Introduction
Colorectal cancer (CRC) is one of the most common
cancers in China, and its incidence rate has increased rapidly in
both men and women in recent years (1). Various factors affect the expression
of colorectum-specific genes, which contribute to the development
and progression of CRC. Mutations and microRNAs (miRNAs) play
important roles in the development and progression of CRC.
KRAS mutations, particularly the activated KRAS
mutation, induces cell invasion and maintains metastases in CRC,
which is considered a landmark event in CRC (2), and patients with KRAS mutations
have lower survival rates (3).
Therefore, targeting KRAS is a promising treatment strategy
for CRC.
miRNAs play critical roles in CRC tumorigenesis
(4). In particular, the following
miRNA genes are important: the miR-17-92 cluster consisting of 6
miRNA genes (miR-17, miR-18a, miR-19a,
miR-20a, miR-19b-1 and miR-92a-1), the
miR-106b-25 and the miR-106a-363 cluster (5). Previous studies have shown that the
miR-17-92 cluster is upregulated in various human cancers,
including CRC (6), and is often
thought to play important roles in the development and progression
of CRC (7,8).
Among the miRNAs within the miR-17-92
cluster, miR-19 functions as a key oncogenic miRNA (9). miR-19 is comprised of
miR-19a and miR-19b, which share a 96% identity,
differing by a single nucleotide at position 11, and are likely to
regulate the same mRNA targets. Indeed, miR-19a expression
has been revealed to be closely related to the development and
progression of several types of tumors (10,11).
For example, miR-19a contributed to the proliferation and
invasion of CRC cells (12).
Additionally, hsa-miR-19a is associated with lymph
metastasis and mediates tumor necrosis factor (TNF)-α-induced
epithelial-to-mesenchymal transition in CRC (13). miR-19 mediated inhibition of
transglutaminase-2 led to enhanced invasion and metastasis in CRC
(14). However, several studies
have revealed that miR-19a may negatively affect CRC
development. For example, Yu et al demonstrated that
miR-19a can suppress tissue factor expression in
vitro and inhibit colon cancer cell migration and invasion
(15). Moreover, most of the
experiments reported have been based on in vitro cell
culture, and few experiments have ascertained the actual role of
miR-19a in colorectal solid tumors in vivo. Thus, the
specific role of miR-19a in CRC remains unclear.
We identified KRAS as one of the target genes
of miR-19a, using a site-prediction web site, and were
interested in investigating the effect of miR-19a
overexpression in CRC.
In the present study, we aimed to elucidate the
precise role and underlying mechanisms of miR-19a in CRC,
using both in vitro assays and an in vivo mouse tumor
model. We believe that our findings provide important insights into
CRC biology and highlight the possibility of using miR-19a
as a therapeutic agent for CRC in future.
Materials and methods
Study approval
Animal experiments were performed in compliance with
the rules of the Animal Use Committee of Fujian Medical University.
The present study was approved by the Institutional Ethics Board of
Fujian Medical University Union Hospital.
Northern blotting
Northern blotting was performed to detect miRNAs as
previously described (16).
Briefly, we used RNAiso Plus to extract 10–20 µg total RNA from
cells. The RNA was then separated on 10% denaturing polyacrylamide
gels and electrotransferred to nylon membranes. DNA
oligonucleotides that were antisense to mature miRNAs and
end-labeled with 32P were used as probes. U6 snRNA was
used as an internal control. A miR-19a probe (Pr009177336,
5′-UCAGUUUUGCAUAGAUUUGCACA-3′) was used to detect
miR-19a.
Cell culture and transfection
The human CRC cell line HCT116 (a gift from
Professor Pan Jinshui) was cultured in Dulbecco's modified Eagle's
medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco
Life Technologies, Gaithersburg, MD, USA) and was maintained at
37°C in a humidified 5% CO2 atmosphere. The
miR-19a mimic (5′-UGUGCAAAUCUAUGCAAAACUGA-3′), inhibitor and
negative control were purchased from RiBoBio (Guangzhou, China).
For transfection, cells (3×105/well) were cultured in a
six-well plate until they reached 40–50% confluency, and the mimics
were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad,
CA, USA) according to the manufacturer's instructions.
Histology and
immunohistochemistry
Hematoxylin and eosin (H&E) staining was
performed using standard protocols. Briefly, deparaffinized
rehydrated sections were stained with hematoxylin for 5 min,
incubated in 0.1% acid alcohol, and then counterstained in 0.5%
eosin Y solution. Sections were finally dehydrated and examined by
a skilled pathologist. For immunohistochemistry, sections were
deparaffinized and quenched with 3% H2O2.
After the antigen was retrieved in 10 mM sodium citrate buffer and
blocked with goat serum (ZLI-9022; ZSGB-BIO, Xicheng, Beijing,
China), the sections were incubated with antibodies recognizing
CD31 (ab28364; Abcam, Cambridge, UK) or KRAS (12063–1-AP;
Proteintech, Chicago, IL, USA) and then with secondary antibodies
conjugated with horseradish peroxidase (PV-6101; ZSGB-BIO).
