Introduction
Breast cancer susceptibility gene 1 (BRCA1)
is a tissue-specific tumor suppressor involved in basic cellular
functions necessary for cell replication and DNA synthesis
(1–3). A large body of evidence (4–6)
indicates that BRCA1 is involved in several important
cellular pathways, including DNA damage repair, chromatin
remodeling, and checkpoint activation. BRCA1 affects the
outcomes of chemotherapy or radiotherapy for the treatment of
breast cancer (7), although whether
it increases or decreases the chemo/radiosensitivity of different
cancers remains unclear.
Malignant gliomas, including multitype and mixed
gliomas, are the most common and deadly malignant primary brain
tumors and are associated with poor prognosis and rapid invasion.
The median survival of patients with the most aggressive type of
malignant glioma is 12–14 months, despite the availability of
treatments such as surgery, chemotherapy and/or radiotherapy
(8,9). Therefore, new strategies to enhance
the therapeutic effect of chemo/radiotherapy are urgently
needed.
MicroRNAs (miRNAs) are small-non-coding RNAs that
regulate gene expression at the post-transcriptional level through
complete or incomplete complementary binding to the 3′-untranslated
region (3′-UTR) of target genes (10). miRNAs play an important role in the
regulation of gene expression, and they mostly function as negative
regulators (11), with involvement
in positive gene regulation in fewer cases (12,13).
miRNAs regulate approximately 60% of protein-coding genes and
participate in several biological processes at multiple steps. They
play an important role in tumor development by affecting tumor cell
growth, differentiation, and apoptosis, and the cell cycle. miRNAs
can function as tumor suppressors or oncogenes in cancer depending
on the genes or pathways that they regulate. Growing evidence
(9,14,15)
indicates that miRNAs affect the therapeutic response to
chemotherapy or radiotherapy, which opens new avenues for the
diagnosis and prognosis of tumors and identifies potential
therapeutic targets to improve patient survival. miR-212, which is
located at chromosome 17p13.3, is overexpressed in many cancers,
including non-small cell lung cancer (16) and oral carcinoma (17), whereas it is downregulated in other
tumors such as hepatocellular carcinoma (18), gastric cancer (19,20),
colorectal cancer (21), and
prostate cancer (22). In the
present study, we explored the role of miR-212 in the
radiosensitivity of the U251 human glioma tumor cell line. miR-212
was identified as a radiation-induced miRNA by gene chip screening
and confirmed by fluorescence quantitative PCR. We identified
BRCA1 as a target of miR-212, and examined the role of the
association between BRCA1 and miR-212 in the
radiosensitivity of glioma cells. Gain- and loss-of-function assays
showed that miR-212 contributes to the radioresistance of glioma
cells. To the best of our knowledge, this is the first study
reporting the role of miR-212 in the radiosensitivity of glioma
tumor cells and the possible underlying mechanism. Our findings may
help to develop a novel miRNA-based therapeutic strategy which
combines miR-212/BRCA1 interaction with radiotherapy for
glioma.
Materials and methods
Cell lines and γ-irradiation
Human glioma U251, U-118MG and SHG-44 cell lines
were purchased from the American Type Culture Collection (ATCC)
(Manassas, VA, USA) and cultured in DMEM supplemented with 10%
dialyzed fetal bovine serum (FBS) and 1% PS (100 U/ml penicillin
and 100 µg/ml streptomycin). Cells were maintained in a humidified
incubator with 5% CO2 at 37°C. The cells were
transfected with pcDNA3/hsa-miR-212 or the pcDNA3 control,
antisense oligonucleotide (ASO)-miR-212 or ASO-ctrl, pcDNA3/EGFP,
pcDNA3/EGFP-BRCA1, or pcDNA3/EGFP-BRCA1 3′-UTR mut using Invitrogen
Lipofectamine™ 2000 transfection reagent (Thermo Fisher Scientific,
Waltham, MA, USA) according to the manufacturer's protocol. Cells
were exposed to different doses of 137Cesium
γ-irradiation at a rate of 0.8 Gy/min.
