 expression was upregulated at the protein
level by up to 200% in the KYSE150 cells (Fig. 2B).
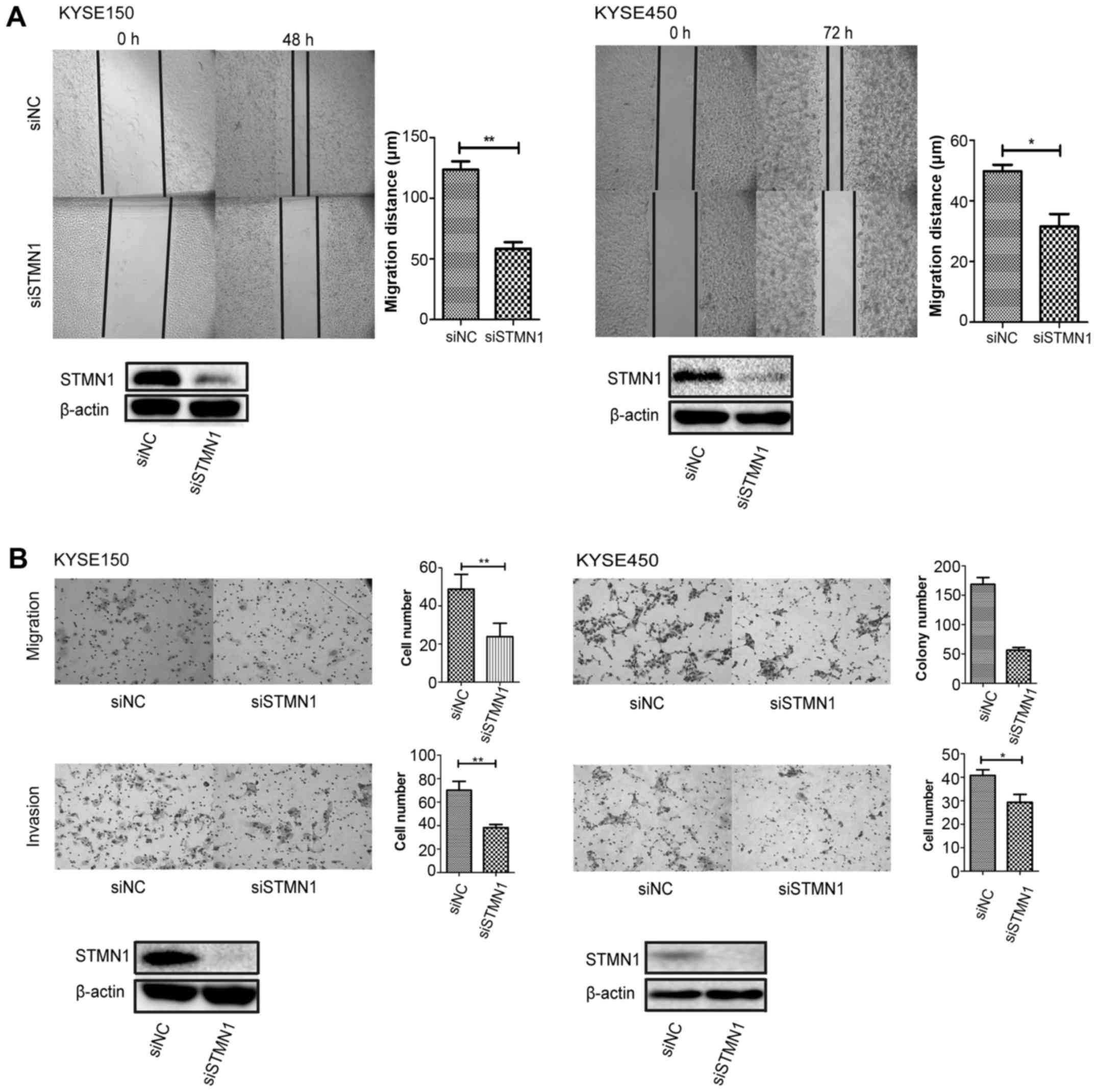
Knockdown of STMN1 expression inhibits
ESCC cell migration and invasion
Wound-healing assay was performed in both KYSE150
and KYSE450 cells. The results showed that cell migration was
inhibited after the knockdown of STMN1 expression (Fig. 3A). Furthermore, migration and
invasion assays were performed in Transwell chambers. The migration
and invasion abilities were both inhibited after downregulation of
STMN1 expression in the KYSE150 and KYSE450 cells (Fig. 3B).