Sections were processed according to the manufacturer's
instructions and counterstained with hematoxylin.
Western blotting and antibodies
We performed sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE) to separate total cell lysates, and
then transferred the proteins to polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). Western blotting was
performed with the appropriate antibodies, which were visualized by
enhanced chemiluminescence in accordance with the manufacturer's
instructions (ECL; Millipore).
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Reverse transcription of total RNA was performed
using 4 µg total RNA from HCT116 cells. M-MLV reverse transcriptase
(BGI, Shenzhen, China) was used to generate cDNAs. The mRNA level
of KRAS was assessed by qRT-PCR using SYBR-Green I on a
CFX96 real-time RT-PCR detection system (Bio-Rad, Hercules, CA,
USA). The sequences of the primers used in the present study are
listed in Table I. PCR was
performed for 45 cycles using the following conditions:
denaturation at 95°C for 20 sec, annealing at 58°C for 20 sec, and
elongation at 72°C for 20 sec. All values were normalized to the
expression of glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) mRNA, and the relative expression was calculated
according to the ΔΔCT method (17),
where ΔΔCT = ΔCTsample - ΔCTGAPDH.
 | Table I.Sequences of primers used in the
present study. |
Table I.
Sequences of primers used in the
present study.
| Primer | Sequence |
|---|
| miR-19a |
|
|
Probe |
5′-UCAGUUUUGCAUAGAUUUGCACA-3′ |
| KRAS |
|
|
qRT-PCR |
5′-ACAGAGAGTGGAGGATGCTTT-3′ (forward) |
|
|
5′-TTTCACACAGCCAGGAGTCTT-3′ (reverse) |
|
hKRAS-3′UTR |
5′-CCGTGTAATTCTAGAGCTTATTTTAAAATGACAGTGGAAGT-3′
(forward, F1) |
|
|
5′-CGCCCCGACTCTAGAGGATAGGGTTCTGTCTATTCATACC-3′
(reverse, R1) |
|
hKRAS-3′UTR-M |
5′-TTGGATAGCTCAACAAGATACAATCTCACTCTGTGG-3′
(forward, F2) |
|
|
5′-TGTTGAGCTATCCAAGTCCCTCCCCATTTTGACTA-3′
(reverse, R2) |
|
sh-KRAS-1# |
5′-AAAAGGATTCCTACAGGAAGCAAGTTTGGATCCAAACTTGCTTCCTGTAGGAATCC-3′ |
|
sh-KRAS-2# |
5′-AAAAGGACTTAGCAAGAAGTTATGGTTGGATCCAACCATAACTTCTTGCTAAGTCC-3′ |
|
sh-control |
5′-AAAATACAACAGCCACAACGTCTATTTGGATCCAAATAGACGTTGTGGCTGTTGTA-3′ |
| GAPDH |
5′-AGAAGGCTGGGGCTCATTTG-3′ (forward) |
|
|
5′-AGGGGCCATCCACAGTCTTC-3′ (reverse) |
| U6 probe |
5′-CTCGCTTCGGCAGCACA-3′ |
In vitro tube formation assay
The tube formation efficiency of human umbilical
vein endothelial cells (HUVECs) was assessed by an angiogenesis
assay on Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). First,
we prepared the GFP-labeled HUVECs, as described by Guo et
al (18), and then 36 h after
transfection with miR-19a, KRAS/sh-KRAS-2# or
VEGFA expression constructs, the HUVECs (1×104
cells) were added to a 24-well plate coated with 100 µl Matrigel
basement membrane matrix (BD Biosciences), which was derived from
an Engelbreth-Holm-Swarm tumor. After culturing for 12 h, we
recorded tube formation with an IX71 inverted fluorescence
microscope (Olympus, Tokyo, Japan) at a low magnification of (×10).
Four wells for each treatment were photographed. The images were
saved as JPEG files and analyzed using TCS Cellworks AngioSys 1.0
software (Botolph Claydon, Buckingham, UK) to quantify the effects
on angiogenesis. Data were analyzed using unpaired Student's t-test
of GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA).
Packaging of lentivirus
For lentivirus packaging, 1.5 µg lentivirus vector
was mixed with 1.5 µg packaging plasmids (0.75 µg pMDL, 0.45 µg
VSV-G and 0.3 µg REV) in six-well plates. The mixed plasmids were
transfected into 293T cells at a density of ~80% confluence using
calcium phosphate precipitation. Lentivirus particles were
harvested 48 h after transfection. The plasmid pBOB-EGFP was used
as a positive control.
Tumorigenesis assay
HCT116 cells were infected using the appropriate
amount of lentivirus, and then subjected to amplification. Infected
cells were trypsinized and suspended in FBS-free DMEM-F12 medium at
a density of 6×106 cells/0.1 ml. The suspended cells
were then implanted into the backs of 27 five-week-old BALB/c
female nude mice divided into 3 groups, with each group containing
nine mice for initiating tumor xenografts. Tumor diameters were
measured every 3 days.
