Introduction
The Hedgehog (Hh) signaling pathway is an
evolutionarily conserved developmental pathway occurring during
embryonic development that regulates cell proliferation and
differentiation (1,2). In the absence of extracellular Hh
ligands, Patched (PTCH) inhibits the activity of Smoothened (Smo).
Signal transduction is activated when the Hh ligand binds to PTCH,
relieving its inhibition of Smo, and uninhibited Smo transmits Hh
signals to the nucleus through the activation of the Gli family of
transcription factors. The Gli family consists of Gli1, Gli2 and
Gli3; Gli1 induces Hh-target genes, Gli2 acts as an activator or a
partial repressor, Gli3 mainly acts as a repressor (3–7). The
Gli1 and Gli2 mRNA levels are relevant indicators of Hh pathway
activity. Other than during embryonic development, the Hh pathway
is also activated in mammalian adult tissues, such as the brain and
bladder (8,9), where it contributes to the maintenance
of tissue homeostasis and facilitates tissue repair (10). Moreover, the aberrant augmented
activation of Hh signaling has recently been shown to play a
causative or promoting role in the development and progression of
various human malignancies, such as lung cancer (11,12),
pleural mesothelioma (13), colon
cancer (14), melanoma (14) and neuroblastoma (15).
Large cell neuroendocrine carcinoma (LCNEC) of the
lung is a highly aggressive tumor (16–18).
Although patients with early-stage disease are treated surgically,
a standard therapeutic regimen for advanced-stage disease has not
been established, leading to a poor therapeutic outcome. Thus, new
approaches that target LCNEC are urgently needed. The Hh signaling
pathway plays an important role in the growth and maintenance of a
malignant phenotype in small cell lung cancer, which also has
neuroendocrine features (19,20).
However, the roles of the Hh pathway in LCNEC remain unconfirmed.
The objective of the present study was to determine whether the Hh
pathway is activated in LCNEC cells and whether it contributes to
cell proliferation. In addition, the potential of the Hh pathway as
a therapeutic target for LCNEC of the lung was investigated.
Materials and methods
Reagents
The Smo antagonists GDC-0449 (Selleckchem, Houston,
TX, USA) and BMS-833923 (Selleckchem) and the Gli antagonist GANT61
(Wako Pure Chemical Industries Ltd., Osaka, Japan) were dissolved
in dimethyl sulfoxide (DMSO) in aliquots of 10 mM stock solution
and stored at −20°C. Cisplatin was purchased from Wako Pure
Chemical Industries and was dissolved in DMSO in aliquots of 10 mM
stock solution and stored at 4°C.
Cell lines and cell culture
Three cell lines derived from human LCNEC cells
(H460, H1299 and H810) and one cell line derived from human lung
adenocarcinoma cells (A549) were obtained from the American Type
Culture Collection (ATCC, Manassas, VA, USA). H460, H1299 and A549
cells were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis,
MO, USA) supplemented with 10% fetal bovine serum (FBS). H810 cells
were cultured in HITES medium with 5% FBS that contained DMEM/F12,
insulin (5 µg/ml), transferrin (10 µg/ml), sodium selenite (30 nM),
hydrocortisone (10 nM), β-estradiol (10 nM), and L-glutamine (4.5
mM). All the cells were cultured in a 37°C humidified atmosphere
containing 5% CO2. According to the ATCC database, H1299
was confirmed as an LCNEC cell line. A previous report revealed
that H460 and H810 cells express N-CAM and neuroendocrine markers
such as synaptophysin or chromogranin A (21). Moreover, another report revealed
that H460 and H810 cells express a high level of neuron-specific
enolase (NSE) (22). The cell lines
of H460, H1299 and H810 were used in several studies as LCNEC
(23–25). We used A549 in this study as the
most adequate positive cellular model of the Hh pathway in lung
cancer. A549 cells have a higher level of expression of Hh pathway
components than other lung cancer cell lines such as A427, NER51 or
INER37 (26). Furthermore,
inhibition of the Hh pathway has revealed an antitumor effect on
A549 in some previous reports (15,27).
Evaluation of cell viability
Cell viability was determined using the MTT dye
reduction method. A total of 5,000 cells/well were seeded in
96-well culture plates. The cells were treated with various
concentrations of GDC-0449, BMS-833923 or GANT61 for 48 h. To each
well, 15 µl of dye solution (cat. no. G402A; Promega, Madison, WI,
USA) was added, and the cells were further incubated at 37°C for 4
h, followed by the addition of 100 µl of stop solution (cat. no.
G4001; Promega) and an additional 1 h of incubation. The absorbance
at 570 nm of the resulting solution was measured using Infinite 200
PRO (FPRO-T; Tecan, Seestrasse, Switzerland). Cell viability was
determined by dividing the absorbance value of the treated cells by
that of the untreated cells.
Small interfering RNA (siRNA)
transfection
Predesigned siRNAs targeting human Gli1 and Gli2
were purchased from Thermo Fisher Scientific (Waltham, MA, USA). As
a non-specific control siRNA, scrambled siRNA duplex (Thermo Fisher
Scientific) was used. Transfections were performed using
Lipofectamine RNAiMAX Transfection reagent (Thermo Fisher
Scientific) according to the manufacturer's instructions. Gli1 and
Gli2 were silenced by siRNA for 48 h prior to assay or
treatment.
Quantitative real-time PCR
(qRT-PCR)
The expression of mRNA was quantified using RT-PCR
with the TaqMan Gene Expression assays, Step One Plus Real-Time PCR
system, and Step One Software (Thermo Fisher Scientific). After
culturing cells at 80% confluence in 6-well culture plates, the
total RNA was extracted using the RNeasy Mini kit (cat. no. 74104;
Qiagen, Venlo, Limburg, The Netherlands) and the cDNA was
immediately synthesized using SuperScript First-Strand Synthesis
for RT-PCR (cat. no. 11904–018; Thermo Fisher Scientific) for
RT-PCR according to the manufacturer's instructions. The gene
expression was quantified relative to the
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level. TaqMan
probes for GAPDH, Smo, Gli1 and Gli2 were obtained by ordering from
Thermo Fisher Scientific (assay identification nos. Hs02758991_g1,
Hs01090242_m1, Hs00171790_m1 and Hs01119974_m1).