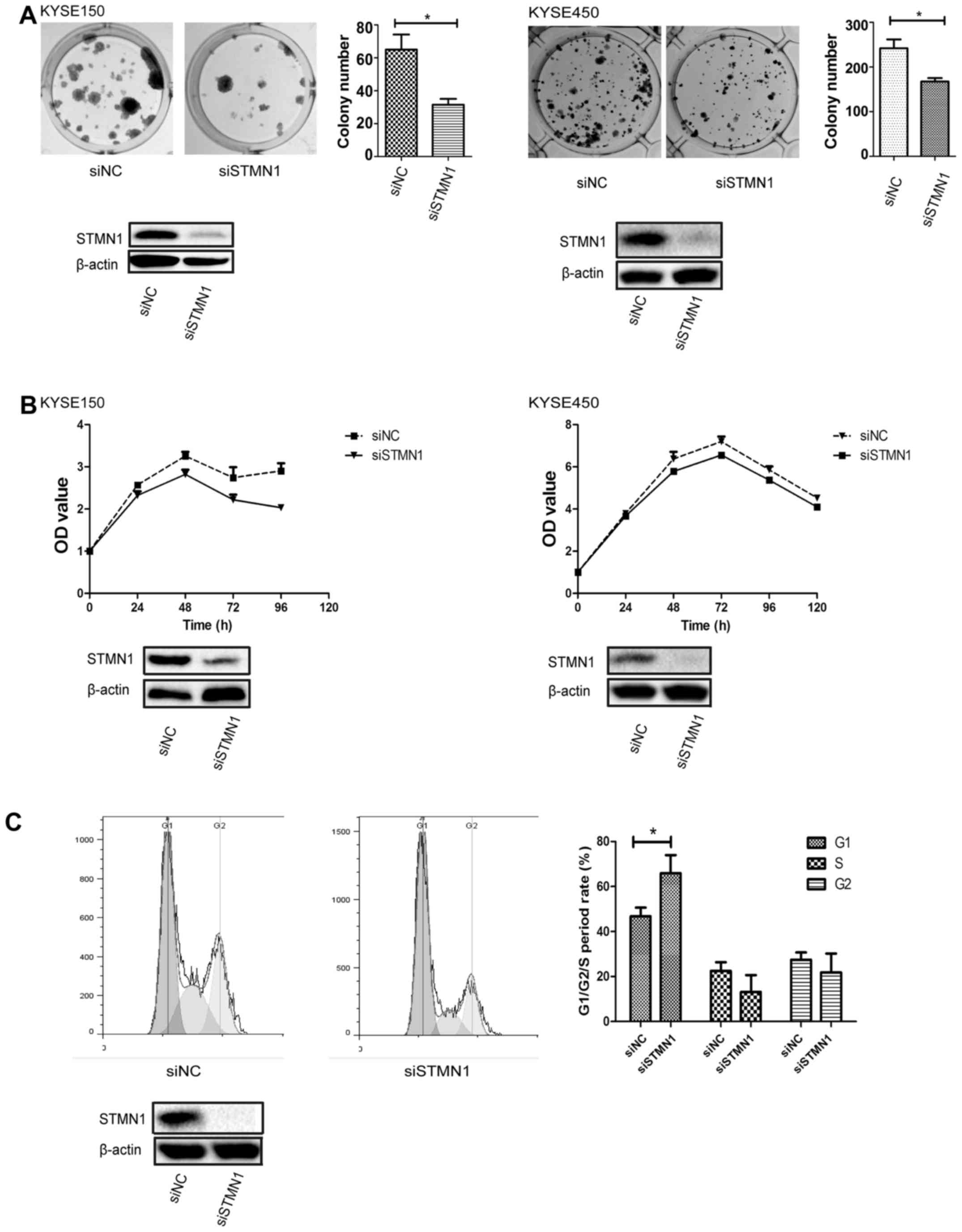
Knockdown of STMN1 expression inhibits
ESCC cell proliferation and induces cell cycle arrest in the G1
phase
Colony formation and cell proliferation assays were
performed to investigate the influence of the knockdown of STMN1
expression on ESCC cells. The results showed that STMN1
downregulation significantly decreased the number of cell colonies
(Fig. 4A). The same results were
also seen in the proliferation assay by MTS in both the KYSE150 and
KYSE450 cells (Fig. 4B).