DNA constructs
The plv vector carrying a copy of miR-19a or
VEGFA was a kind gift from Dr Lixin Hong (School of Life
Science, Xiamen University, Fujian, China). The pGL3 control vector
carrying a 1,923-bp fragment of the 3′UTR of human VEGFA
mRNA was directly purchased from RiBoBio. The KRAS 3′UTR
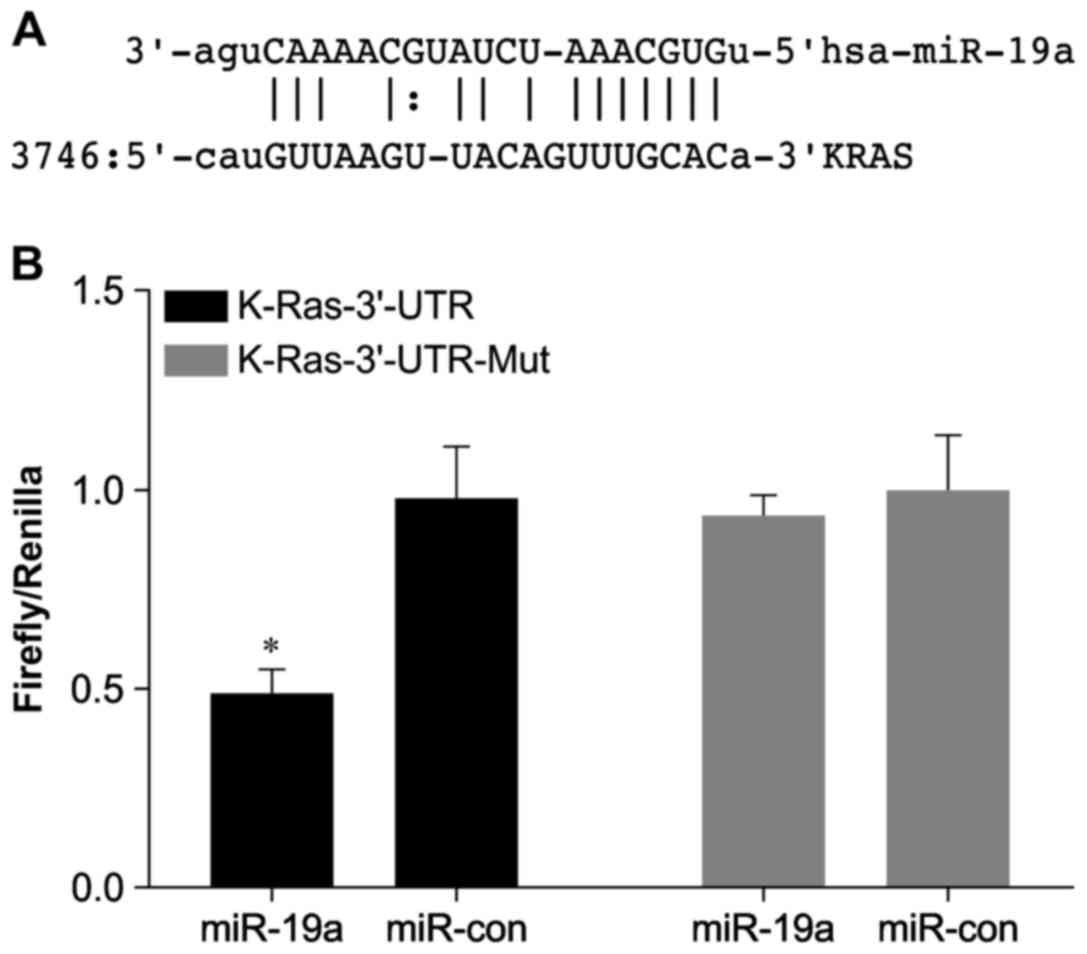
harboring predicted target sites for miR-19a (Fig. 1A) was PCR-amplified using primers
containing XbaI sites and cloned into the
XbaI-digested pGL3 control vector. The recombinant plasmids
were constructed using ligation-independent combination (LIC). F1
and R1 primers were used to generate the KRAS wild-type
3′UTR. To construct 3′UTRs with deletion mutations, two sets of PCR
were performed using the primer pairs F1/R2 and F2/R1 to generate
two fragments. Then, the two fragments were cloned into the
XbaI-digested pGL3 control vector using LIC.
The DNA oligos encoding shRNA sequences were
designed and cloned into the expression vector pLV-H1-EF1α-puro
using single oligonucleotide RNAi technology developed by Biosettia
(San Diego, CA, USA). All lentiviral-shRNA vectors were constructed
following the manufacturer's protocol. The primers used in the
present study are listed in Table
I.