Apoptosis assay
To determine caspase 3 and 7 activities, the cells
were seeded in 96-well plates in triplicate. At 48 h after
treatment with cisplatin, the caspase 3 and 7 activities were
determined using a Caspase-Glo 3/7 kit (cat. no. G8090; Promega),
which measures caspase 3 and 7 levels in a single assay, according
to the manufacturer's instructions. For the fluorescent
immunohistochemical evaluation of apoptotic cells, we used the
DeadEnd™ Fluorometric TUNEL system (Promega) according to the
manufacturer's instructions. A total of 1×105 cells in 1
ml of medium was distributed in each well of a Lab-Tek II Chamber
Slide (Thermo Fisher Scientific). The cells were cultured overnight
and treated with 10 µM of cisplatin for 48 h. A small drop of DAPI
(1 µg/ml) in Vectashield anti-fade mounting medium (Vector Labs,
Peterborough, UK) was placed on each slide, and the specimens were
covered with a coverslip. The number of fluorescent-positive cells
was counted using the cell counter plugin for ImageJ software
(National Institutes of Health; http://imagej.nih.gov/ij/).
Statistical analyses and ethical
considerations
The data are presented as the means ± standard
errors, and differences between groups were evaluated using the
Student's t-test. Values of P<0.05 (2-tailed) were considered
statistically significant. All the experiments were conducted in
close adherence to institutional regulations.
Results
Expression of Hh pathway signaling
components
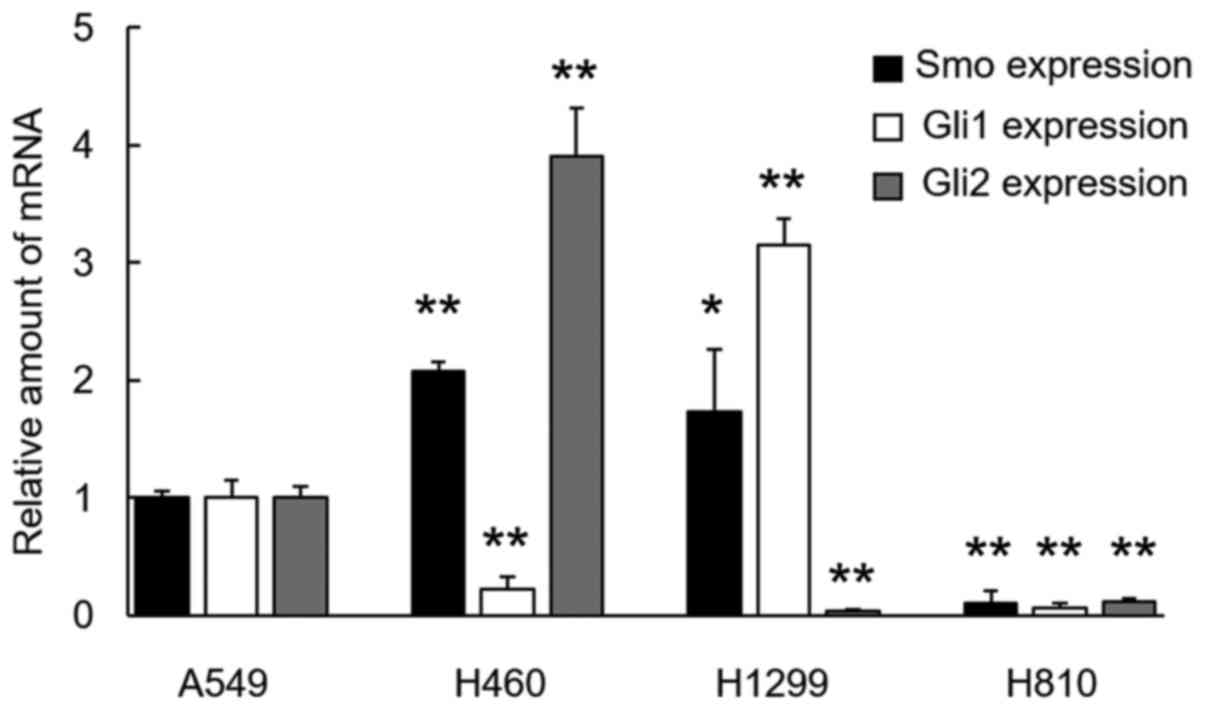
As markers for Hh signal activation, the expression
levels of Gli1, Gli2 and Smo were evaluated using qRT-PCR. Gli2 and
Smo in H460 cells and Gli1 and Smo in H1299 cells were
overexpressed, compared with the level in the A549 cells. None of
the transcription factors were overexpressed in the H810 cells
(Fig. 1).
Suppression of Hh pathway signaling
and cell viability by Smo or Gli antagonists
A Smo antagonist, GDC-0449, failed to exert
cytotoxicity in the 3 cell lines, whereas another Smo antagonist,
BMS-833923, exerted significant cytotoxicity in all 3 cell lines in
a dose-dependent manner, being more pronounced in the H460 and H810
cells than in the H1299 cells (Fig.
2A). GDC-0449 at a concentration of 10 µM failed to suppress
Gli1 expression in all 3 cell lines, while it suppressed Gli2
expression solely in H810 cells. In contrast, BMS-833923 at a
concentration of 5 µM (this dose was determined based on the
cytotoxicity effect shown in Fig.