Furthermore, a flow cytometric assay was performed
to measure the function of STMN1 in cell cycle distribution.
Upregulation of STMN1 expression induced cell cycle arrest in the
G1 phase in the KYSE450 cells (Fig.
4C). In cells transfected with siSTMN1, the percentage of cells
in the G1 phase was 65.9±4.6%, which was significantly higher than
that noted in the control cells (46.8±2.2%) (P<0.05). Moreover,
the percentage of siSTMN1 cells in the S phase was 13.1±4.2%, which
was lower than that noted in the control cells (22.5±2.2%).
However, the results did not achieve a significant difference
(P>0.05).
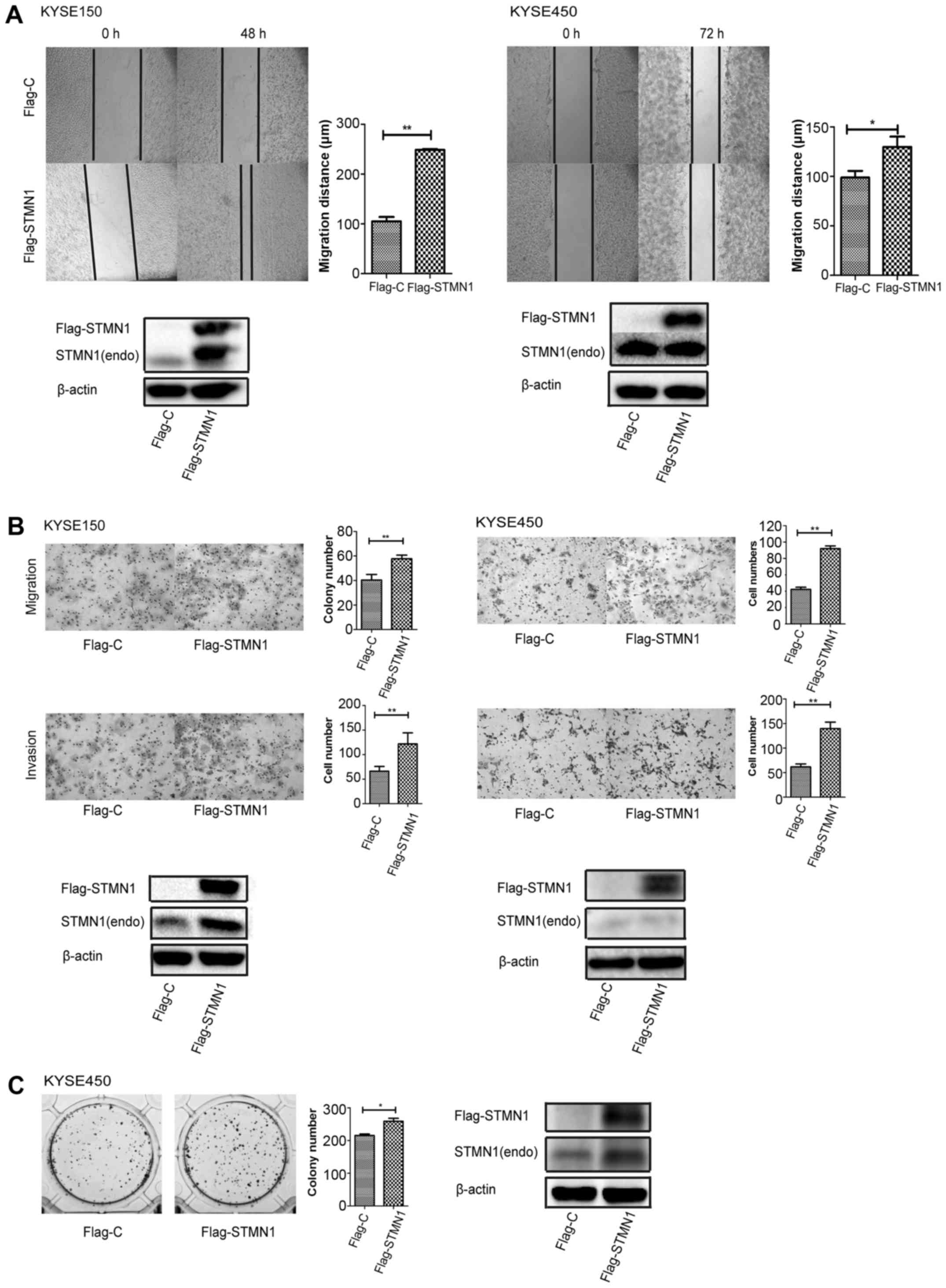
Upregulation of STMN1 expression
promotes the migration, invasion and proliferation of ESCC
cells
Finally, we upregulated STMN1 expression to identify
its function in cell motility and proliferation. Wound-healing
assay (Fig. 5A) and migration assay
(Fig. 5B) showed that
overexpression of STMN1 significantly increased cell migration in
both the KYSE150 and KYSE450 cells. The same results were also
observed in the invasion assay (Fig.