Dual-luciferase reporter assay
HCT116 cells were plated in six-well culture plates
24 h before transfection. The cells were then transfected with the
recombinant pGL3 vector (1.6 µg), plv-miR-19a (0.2 µg) and
the pRLTK vector (0.2 µg) for normalization of transfection
efficiency. Cell lysates were collected and assayed 48 h after
transfection. Firefly and Renilla luciferase activities were
determined using a Dual-Luciferase Reporter Assay System (Promega,
Madison, WI, USA). Firefly luciferase activities were calculated as
the mean ± standard deviation (SD) after normalization to
Renilla luciferase activity. Three independent experiments
were performed.
Statistical analysis
Mann-Whitney tests were performed to compare the
differences between two groups when variances were unequal.
Analysis of variance with post hoc tests were performed when
comparing >2 groups. For qualitative data, non-parametric tests
such as Kruskal-Wallis tests were performed.
Results
miR-19a suppresses KRAS expression in
HCT116 cells
We identified the conserved miR-19a-binding
site in the 3′UTR region of human KRAS using the microRNA.org website (Fig.
1A). The KRAS 3′UTR-Mut was generated by deleting the
predicted binding site. We observed that a reporter gene containing
the KRAS 3′UTR was suppressed by miR-19a, while
removal of the predicted miR-19a-binding site resulted in
insensitivity of the reporter gene to miR-19a in HCT116
cells (Fig. 1B).
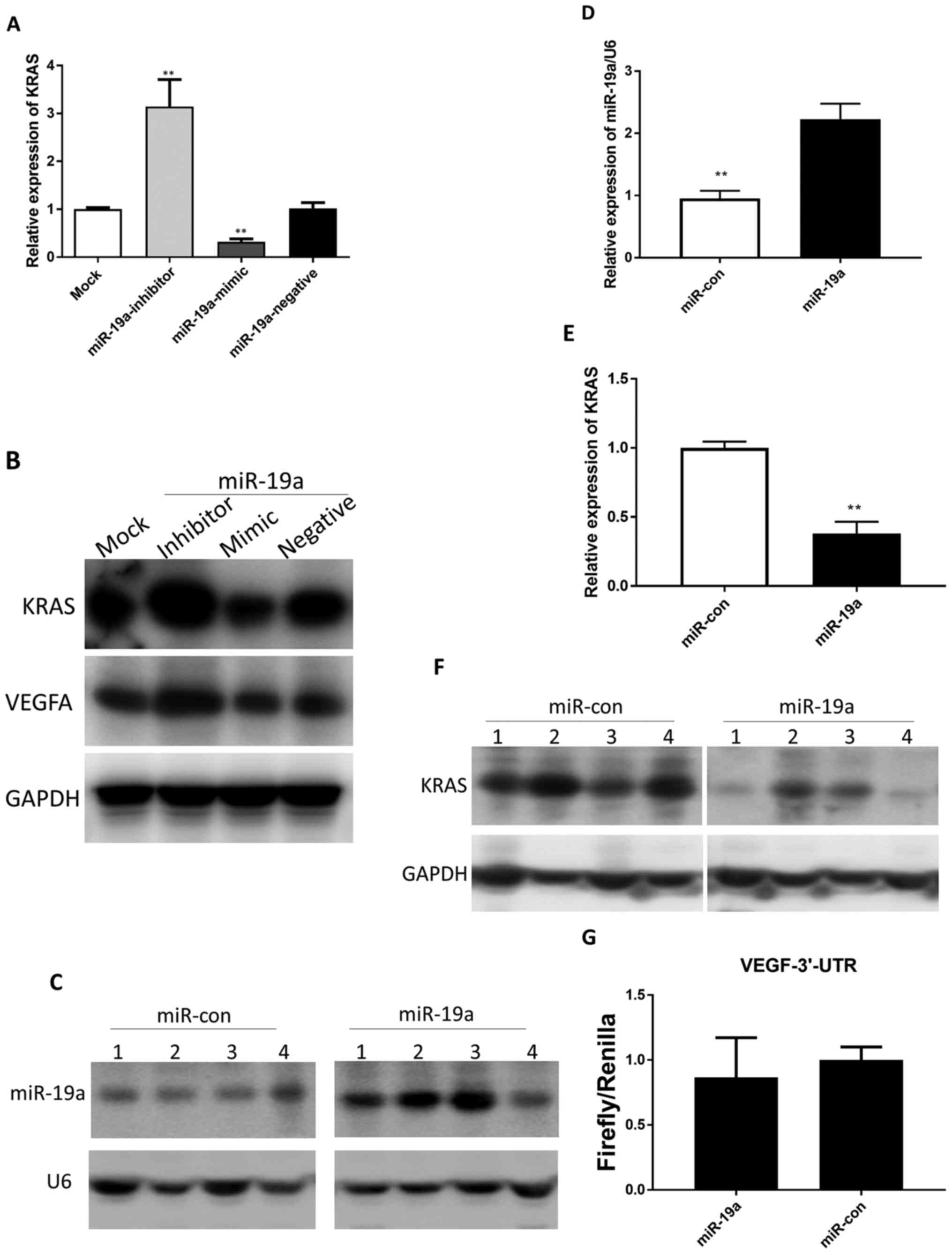
To ascertain the effect of miR-19a on
KRAS expression in HCT116 cells, the mature miR-19a
mimic/inhibitor was transfected in HCT116 cells. The results
revealed that the endogenous KRAS mRNA and protein levels
were significantly downregulated in the miR-19a mimic cells
and upregulated in the miR-19a inhibitor cells compared with
those in the untransfected cells (Fig.