2A) did suppress Gli1 expression in all 3 cell lines and Gli2
expression in the H460 and H810 cell lines (Fig. 2B). A Gli antagonist, GANT61, failed
to show significant cytotoxicity in all 3 cell lines, except for
within a high dose range of 50–100 µM (Fig. 2A). When administered at a dose as
high as 50 µM, this agent suppressed Gli1 expression in H460 and
H810 cells, whereas it suppressed Gli2 expression solely in the
H810 cells (Fig. 2B).
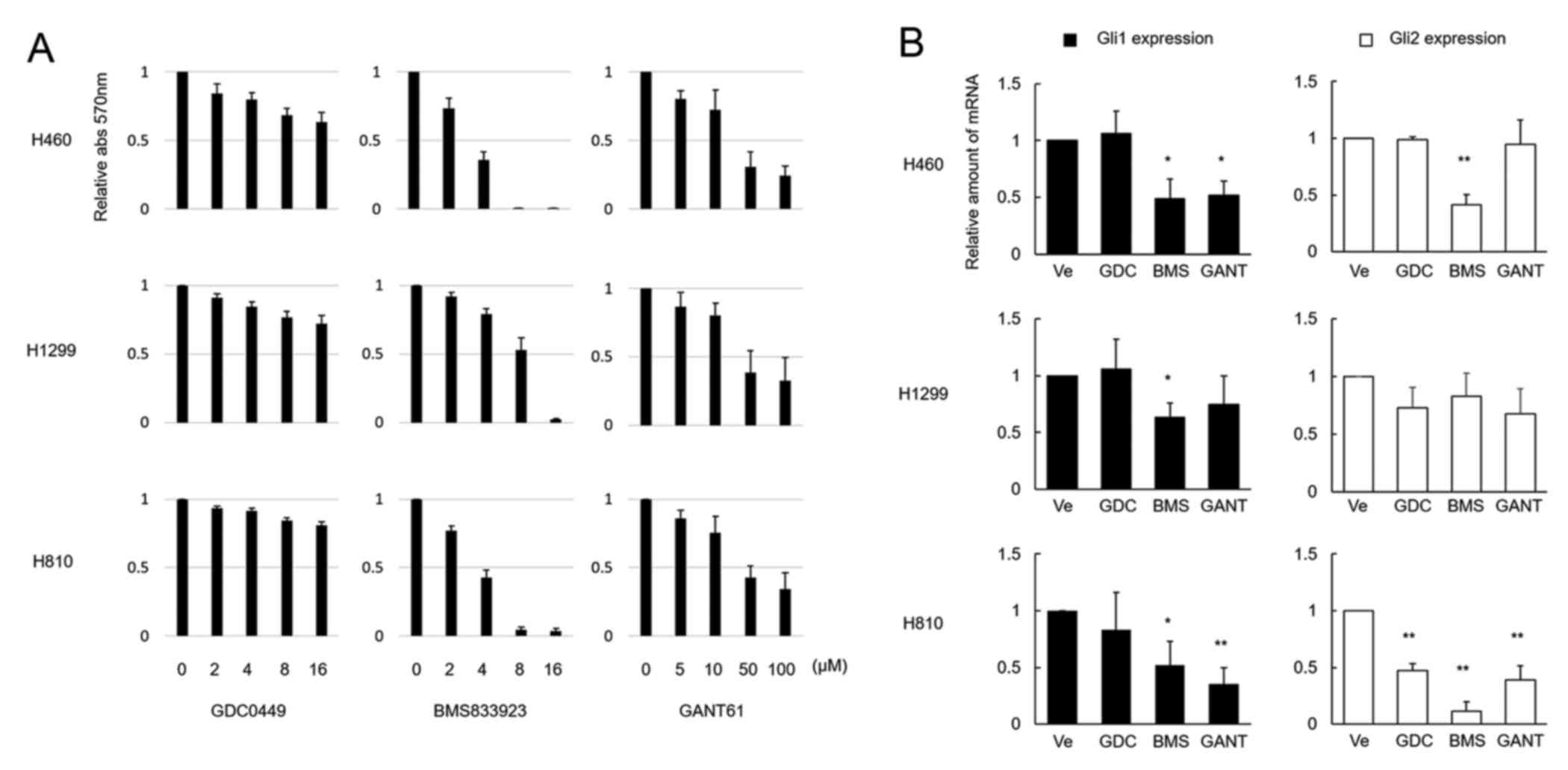 | Figure 2.Effects of Smo and Gli inhibitors on
LCNEC cell lines. (A) H460, H1299 and H810 cell lines were treated
with various concentrations of Smo inhibitors GDC-0449 and
BMS-833923 and the Gli antagonist GANT61 for 48 h. GDC-0449
exhibited limited cytotoxicity in all 3 cell lines, whereas
BMS-833923 exhibited significant effects in all 3 cell lines.
GANT61 was not effective enough in suppressing cell viability, as a
high concentration of the agent up to 100 µM was required to
achieve cell viability supression. The columns and bars represent
the means and SEs (n=3), respectively. (B) Downregulation of Gli1
and Gli2 mRNA by the 3 inhibitors, normalized by mRNA expression
levels in the untreated cells, are shown. In this experiment, the
concentrations of the 3 agents were determined according to the
cell viability experiment shown in (A) and were 10 µM for GDC-0449
(GDC), 5 µM for BMS-833923 (BMS), and 50 µM for GANT61 (GANT).
Except for the expression of Gli2 mRNA in the H810 cells, GDC did
not reduce the mRNA expression levels. BMS exerted significant
effects on both Gli1 and Gli2 mRNA, except for the expression of
Gli2 mRNA in H1299 cells. GANT suppressed Gli1 mRNA in the H460 and
H810 cell lines and Gli2 mRNA in the H810 cell line. The columns
and bars represent the means and SEs (n=3), respectively. Ve
represents the vehicle alone, without the addition of any agents.