RNA isolation, miRNA chip array, and
real-time quantitative RT-PCR assay
RNA isolation and miRNA chip array were performed at
30 min after γ-ray exposure. Total RNA was isolated with TRIzol
(cat. no. 15596–026, Invitrogen; Thermo Fisher Scientific) and
pretreated with a wash buffer kit (cat. no. 208021, Exiqon, Inc.,
Woburn, MA, USA) according to the manufacturer's protocol. The
miRCURY™ Array (Exiqon) was performed with a GenePix 4000B scanner
and the GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA,
USA) using the following parameters: wavelength, 532 nm, Cy3; 532
PMT gain, 650 (default, modified as indicated); power, 33%; pixel
size, 10 µm; lines to average, 1; and focus position, 0 µm.
Specific primers were used for the miRNA reverse transcription (RT)
reaction, whereas the cDNA of genes was reverse transcribed using
oligo(dT) primers. The PCR assay was performed using the SYBR Green
PCR Master Mix (cat. no. 4367659, ABI) and analyzed using the ABI
Prism 7900 (both from Thermo Fisher Scientific). The reaction
conditions were as follows: 95°C for 5 min, 40 cycles of 95°C for
15 sec, 65°C for 15 sec, and 72°C for 32 sec. Melting curve
analysis was performed at 60–95°C. The primers for RT and PCR are
shown in Table I. The U6 small
nuclear B non-coding RNA (RNU6B) was used as the endogenous control
to normalize the level of miR-212.
 | Table I.PCR primer sequences. |
Table I.
PCR primer sequences.
| Primers | Sequences |
|---|
| hsa-miR-212-5p
RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGTAAGCA-3′ |
| hsa-miR-212-5p
forward |
5′-TGCGGACCTTGGCTCTAGACT-3′ |
| hsa-miR-96-5p
RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGCAAAAA-3′ |
| hsa-miR-96-5p
forward |
5′-TGCGGTTTGGCACTAGCACAT-3′ |
| hsa-miR-21-3p
RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACTCAACAT-3′ |
| hsa-miR-21-3p
forward |
5′-TGCGGUAGCUUAUCAGACUG-3′ |
| hsa-miR-423-5p
RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAGTCTC-3′ |
| hsa-miR-423-5p
forward |
5′-TGCGGUGAGGGGCAGAGAGCG-3′ |
| U6 RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG-3′ |
| U6 forward |
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ |
| U6 reverse |
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| BRCA1 sense |
5′-GATGGATCCTAACGGAGAAGCACAGGTC-3 |
| BRCA1
antisense |
5′-GCGGAATTCACTATCACAATCAATCAATAG-3′ |
| BRCA1 mut
sense |
5′-GCCCCAGCCCGACAGTGATAAATC-3′ |
| BRCA1 mut
antisense |
5′-GATTTATCACTGTCGGGCTGGGGC-3′ |
| β-actin sense |
5′-CGTGACATTAAGGAGAAGCTG-3′ |
| β-actin
antisense |
5′-CTAGAAGCATTTGCGGTGGAC-3′ |
Plasmid construction
The primary miR-212 sequence was amplified from
genomic DNA and cloned into the pcDNA3/EGFP vector at BamHI
and EcoRI sites. The gene coding BRCA1 was amplified
from the cDNA of MCF-7 cells and cloned into the pcDNA3 vector at
NheI and ApalI sites. The shRNA of BRCA1 was
annealed and cloned into the pcDNA3 vector at BamHI and
HindIII sites. The 3′-UTR of BRCA1 (containing the
binding sites for miR-212) was amplified from cDNA of MCF-7 cells.