2A and B). Accordingly, the VEGFA protein level was also
downregulated in the miR-19a mimic cells and upregulated in
the miR-19a inhibitor cells (Fig. 2B).
Next, in order to construct stable
miR-19a-overexpressing HCT116 cell lines for subsequent
studies, we transfected HCT116 cells with a lentivirus expressing
miR-19a. Then, via northern blotting, we confirmed
miR-19a overexpression in HCT116 cells (Fig. 2C and D). Next, we compared the
levels of KRAS expressed in HCT116 cells with and without
lentiviral transfection for overexpression of miR-19a. The
resulting in vitro data revealed that overexpression of
miR-19a decreased KRAS mRNA and protein levels in
HCT116 cells compared with that in the control (miR-Con; Fig. 2E and F).
TargetScan analysis revealed that VEGFA was
not a miR-19a target. Since the sites that match the miRNA
seed sequence are located in 3′UTRs in most mammalian mRNAs, we
performed the dual-luciferase reporter assay to detect whether
miR-19a bound to the 3′UTR of VEGFA. The results
revealed that the reporter gene containing the VEGFA 3′UTR
was not suppressed by miR-19a, which confirmed that
VEGFA was not directly regulated by miR-19a in HCT116
cells (Fig. 2G).
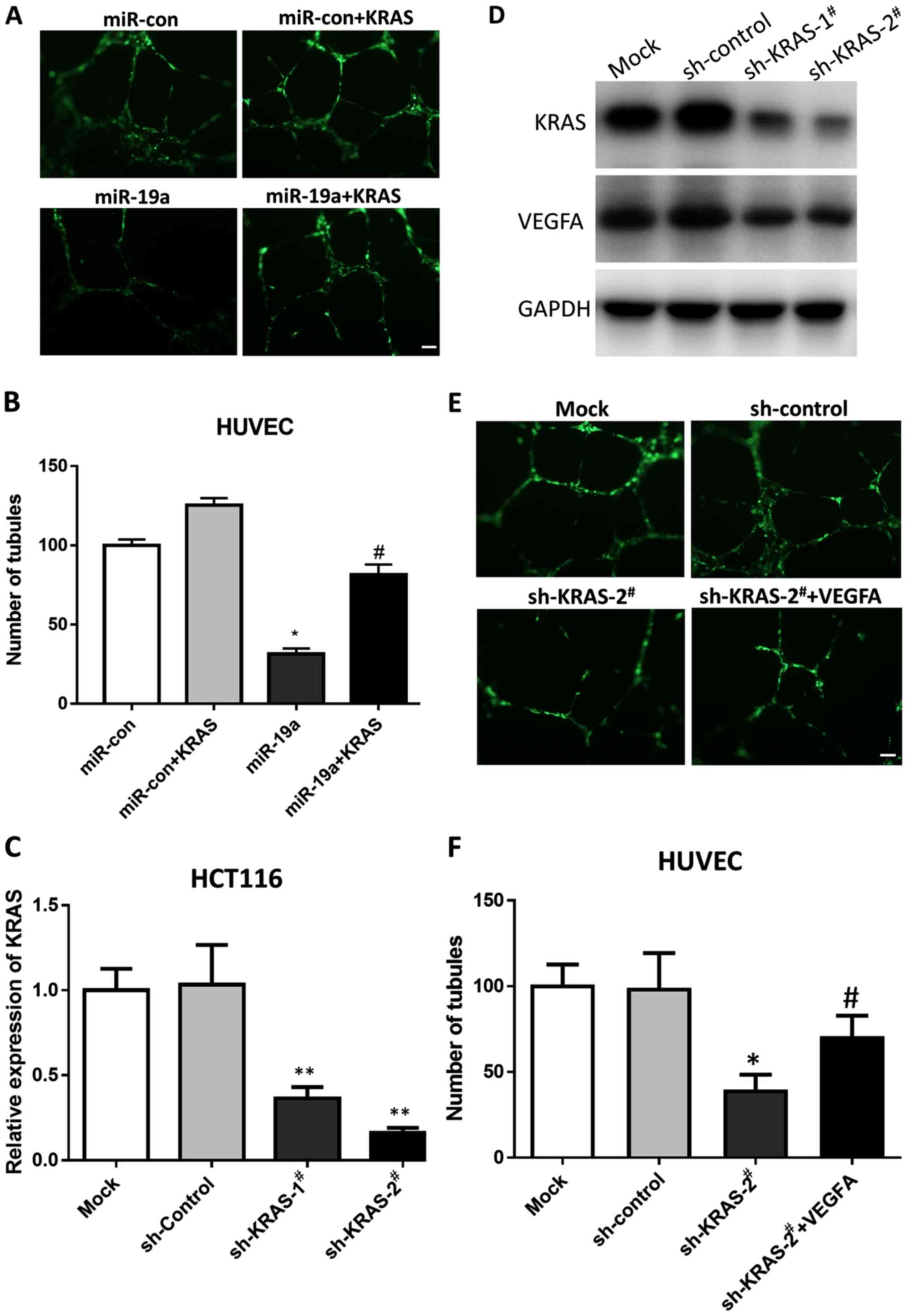
miR-19a suppresses tube formation by
targeting KRAS in vitro
Since high levels of miR-19a downregulates
KRAS, we hypothesized that miR-19a may affect
angiogenesis. Hence, we performed tube formation assays using
HUVECs, which differentiate and form capillary-like structures on
Matrigel, to mimic the process via which endothelial cells form
capillaries in vivo (19).