Statistically significant differences, *P<0.05 and
**P<0.01. |
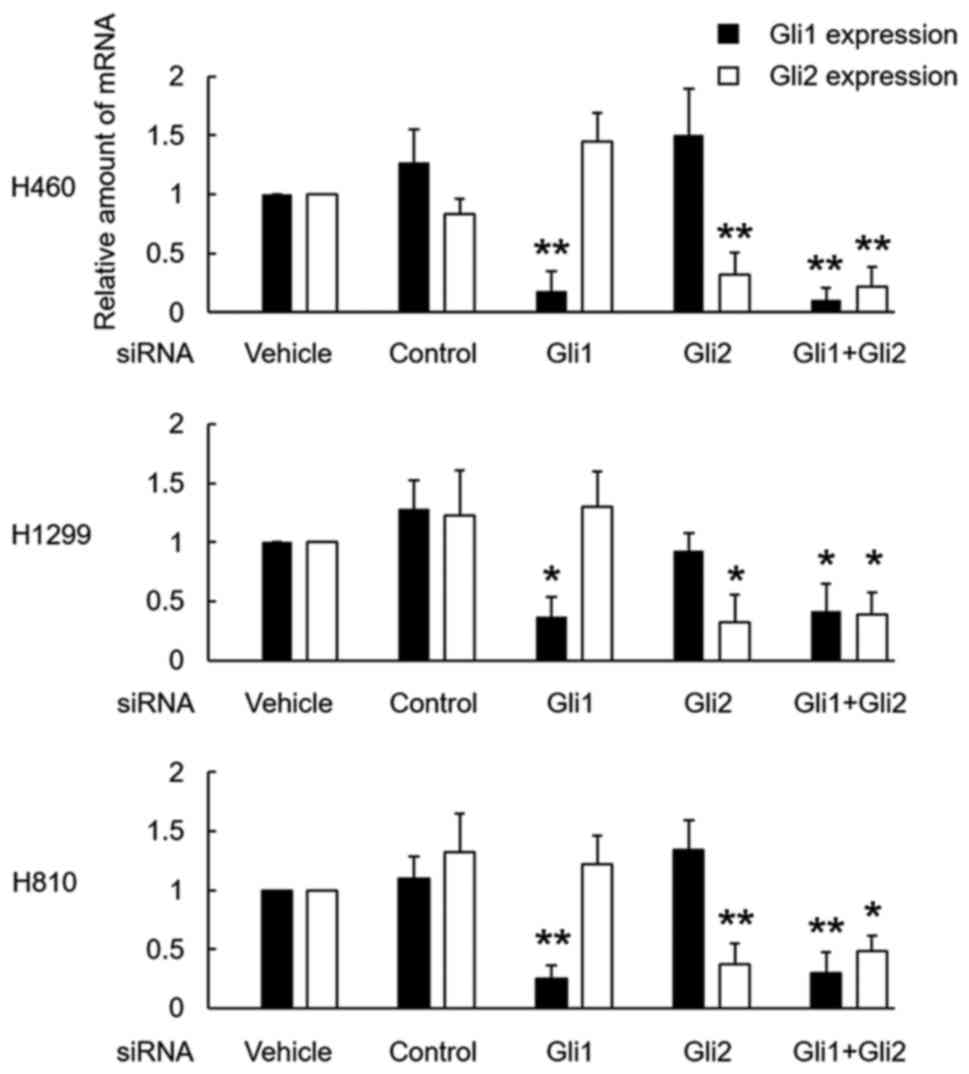
Cytotoxicity of the silencing of Gli
using siRNA
The treatment of each cell line with siRNA for Gli1
and Gli2 successfully downregulated the expression of Gli1 and
Gli2, respectively. A combination of both siRNAs also successfully
downregulated both factors in all 3 cell lines (Fig. 3).
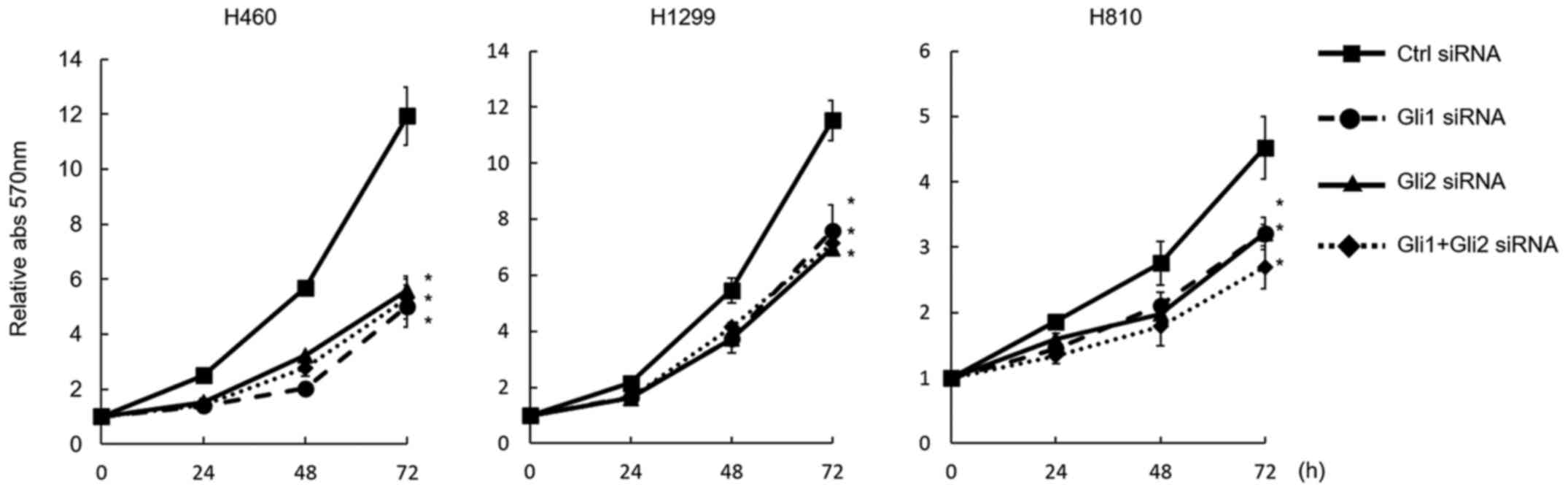
The downregulation of Gli1 and Gli2 equally and
significantly suppressed cell proliferation in all 3 cell lines.