5B). Colony formation assay with STMN1 overexpression also
showed a higher colony number (Fig.
5C). The results indicated that STMN1 overexpression promoted
ESCC cell motility and proliferation.
Discussion
In the present study, we found that STMN1
overexpression indicated a poorer prognosis and the expression of
STMN1 was correlated with tumour grade. In addition, according to
the univariate and multivariate analyses, STMN1 expression was
found to be an independent prognostic factor. KYSE150 and KYSE450
cells were used to investigate the biological function of STMN1 in
ESCC. Knockdown of STMN1 expression inhibited the motility and
invasion ability of the ESCC cells, as well as inhibited tumour
cell proliferation and induced cell cycle arrest in the G1 phase.
Upregulation of STMN1 expression promoted cell migration, invasion
and proliferation.
In previous studies, STMN1 was reported to be highly
expressed in many malignancies (7).
The present study showed that overexpression of STMN1 indicated a
poorer prognosis and was an independent prognostic factor for ESCC.
Similar results were also reported in other malignancies, such as
oral squamous cell carcinoma (11),
breast (24), pancreatic (25) and gastric cancer (26), hepatoma (27) and ovarian carcinoma (8). We also found that the expression of
STMN1 was correlated with tumour grade and a higher proportion of
cells with STMN1 overexpression was found in cells with poor
differentiation (Table I). STMN1
expression was closely related to the state of cell
differentiation, not only in erythropoietic and neurite
differentiation (28), but also in
some malignant cells (11,29–31).
These results indicate that STMN1 expression is an important
biomarker for ESCC.
We also performed a series of assays to investigate
further the biological function of STMN1 in ESCC cells. According
to wound-healing, and Transwell migration and invasion assays, we
found that the ability of cell migration and invasion was impaired
by deficient STMN1 (Fig. 3). The
same result was observed in the KYSE30 and KYSE410 cells. Similar
results were reported by Liu et al (31) in ESCC, and also observed in other
malignancies (5,29). Moreover, the opposite phenomenon was
presented after upregulating STMN1 expression (Fig. 5). These findings suggest that STMN1
may play a crucial role in the process of tumour metastasis by
regulating microtubule dynamics. However, this mechanism needs to
be further explored.
The regulation of microtubule depolymerization by
STMN1 is essential in the cell cycle. STMN1 appears to be involved
in the G1/S and G2/M checkpoints to regulate cell cycle
proliferation by interaction with other cell cycle control proteins
such as p53 and Rb (28). Our
results also support this idea; we found cell cycle arrest in the
G1/S phase following the knockdown of STMN1 expression. The same
results were also reported in hypopharyngeal squamous cell
carcinoma by Chen et al (32).