The results revealed that overexpression of miR-19a
significantly (P<0.01) inhibited angiogenesis in the Matrigel
assay, and that rescuing KRAS expression could restore
angiogenesis inhibited by miR-19a (Fig. 3A and B). This confirmed our
hypothesis that miR-19a suppressed angiogenesis by targeting
KRAS.
KRAS is a key regulator in
VEGF-related angiogenesis
A previous study revealed that the vascular
endothelial growth factor receptor (VEGFR) signaling pathway plays
a major role in angiogenesis (20)
and that KRAS plays a key role in VEGF signaling (21). To ascertain these results, we
knocked down KRAS using shRNA and determined the expression
level of VEGFA in HCT116 cells. The results revealed that two
KRAS shRNAs reduced KRAS mRNA and protein levels
significantly, and consequently, VEGFA was also downregulated
(Fig. 3C and D). Furthermore, we
knocked down KRAS using shRNA in HUVECs and performed the
tube formation assay. As shown in Fig.
3E, KRAS knockdown disrupted tube formation, whereas
re-expression of VEGFA rescued the defect. Thus, the results
demonstrated that KRAS was a key regulator of VEGFA
expression and angiogenesis.
miR-19a suppresses CRC tumor growth in
nude mice
The above data indicated that miR-19a
overexpression could target KRAS in HCT116 cells and inhibit
tube formation. Therefore, we next examined whether overexpression
of miR-19a could suppress CRC progression by blocking
angiogenesis via targeting of KRAS using a xenograft
model.
We transplanted the transfected cells
into the flanks of nude mice
The results revealed that the sizes of xenograft
tumors that developed from HCT116 cells expressing miR-19a
were much smaller than those from HCT116 cells transfected with the
control lentivirus (Fig. 4A and B).
In addition, tumors that developed from miR-19a-expressing
lentivirus-transfected cells tended to be paler in color than
xenograft tumors developed from control lentivirus-transfected
cells, indicating decreased blood flow in miR-19a-expressing
tumors (Fig. 4A).
We performed H&E staining to
examine the blood vessels in these tumors
We observed that tumors that developed from
miR-19a-expressing cells had fewer blood vessels than the
control tumors. Immunohistochemical staining for CD31, a marker of
vascular endotheliocytes, also revealed that the blood vessels in
tumors that developed from miR-19a-expressing cells were
smaller than those in the control group (Fig. 4D). Moreover, immunohistochemical
staining for KRAS and western blotting analysis confirmed
that the tumors that developed from miR-19a-expressing cells
exhibited reduced KRAS expression compared with those in the
control group (Fig. 4C and D).
Moreover, western blotting analysis revealed that the expression
level of VEGFA in tumors that developed from
miR-19a-expressing cells was also reduced (Fig. 4C). These results indicated that
overexpression of miR-19a inhibited KRAS expression
in CRC, which may reduce the production of VEGFA and suppress tumor
angiogenesis, thereby delaying tumor growth.
Discussion
In the present study, we evaluated the role of
miR-19a in colorectal cancer (CRC) development and
progression, and observed that overexpression of miR-19a
resulted in the inhibition of CRC angiogenesis owing to reduced
expression of KRAS, a known oncogene that has been
demonstrated to be a target of miR-19a in chronic myeloid
leukemia (22).
A study in 2010 revealed that overexpression of
miR-17, miR-18a, miR-19a and miR-20a
considerably inhibited three-dimensional spheroid sprouting in
vitro (23). Additionally,
Landskroner-Eiger et al suggested that miR-19 of the
miR-17-92 family may negatively affect the formation of
arterial blood vessels (13).
Hence, we hypothesized that miR-19a inhibited angiogenesis.