Although the combination of Gli1 and Gli2 downregulation
significantly suppressed cell growth, no additive effect was
observed by the combination, compared with the downregulation of a
single gene (Fig. 4).
Cisplatin sensitivity and
downregulation of Gli expression
The MTT assay demonstrated that the downregulation
of Gli1, Gli2, and a combination of Gli1 and Gli2 significantly
enhanced the sensitivity to cisplatin in all 3 cell lines (Fig. 5A). When treated with cisplatin, the
downregulation of Gli1 and Gli2 induced significantly higher
caspase 3 and 7 activity levels, compared with the control, in all
3 cell lines (Fig. 5B). A TUNEL
assay also disclosed enhanced apoptosis with cisplatin treatment
after Gli downregulation in the H460 and H1299 cell lines (Fig. 5C). As repeated TUNEL assays failed
to show clear TUNEL-positive cells among H810 cells, the results
for the H810 cell line are not shown.
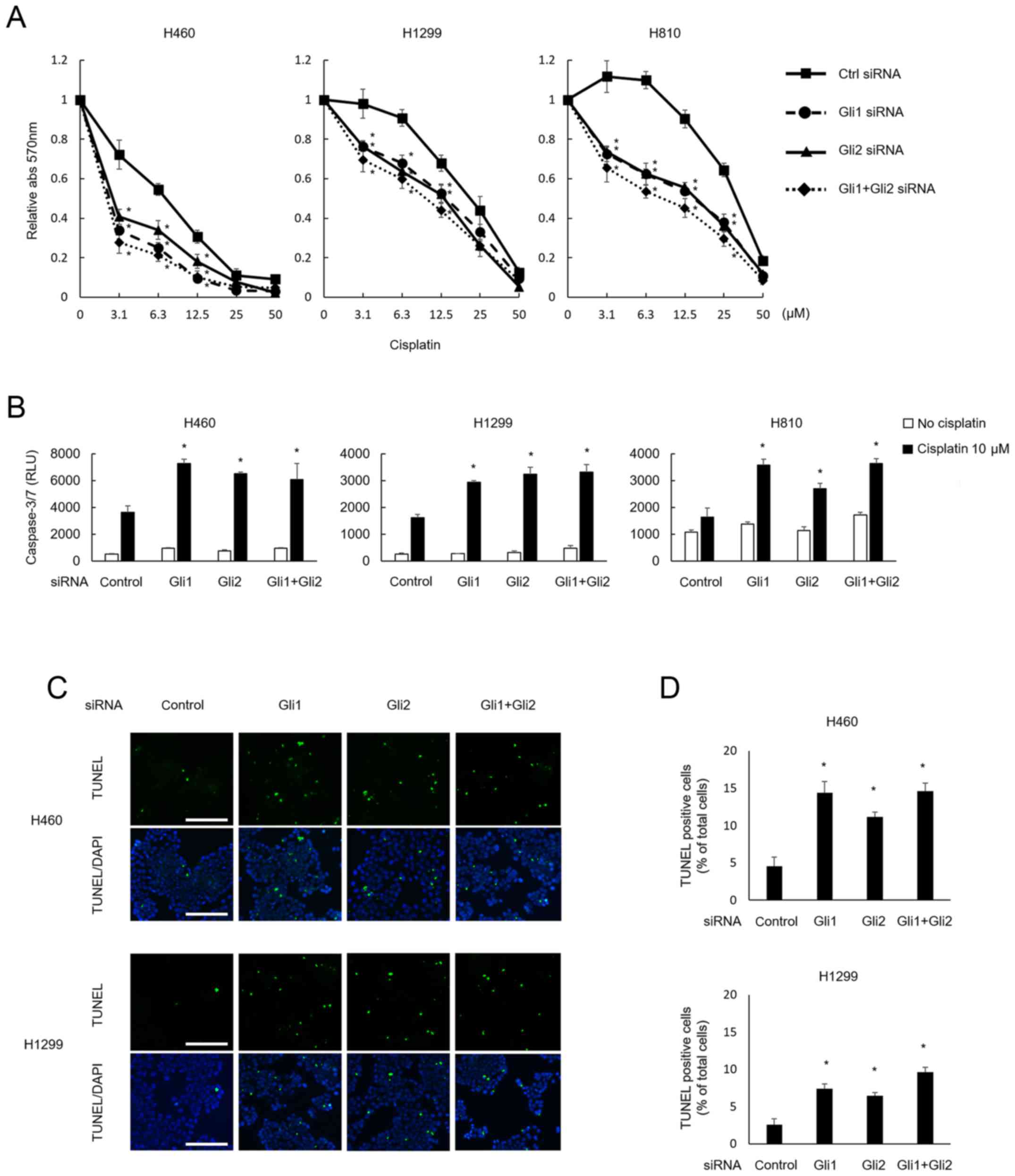 | Figure 5.Cisplatin sensitivity and
downregulation of Gli expression. (A) After treatment with siRNA,
the cells were next treated with various concentrations of
cisplatin for 48 h. Treatment with siRNA for Gli1, Gli2, or both
made all 3 cell lines significantly (*P<0.05) more sensitive to
cisplatin. To verify this phenomenon, induction of apoptosis was
evaluated using a caspase 3/7 assay (B) and a TUNEL assay (C and
D). (B) Caspase 3/7 activity was assessed at 48 h post-treatment
with cisplatin at 10 µM, and the activity was significantly
enhanced in cells treated with siRNA for Gli1, Gli2, or both,
whereas apoptosis was not induced in cells treated without
cisplatin and with siRNA treatment alone. Again, no additive effect
was observed for the combination of both siRNAs. (C) Apoptotic
cells after treatment with cisplatin were evaluated and visualized
using a TUNEL assay. H460 and H1299 cells treated with Gli1 and/or
Gli2 siRNA were further treated with 10 µM of cisplatin for 48 h.