A proliferation assessment was also performed using
MTS agent and colony formation assay. A significantly decreased
proliferative rate was shown in both KYSE150 and KYSE450 cells
after STMN1 knockdown (Fig. 4). Our
findings are consistent with previous studies in hepatoma (27) and cutaneous squamous cell carcinoma
(29). In some solid tumours, such
as breast and ovarian cancer, it was shown that tumours with high
proliferative potential generally express higher levels of STMN1
than less proliferative tumours (4). These observations suggest a strong
correlation between STMN1 expression and cellular proliferation in
malignant cells.
STMN1 expression was significantly associated with
prognosis and tumour differentiation in ESCC, indicating that STMN1
expression is an independent prognostic factor for ESCC and could
be a potential biomarker of ESCC. Regulation of STMN1 expression
could influence tumour cell motility, invasion, and proliferation.
In conclusion, our data indicate that STMN1 plays an important role
in cancer progression and may be a new therapeutic target for the
treatment of ESCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 30770982, 81360331,
81472613 and 81572341), the Natural Science Foundation of
China-Guangdong Joint Fund (grant nos. U1301227 and U0932001), and
the Science and Technology Planning Project of Guang Dong Province
(grant no. 2014A030304060), and the Department of Education,
Guangdong Government under the Top-Tier University Development
Scheme for Research and Control of Infectious Diseases.
References
|
1
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kollarova H, Machova L, Horakova D,
Janoutova G and Janout V: Epidemiology of esophageal cancer - an
overview article. Biomed Pap Med Fac Univ Palacky Olomouc Czech
Repub. 151:17–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rubin CI and Atweh GF: The role of
stathmin in the regulation of the cell cycle. J Cell Biochem.
93:242–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tian FJ, Qin CM, Li XC, Wu F, Liu XR, Xu
WM and Lin Y: Decreased stathmin-1 expression inhibits trophoblast
proliferation and invasion and is associated with recurrent
miscarriage. Am J Pathol. 185:2709–2721. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Machado-Neto JA, Saad STO and Traina F:
Stathmin 1 in normal and malignant hematopoiesis. BMB Rep.
47:660–665. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rana S, Maples PB, Senzer N and Nemunaitis
J: Stathmin 1: A novel therapeutic target for anticancer activity.
Expert Rev Anticancer Ther. 8:1461–1470. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei SH, Lin F, Wang X, Gao P and Zhang HZ:
Prognostic significance of stathmin expression in correlation with
metastasis and clinicopathological characteristics in human ovarian
carcinoma. Acta Histochem. 110:59–65. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghosh R, Gu G, Tillman E, Yuan J, Wang Y,
Fazli L, Rennie PS and Kasper S: Increased expression and
differential phosphorylation of stathmin may promote prostate
cancer progression. Prostate. 67:1038–1052. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baquero MT, Hanna JA, Neumeister V, Cheng
H, Molinaro AM, Harris LN and Rimm DL: Stathmin expression and its
relationship to microtubule-associated protein tau and outcome in
breast cancer. Cancer. 118:4660–4669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma HL, Jin SF, Tao WJ, Zhang ML and Zhang
ZY: Overexpression of stathmin/oncoprotein 18 correlates with
poorer prognosis and interacts with p53 in oral squamous cell
carcinoma. J Craniomaxillofac Surg. 44:1725–1732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gan L, Guo K, Li Y, Kang X, Sun L, Shu H
and Liu Y: Upregulated expression of stathmin may be associated
with hepatocarcinogenesis. Oncol Rep. 23:1037–1043. 2010.PubMed/NCBI
|
|
13
|
Zhang HZ, Gao P, Yan L and Lin F:
Significance of stathmin gene overexpression in osteosarcoma cells.
Ai Zheng. 23:493–496. 2004.(In Chinese). PubMed/NCBI
|
|
14
|
Sun R, Liu Z, Wang L, Lv W, Liu J, Ding C,
Yuan Y, Lei G and Xu C: Overexpression of stathmin is resistant to
paclitaxel treatment in patients with non-small cell lung cancer.