In the present study, we revealed that overexpression of
miR-19a reduced KRAS expression in the HCT116 cell
line and inhibited angiogenesis, as determined by an in
vitro tube formation assay. This inhibition, which could be
rescued by KRAS, consequently slowed down xenograft tumor
growth. Hence, we believe that miR-19a may be closely
related to CRC angiogenesis, and that the anti-angiogenesis effect
results from the targeting of KRAS by a microRNA. To the
best of our knowledge, this is the first study to investigate the
primary function of miR-19a in solid CRC tumors and to
provide a theoretical foundation for the application of miRNA
treatment to CRC.
Vascular supply is key for tumor development
(24) in various types of cancers,
including CRC. Vascular endothelial growth factor (VEGF) and the
VEGF receptor pathway play key roles in tumor angiogenesis
(25). VEGF/VEGF receptor
activation can initiate a signaling cascade that promotes
endothelial cell proliferation, migration and differentiation
(26). VEGF can also be secreted by
CRC cells (27), and it induces
angiogenesis. KRAS and VEGF have a close relationship.
KRAS activation significantly enhances the production of
angiogenic factors, including CXC chemokines and VEGF (28), and KRAS mutations
significantly increase VEGF production (29). Therefore, KRAS and VEGF may
be involved in creating tumor vascular networks.
Moreover, previous studies have shown a pivotal role
of KRAS in human cancer development (30) and demonstrated that KRAS
mutations were indicative of poor prognosis in CRC (31). Therefore, targeting KRAS is a
promising therapeutic strategy for CRC treatment, particularly for
patients with mutations in KRAS, which are common in CRC
(32). Use of miRNAs for degrading
KRAS holds promise. For most mammalian mRNAs, the sites that
match the miRNA seed sequence (nucleotides 2–7), particularly those
in 3′UTRs, are preferentially conserved (33). Therefore, miRNA therapy is unlikely
to be affected by gene variations.
Our findings in the present study provided
important insights into the role of miR-19a in cancer and
laid the foundation for future research in this area. High
miR-19a levels reduced KRAS expression and affected
angiogenesis in CRC. Thus, miR-19a may have applications as
a therapeutic agent. However, the present study has one limitation.
The in vitro analyses were only performed in HCT116 cells,
and additional studies using other CRC cell lines are required.
With greater and improved understanding of the role of
miR-19a in CRC, the usefulness of this microRNA in the
diagnosis and treatment of CRC may be maximized.
Acknowledgements
The plv vector carrying copies of miR-19a
and VEGFA was a kind gift from Dr Lixin Hong (School of Life
Science, Xiamen University, Fujian, China). We are grateful to
Professor Pan Jinshui (Xiamen University Zhongshan Hospital,
Fujian, China) for kindly providing the HCT116 cell line. The
present study was supported by the Key Clinical Specialty
Discipline Construction program of Fujian, P.R. China (grant no.
2012-149).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boutin AT, Liao WT, Wang M, Hwang SS,
Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, et al:
Oncogenic Kras drives invasion and maintains metastases in
colorectal cancer. Genes Dev. 31:370–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taieb J, Le Malicot K, Shi Q, Penault
Lorca F, Bouché O, Tabernero J, Mini E, Goldberg RM, Folprecht G,
Luc Van Laethem J, et al: Prognostic value of BRAF and KRAS
mutations in MSI and MSS stage III colon cancer. J Natl Cancer
Inst. 109:pii: djw2722016. View Article : Google Scholar
|
|
4
|
Yi R, Li Y, Wang FL, Miao G, Qi RM and
Zhao YY: MicroRNAs as diagnostic and prognostic biomarkers in
colorectal cancer. World J Gastrointest Oncol. 8:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lanza G, Ferracin M, Gafà R, Veronese A,
Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM and Negrini M:
mRNA/microRNA gene expression profile in microsatellite unstable
colorectal cancer. Mol Cancer. 6:542007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Diosdado B, van de Wiel MA, Terhaar Sive
Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B and Meijer
GA: MiR-17-92 cluster is associated with 13q gain and c-myc
expression during colorectal adenoma to adenocarcinoma progression.
Br J Cancer. 101:707–714. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma H, Pan JS, Jin LX, Wu J, Ren YD, Chen
P, Xiao C and Han J: MicroRNA-17~92 inhibits colorectal cancer
progression by targeting angiogenesis. Cancer Lett. 376:293–302.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Olive V, Bennett MJ, Walker JC, Ma C,
Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ and He L: miR-19
is a key oncogenic component of mir-17-92. Genes Dev. 23:2839–2849.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamamoto K, Ito S, Hanafusa H, Shimizu K
and Ouchida M: Uncovering direct targets of miR-19a involved in
lung cancer progression. PLoS One. 10:e01378872015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu WD, Zuo Y, Xu Z and Zhang M: MiR-19a
promotes epithelial-mesenchymal transition through PI3K/AKT pathway
in gastric cancer. World J Gastroenterol. 21:4564–4573. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Xiao Z, Lai D, Sun J, He C, Chu
Z, Ye H, Chen S and Wang J: miR-21, miR-17 and miR-19a induced by
phosphatase of regenerating liver-3 promote the proliferation and
metastasis of colon cancer. Br J Cancer. 107:352–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Landskroner-Eiger S, Qiu C, Perrotta P,
Siragusa M, Lee MY, Ulrich V, Luciano AK, Zhuang ZW, Corti F,
Simons M, et al: Endothelial miR-17~92 cluster negatively regulates
arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl
Acad Sci USA. 112:pp. 12812–12817. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cellura D, Pickard K, Quaratino S, Parker
H, Strefford JC, Thomas GJ, Mitter R, Mirnezami AH and Peake NJ:
miR-19-mediated inhibition of transglutaminase-2 leads to enhanced
invasion and metastasis in colorectal cancer. Mol Cancer Res.