The green and blue fluorescence of DAPI shows apoptotic cells and
nuclei, respectively. Scale bar, 100 µm. (D) The proportions of
apoptotic cells were quantified for comparison among the treatment
groups. The number of TUNEL-positive cells was significantly higher
after treatment with siRNA for Gli1 and/or Gli2. Statistically
significant differences, relative to the control (*P<0.05) in
(A), (B) and (D). The columns and bars represent the means and SEs
(n=3), respectively, in (B) and (D). |
Discussion
The present study clearly demonstrated a close
relationship between the downregulation of Gli and cell growth
inhibition in 3 human LCNEC cell lines of the lung. That is, the
Smo inhibitor BMS-833923 and the Gli inhibitor GANT61 significantly
suppressed the expression of Gli1 and/or Gli2, leading to cell
growth inhibition as assessed using an MTT assay. The
downregulation of Gli1 and/or Gli2 by treatment with siRNA for each
gene also led to cell growth inhibition. On the other hand, another
Smo inhibitor, GDC-0449, failed to downregulate Gli1 and Gli2,
except for Gli2 inhibition in H810 cells, leading to
non-significant growth inhibition in the 3 cell lines. Although the
reason is unknown, GDC-0449 was ineffective for inhibiting the cell
growth of LCNEC cells because it failed to suppress Gli expression.
The action mechanisms of GDC-0449 and GANT61 are well documented.
GDC-0449 binds to the transmembrane domain of Smo protein inducing
a conformational change in Smo, which results in blocking the
signals of normal Hh signaling (28), while the action mechanisms of
BMS-833923 has not been elucidated. On the other hand, GANT61 acts
in the nucleus interfering with transcriptional factor Gli binding
to DNA (29). In contrast, these
chemical inhibitors non-specifically suppressed the expression of
both Gli1 and Gli2, and siRNA for each gene specifically suppressed
the expression of either Gli1 or Gli2. Interestingly, the
suppression of either Gli1 or Gli2 was shown to be sufficient to
inhibit cell growth, and an additive effect was not observed for
the double downregulation of both genes. Whether Gli1, Gli2 or
their combination is required for Hh-related tumorigenesis remains
unclear. A previous report demonstrated that Gli1 and Gli2 act as
compensation for each other in mouse models (30). In some cell lines, inhibiting only
Gli1 was not enough to suppress tumor growth as Gli2 can behave
like Gli1 in mice (31) and Gli2
also mediates the Hh signaling pathway (32). Our results also showed that mRNA of
Gli2 increased when Gli1 was suppressed by siRNA and vice versa,
implicating those factors act in a compensational manner. On the
other hand, several studies have shown that Gli1, but not Gli2,
plays a central role in mediating the oncogenic Hh signaling
(33–35). Interestingly, the relationship
between Hh signaling and tumor proliferation was observed even in
H810 cells despite the absence of the overexpression of these genes
in this cell line, as shown in Fig.
1. We showed that Hh inhibitors and siRNAs effectively
suppressed these baseline expression levels of Gli1/2 together with
suppression of cell growth (Figs.
2–4) in H810 cells. It might be
speculated that Gli1/2 are essential for cell growth independent of
baseline expression. Similar phenomena were also observed: Gli1
downregulation in H460 cells led to cell growth inhibition despite
the fact that these cells did not overexpress Gli1, while Gli2
downregulation in H1299 cells led to cell growth inhibition despite
the fact that these cells do not overexpress Gli2. These facts
suggest that both Gli1 and Gli2 independently have critical roles
in maintaining cell growth ability irrespective of their baseline
expression levels. In a previous report regarding malignant pleural
mesothelioma cells, a single treatment with Gli1 or Gli2 siRNA did
not produce significant inhibitory effects on cell growth, whereas
the double downregulation of Gli1 and Gli2 significantly inhibited
cell proliferation (36).
Therefore, the independence of Gli1 and Gli2 might vary among tumor
types.
The present study also demonstrated a close
relationship between Gli downregulation and an increased
sensitivity to cisplatin. That is, the downregulation of Gli1
and/or Gli2 made the cells more sensitive to cisplatin, possibly
through an increase in apoptosis as was assessed using caspase 3/7
and TUNEL assays. A previous study also revealed that Gli2
knockdown using an antisense oligonucleotide led to enhanced
chemosensitivity to paclitaxel in prostate cancer (37). Another study showed that treatment
with GANT61 enhanced the cytotoxicity of cytarabine for acute
myeloid leukemia (38). As to the
underlying mechanism, Sims-Mourtada et al reported that Gli1
regulates the ATP-binding cassette transporter family of proteins
that is required for drug efflux (39). Amable et al showed that Gli1
plays a role in the cellular accumulation of cisplatin through the
regulation of multiple transport proteins including octamer-binding
protein (OCT)1, OCT2, OCT3, the copper transporter CTR1 and the
ATPase copper transporter ATP7B (40). On the other hand, cisplatin, in
turn, affects the Hh pathway. After treatment with cisplatin, DNA
repair mechanisms, including nucleotide excision repair, mismatch
repair and DNA double strand break repair, are upregulated in
cancer cells to avoid apoptosis. Upregulated Hh signaling helps the
DNA repair mechanisms. Suppression of Gli1 by GANT61 downregulated
the genes related to double-strand break repair by homologous
recombination (41) and nucleotide
excision repair (42). Therefore,
there is possibility of synergistic effects of concurrent exposure
to cisplatin and Gli knockdown. The shortcomings of this study
contain the lack of protein expression analysis, the lack of
confirmation with in vivo experiments and the lack of
elucidating precise mechanism due to its preliminary nature.