Tumour Biol. 36:7195–7204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nie W, Xu MD, Gan L, Huang H, Xiu Q and Li
B: Overexpression of stathmin 1 is a poor prognostic biomarker in
non-small cell lung cancer. Lab Invest. 95:56–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang F, Wang LX, He W, Zhu LN, Zhao PR and
Fan QX: Expression of stathmin in esophageal squamous cell
carcinoma and its biological significance. Nan Fang Yi Ke Da Xue
Xue Bao. 30:1552–1557. 2010.(In Chinese). PubMed/NCBI
|
|
17
|
Akhtar J, Wang Z, Jiang WP, Bi MM and
Zhang ZP: Stathmin overexpression identifies high risk for
lymphatic metastatic recurrence in pN0 esophageal squamous
cell carcinoma patients. J Gastroenterol Hepatol. 29:944–950. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rice TW, Blackstone EH and Rusch VW: 7th
edition of the AJCC Cancer Staging Manual: Esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fléjou JF: WHO Classification of Digestive
Tumors: 4th edition. Ann Pathol. 31 Suppl 5:S27–S31. 2011.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie JJ, Xu LY, Wu ZY, Zhao Q, Xu XE, Wu
JY, Huang Q and Li EM: Prognostic implication of ezrin expression
in esophageal squamous cell carcinoma. J Surg Oncol. 104:538–543.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun LL, Holowatyj A, Xu XE, Wu JY, Wu ZY,
Shen JH, Wang SH, Li EM, Yang ZQ and Xu LY: Histone demethylase
GASC1, a potential prognostic and predictive marker in esophageal
squamous cell carcinoma. Am J Cancer Res. 3:509–517.
2013.PubMed/NCBI
|
|
22
|
Xie JJ, Zhang FR, Tao LH, Lü Z, Xu XE,
Jian S, Xu LY and Li EM: Expression of ezrin in human embryonic,
fetal, and normal adult tissues. J Histochem Cytochem.
59:1001–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du
ZP, Lv Z, Gu W, Pan F, Xu XE, et al: Involvement of CYR61
and CTGF in the fascin-mediated proliferation and
invasiveness of esophageal squamous cell carcinomas cells. Am J
Pathol. 176:939–951. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuang XY, Chen L, Zhang ZJ, Liu YR, Zheng
YZ, Ling H, Qiao F, Li S, Hu X and Shao ZM: Stathmin and
phospho-stathmin protein signature is associated with survival
outcomes of breast cancer patients. Oncotarget. 6:22227–22238.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu Y, Liu C, Cheng H, Xu Y, Jiang J, Xu J,
Long J, Liu L and Yu X: Stathmin, interacting with Nf-κB, promotes
tumor growth and predicts poor prognosis of pancreatic cancer. Curr
Mol Med. 14:328–339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ke B, Wu LL, Liu N, Zhang RP, Wang CL and
Liang H: Overexpression of stathmin 1 is associated with poor
prognosis of patients with gastric cancer. Tumour Biol.
34:3137–3145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hsieh SY, Huang SF, Yu MC, Yeh TS, Chen
TC, Lin YJ, Chang CJ, Sung CM, Lee YL and Hsu CY: Stathmin1
overexpression associated with polyploidy, tumor-cell invasion,
early recurrence, and poor prognosis in human hepatoma. Mol
Carcinog. 49:476–487. 2010.PubMed/NCBI
|
|
28
|
Sherbet GV and Cajone F: Stathmin in cell
proliferation and cancer progression. Cancer Genomics Proteomics.
2:227–237. 2005.
|
|
29
|
Li X, Wang L, Li T, You B, Shan Y, Shi S,
Qian L and Cao X: STMN1 overexpression correlates with biological
behavior in human cutaneous squamous cell carcinoma. Pathol Res
Pract. 211:816–823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Hu G, Kong F, Wu K, Song K, He J and
Sun W: Elevated STMN1 expression correlates with poor prognosis in
patients with pancreatic ductal adenocarcinoma. Pathol Oncol Res.
21:1013–1020. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu F, Sun YL, Xu Y, Liu F, Wang LS and
Zhao XH: Expression and phosphorylation of stathmin correlate with
cell migration in esophageal squamous cell carcinoma. Oncol Rep.
29:419–424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Y, Zhang Q, Ding C, Zhang X, Qiu X
and Zhang Z: Stathmin1 overexpression in hypopharyngeal squamous
cell carcinoma: A new promoter in FaDu cell proliferation and
migration. Int J Oncol. 50:31–40. 2017. View Article : Google Scholar : PubMed/NCBI
|