13:1095–1105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu G, Li H, Wang X, Wu T, Zhu J, Huang S,
Wan Y and Tang J: MicroRNA-19a targets tissue factor to inhibit
colon cancer cells migration and invasion. Mol Cell Biochem.
380:239–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao C, Calado DP, Galler G, Thai TH,
Patterson HC, Wang J, Rajewsky N, Bender TP and Rajewsky K: MiR-150
controls B cell differentiation by targeting the transcription
factor c-Myb. Cell. 165:10272016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo H, Jia Y, Shang M, Zhang Y, Xie F,
Wang H, Yuan M, Yuan L and Ye J: Comparison of two in vitro
angiogenesis assays for evaluating the effects of netrin-1 on tube
formation. Acta Biochim Biophys Sin. 46:810–816. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ponce ML: Tube formation: An in vitro
matrigel angiogenesis assay. Methods Mol Biol. 467:183–188. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Simons M and Eichmann A: Molecular
controls of arterial morphogenesis. Circ Res. 116:1712–1724. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yeh YW, Cheng CC, Yang ST, Tseng CF, Chang
TY, Tsai SY, Fu E, Chiang CP, Liao LC, Tsai PW, et al: Targeting
the VEGF-C/VEGFR3 axis suppresses Slug-mediated cancer metastasis
and stemness via inhibition of KRAS/YAP1 signaling. Oncotarget.
8:5603–5618. 2017.PubMed/NCBI
|
|
22
|
Chakraborty C, Sharma AR, Patra BC,
Bhattacharya M, Sharma G and Lee SS: MicroRNAs mediated regulation
of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget.
7:42683–42697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Doebele C, Bonauer A, Fischer A, Scholz A,
Reiss Y, Urbich C, Hofmann WK, Zeiher AM and Dimmeler S: Members of
the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic
function in endothelial cells. Blood. 115:4944–4950. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mihalache A and Rogoveanu I: Angiogenesis
factors involved in the pathogenesis of colorectal cancer. Curr
Health Sci J. 40:5–11. 2014.PubMed/NCBI
|
|
25
|
Shinkaruk S, Bayle M, Laïn G and Déléris
G: Vascular endothelial cell growth factor (VEGF), an emerging
target for cancer chemotherapy. Curr Med Chem Anticancer Agents.
3:95–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ikeda N, Nakajima Y, Sho M, Adachi M,
Huang CL, Iki K, Kanehiro H, Hisanaga M, Nakano H and Miyake M: The
association of K-ras gene mutation and vascular endothelial growth
factor gene expression in pancreatic carcinoma. Cancer. 92:488–499.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu Y, Yu H, Shi X, Xu K, Tang Q, Liang B,
Hu S, Bao Y, Xu J, Cai J, et al: microRNA-497 inhibits invasion and
metastasis of colorectal cancer cells by targeting vascular
endothelial growth factor-A. Cell Prolif. 49:69–78. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsuo Y, Campbell PM, Brekken RA, Sung B,
Ouellette MM, Fleming JB, Aggarwal BB, Der CJ and Guha S: K-Ras
promotes angiogenesis mediated by immortalized human pancreatic
epithelial cells through mitogen-activated protein kinase signaling
pathways. Mol Cancer Res. 7:799–808. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren J, Li G, Ge J, Li X and Zhao Y: Is
K-ras gene mutation a prognostic factor for colorectal cancer: A
systematic review and meta-analysis. Dis Colon Rectum. 55:913–923.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsuchida N, Murugan AK and Grieco M:
Kirsten Ras* oncogene: Significance of its discovery in human
cancer research. Oncotarget. 7:46717–46733. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deng Y, Wang L, Tan S, Kim GP, Dou R, Chen
D, Cai Y, Fu X, Wang L, Zhu J, et al: KRAS as a predictor of poor
prognosis and benefit from postoperative FOLFOX chemotherapy in
patients with stage II and III colorectal cancer. Mol Oncol.
9:1341–1347. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gil Ferreira C, Aran V, Zalcberg-Renault
I, Victorino AP, Salem JH, Bonamino MH, Vieira FM and Zalis M: KRAS
mutations: Variable incidences in a Brazilian cohort of 8,234
metastatic colorectal cancer patients. BMC Gastroenterol.
14:732014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|