Despite these limitations, the present study provides evidence
supporting Gli as a therapeutic target for the treatment of LCNEC
of the lung, which is presently an unmet need.
In conclusion, the present study suggests that Gli
activation plays a critical role in LCNEC proliferation and drug
sensitivity. The inhibition of Gli factors has the potential to
become an effective approach to the treatment of LCNEC of the
lung.
Acknowledgements
This study was supported by grants from the Ministry
of Education, Culture, Sports, Science and Technology in Japan of
grant no. 26860599 to S.I., and nos. 26461182 and 17K09647 to
Y.T.
References
|
1
|
Nüsslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in Drosophila.
Nature. 287:795–801. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: Paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen JK, Taipale J, Cooper MK and Beachy
PA: Inhibition of Hedgehog signaling by direct binding of
cyclopamine to Smoothened. Genes Dev. 16:2743–2748. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duman-Scheel M, Weng L, Xin S and Du W:
Hedgehog regulates cell growth and proliferation by inducing Cyclin
D and Cyclin E. Nature. 417:299–304. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lum L and Beachy PA: The Hedgehog response
network: Sensors, switches, and routers. Science. 304:1755–1759.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taipale J, Chen JK, Cooper MK, Wang B,
Mann RK, Milenkovic L, Scott MP and Beachy PA: Effects of oncogenic
mutations in Smoothened and Patched can be reversed by cyclopamine.
Nature. 406:1005–1009. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sasaki H, Nishizaki Y, Hui C, Nakafuku M
and Kondoh H: Regulation of Gli2 and Gli3 activities by an
amino-terminal repression domain: Implication of Gli2 and Gli3 as
primary mediators of Shh signaling. Development. 126:3915–3924.
1999.PubMed/NCBI
|
|
8
|
Alvarez JI, Dodelet-Devillers A, Kebir H,
Ifergan I, Fabre PJ, Terouz S, Sabbagh M, Wosik K, Bourbonnière L,
Bernard M, et al: The Hedgehog pathway promotes blood-brain barrier
integrity and CNS immune quiescence. Science. 334:1727–1731. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shin K, Lee J, Guo N, Kim J, Lim A, Qu L,
Mysorekar IU and Beachy PA: Hedgehog/Wnt feedback supports
regenerative proliferation of epithelial stem cells in bladder.
Nature. 472:110–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Petrova R and Joyner AL: Roles for
Hedgehog signaling in adult organ homeostasis and repair.
Development. 141:3445–3457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bermudez O, Hennen E, Koch I, Lindner M
and Eickelberg O: Gli1 mediates lung cancer cell proliferation and
Sonic Hedgehog-dependent mesenchymal cell activation. PLoS One.
8:e632262013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang L, Walter V, Hayes DN and Onaitis M:
Hedgehog-GLI signaling inhibition suppresses tumor growth in
squamous lung cancer. Clin Cancer Res. 20:1566–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
You M, Varona-Santos J, Singh S, Robbins
DJ, Savaraj N and Nguyen DM: Targeting of the Hedgehog signal
transduction pathway suppresses survival of malignant pleural
mesothelioma cells in vitro. J Thorac Cardiovasc Surg. 147:508–516.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bosco-Clément G, Zhang F, Chen Z, Zhou HM,
Li H, Mikami I, Hirata T, Yagui-Beltran A, Lui N, Do HT, et al:
Targeting Gli transcription activation by small molecule suppresses
tumor growth. Oncogene. 33:2087–2097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wickström M, Dyberg C, Shimokawa T,
Milosevic J, Baryawno N, Fuskevåg OM, Larsson R, Kogner P,
Zaphiropoulos PG and Johnsen JI: Targeting the hedgehog signal
transduction pathway at the level of GLI inhibits neuroblastoma
cell growth in vitro and in vivo. Int J Cancer. 132:1516–1524.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rieber J, Schmitt J, Warth A, Muley T,
Kappes J, Eichhorn F, Hoffmann H, Heussel CP, Welzel T, Debus J, et
al: Outcome and prognostic factors of multimodal therapy for
pulmonary large-cell neuroendocrine carcinomas. Eur J Med Res.
20:642015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Niho S, Kenmotsu H, Sekine I, Ishii G,
Ishikawa Y, Noguchi M, Oshita F, Watanabe S, Nakajima R, Tada H, et
al: Combination chemotherapy with irinotecan and cisplatin for
large-cell neuroendocrine carcinoma of the lung: A multicenter
phase II study. J Thorac Oncol. 8:980–984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Battafarano RJ, Fernandez FG, Ritter J,
Meyers BF, Guthrie TJ, Cooper JD and Patterson GA: Large cell
neuroendocrine carcinoma: An aggressive form of non-small cell lung
cancer. J Thorac Cardiovasc Surg. 130:166–172. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Watkins DN, Berman DM, Burkholder SG, Wang
B, Beachy PA and Baylin SB: Hedgehog signalling within airway
epithelial progenitors and in small-cell lung cancer. Nature.
422:313–317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vestergaard J, Pedersen MW, Pedersen N,
Ensinger C, Tümer Z, Tommerup N, Poulsen HS and Larsen LA: Hedgehog
signaling in small-cell lung cancer: Frequent in vivo but a rare
event in vitro. Lung Cancer. 52:281–290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carbone DP, Koros AM, Linnoila RI, Jewett
P and Gazdar AF: Neural cell adhesion molecule expression and
messenger RNA splicing patterns in lung cancer cell lines are
correlated with neuroendocrine phenotype and growth morphology.
Cancer Res. 51:6142–6149. 1991.PubMed/NCBI
|
|
22
|
Senden N, Linnoila I, Timmer E, van de
Velde H, Roebroek A, Van de Ven W, Broers J and Ramaekers F:
Neuroendocrine-specific protein (NSP)-reticulons as independent
markers for non-small cell lung cancer with neuroendocrine
differentiation. An in vitro histochemical study. Histochem Cell
Biol. 108:155–165. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koyama N, Zhang J, Huqun, Miyazawa H,
Tanaka T, Su X and Hagiwara K: Identification of IGFBP-6 as an
effector of the tumor suppressor activity of SEMA3B. Oncogene.
27:6581–6589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Odate S, Onishi H, Nakamura K, Kojima M,
Uchiyama A, Kato M and Katano M: Tropomyosin-related kinase B
inhibitor has potential for tumor regression and relapse prevention
in pulmonary large cell neuroendocrine carcinoma. Anticancer Res.
33:3699–3703. 2013.PubMed/NCBI
|
|
25
|
Tatematsu T, Sasaki H, Shimizu S, Okuda K,
Shitara M, Hikosaka Y, Moriyama S, Yano M, Brown J and Fujii Y:
Investigation of neurotrophic tyrosine kinase receptor 1 fusions
and neurotrophic tyrosine kinase receptor family expression in
non-small-cell lung cancer and sensitivity to AZD7451 in vitro. Mol
Clin Oncol. 2:725–730. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Armas-López L, Zúñiga J, Arrieta O and
Ávila-Moreno F: The Hedgehog-GLI pathway in embryonic development
and cancer: Implications for pulmonary oncology therapy.
Oncotarget. 8:60684–60703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giroux Leprieur E, Vieira T, Antoine M,
Rozensztajn N, Rabbe N, Ruppert AM, Lavole A, Cadranel J and Wislez
M: Sonic Hedgehog pathway activation is associated with resistance
to platinum-based chemotherapy in advanced non-small-cell lung
carcinoma. Clin Lung Cancer. 17:301–308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Byrne EFX, Sircar R, Miller PS, Hedger G,
Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF,
Rambo RP, et al: Structural basis of Smoothened regulation by its
extracellular domains. Nature. 535:517–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lauth M, Bergström A, Shimokawa T and
Toftgård R: Inhibition of GLI-mediated transcription and tumor cell
growth by small-molecule antagonists. Proc Natl Acad Sci USA.
104:pp. 8455–8460. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Regl G, Kasper M, Schnidar H, Eichberger
T, Neill GW, Philpott MP, Esterbauer H, Hauser-Kronberger C,
Frischauf AM and Aberger F: Activation of the BCL2 promoter in
response to Hedgehog/GLI signal transduction is predominantly
mediated by GLI2. Cancer Res. 64:7724–7731. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bai CB and Joyner AL: Gli1 can rescue the
in vivo function of Gli2. Development. 128:5161–5172.
2001.PubMed/NCBI
|
|
32
|
Roessler E, Du YZ, Mullor JL, Casas E,
Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A
and Muenke M: Loss-of-function mutations in the human GLI2 gene are
associated with pituitary anomalies and holoprosencephaly-like
features. Proc Natl Acad Sci USA. 100:pp. 13424–13429. 2003;
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Regl G, Neill GW, Eichberger T, Kasper M,
Ikram MS, Koller J, Hintner H, Quinn AG, Frischauf AM and Aberger
F: Human GLI2 and GLI1 are part of a positive feedback mechanism in
Basal Cell Carcinoma. Oncogene. 21:5529–5539. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dahmane N, Lee J, Robins P, Heller P and
Ruiz i Altaba A: Activation of the transcription factor Gli1 and
the Sonic hedgehog signalling pathway in skin tumours. Nature.
389:876–881. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bonifas JM, Pennypacker S, Chuang PT,
McMahon AP, Williams M, Rosenthal A, De Sauvage FJ and Epstein EH
Jr: Activation of expression of hedgehog target genes in basal cell
carcinomas. J Invest Dermatol. 116:739–742. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li H, Lui N, Cheng T, Tseng HH, Yue D,
Giroux-Leprieur E, Do HT, Sheng Q, Jin JQ, Luh TW, et al: Gli as a
novel therapeutic target in malignant pleural mesothelioma. PLoS
One. 8:e573462013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Narita S, So A, Ettinger S, Hayashi N,
Muramaki M, Fazli L, Kim Y and Gleave ME: GLI2 knockdown using an
antisense oligonucleotide induces apoptosis and chemosensitizes
cells to paclitaxel in androgen-independent prostate cancer. Clin
Cancer Res. 14:5769–5777. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Long B, Wang LX, Zheng FM, Lai SP, Xu DR,
Hu Y, Lin DJ, Zhang XZ, Dong L, Long ZJ, et al: Targeting GLI1
suppresses cell growth and enhances chemosensitivity in
CD34+ enriched acute myeloid leukemia progenitor cells.
Cell Physiol Biochem. 38:1288–1302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sims-Mourtada J, Izzo JG, Ajani J and Chao
KS: Sonic Hedgehog promotes multiple drug resistance by regulation
of drug transport. Oncogene. 26:5674–5679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Amable L, Fain J, Gavin E and Reed E: Gli1
contributes to cellular resistance to cisplatin through altered
cellular accumulation of the drug. Oncol Rep. 32:469–474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi T, Mazumdar T, Devecchio J, Duan ZH,
Agyeman A, Aziz M and Houghton JA: cDNA microarray gene expression
profiling of hedgehog signaling pathway inhibition in human colon
cancer cells. PLoS One. 5:52010. View Article : Google Scholar
|
|
42
|
Mazumdar T, DeVecchio J, Agyeman A, Shi T
and Houghton JA: The GLI genes as the molecular switch in
disrupting Hedgehog signaling in colon cancer. Oncotarget.
2:638–645. 2011. View Article : Google Scholar : PubMed/NCBI
|