The product was cloned into the pcDNA3-EGFP control vector
(downstream of EGFP). The mutant 3′-UTR of BRCA1 (four
nucleotides were mutated in the binding site) was amplified from
the construct pcDNA3-EGFP/BRCA1 3′-UTR. The primers used for PCR
amplification are listed in Table
I.
EGFP fluorescent reporter assay
U251 cells were co-transfected with pcDNA3/miR-212
or ASO miR-212 and the 3′-UTR of BRCA1 or the mutant 3′-UTR
of BRCA1 or with the control vectors in 48-well plates. At
48 h after transfection, the fluorescence intensity was measured
with the F-4500 fluorescence spectrophotometer (Hitachi, Ltd.,
Tokyo, Japan). The vector pDsRed2-N1 (Clontech Laboratories, Inc.,
Mountainview, CA, USA) expressing RFP was transfected together with
the above vectors and used as the spiked-in control.
Western blot analysis
Cells were washed with PBS and lysed on ice in RIPA
buffer (V900854, Sigma) containing a cocktail of protease
inhibitors (P8340, Sigma) following the manufacturers protocols.
Protein concentration was measured using the BCA assay. The protein
fractions were suspended in loading buffer and denatured at 100°C
for 5 min. Total proteins (20 µg/lane) were separated on 12%
SDS-PAGE and transferred to PVDF membranes, which were blocked in
5% fat free milk in TBST buffer (0.1% Tween-20) for 2 h at room
temperature. BRCA1 levels were analyzed using a mouse monoclonal
anti-BRCA1 antibody (1:1,000; ab16780; Abcam); Bcl-2, Bax,
pro-caspase-3, cleaved-caspase-3, and cytochrome c levels
were detected using the following mouse or rabbit monoclonal
antibodies at a dilution of 1:1,000: ab117115, ab5714, ab13586,
ab32042, and ab13575, respectively (Abcam). The secondary
antibodies used were goat anti-rabbit antibody (1:1,000; Sc2030;
Santa Cruz Biotechnology) and rabbit anti-mouse antibody (1:1,000;
Sc358917; Santa Cruz Biotechnology). GAPDH was used as the
endogenous control to normalize the expression levels of the
proteins of interest. GAPDH levels were detected using
HRP-conjugated mouse anti-GAPDH monoclonal antibody (1:5,000;
HRP-60004, Proteintech). The densitometry scan of the western
blotting was performed by Image Pro Plus 6.0 (Media Cybernetics,
Inc., Rockville, MD, USA).
Colony formation assay
Cells were seeded into 12-well plates at a density
of 1000 cells/well at 24 h after transfection. The medium was
changed every 3 days. When most of the colonies contained at least
50 cells, they were fixed in 100% methanol and stained with crystal
violet and counted. The colony formation rate was calculated as
(colony number)/(seeded cell number).
TdT-mediated dUTP nick-end labeling
(TUNEL) assay
Cells irradiated at doses of 0, 5, and 10 Gy were
seeded into 14-well plates at a density of 2000 cells/well at 48 h
after transfection. After 24 h, the cells were washed with PBS,
fixed with 4% paraformaldehyde, and permeabilized in 0.5% Triton
X-100. Then, the cells were washed with PBS three times, followed
by a 1-h incubation at 37°C with Terminal deoxynucleotidyl
Transferase (TdT), and the cells were stained with fluorescein
isothiocyanate-dUTP. Finally, the cells were stained with DAPI for
5 min at room temperature. The slides were examined with a
fluorescence microscope.
Statistical analysis
Each experiment was repeated at least twice, and
data are shown as the mean ± SD. The statistical analysis between
two groups was performed by two-tailed Student's t-test. For the
statistical analysis of the rescue experiment with four groups, a
one-way analysis of variance test and the least significant
differences t-test were performed. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-212 is downregulated in human
glioma U251 cells following irradiation
The miRNA expression profile of U251 cells exposed
to γ-radiation (at doses of 5 and 10 Gy) was analyzed by chip array
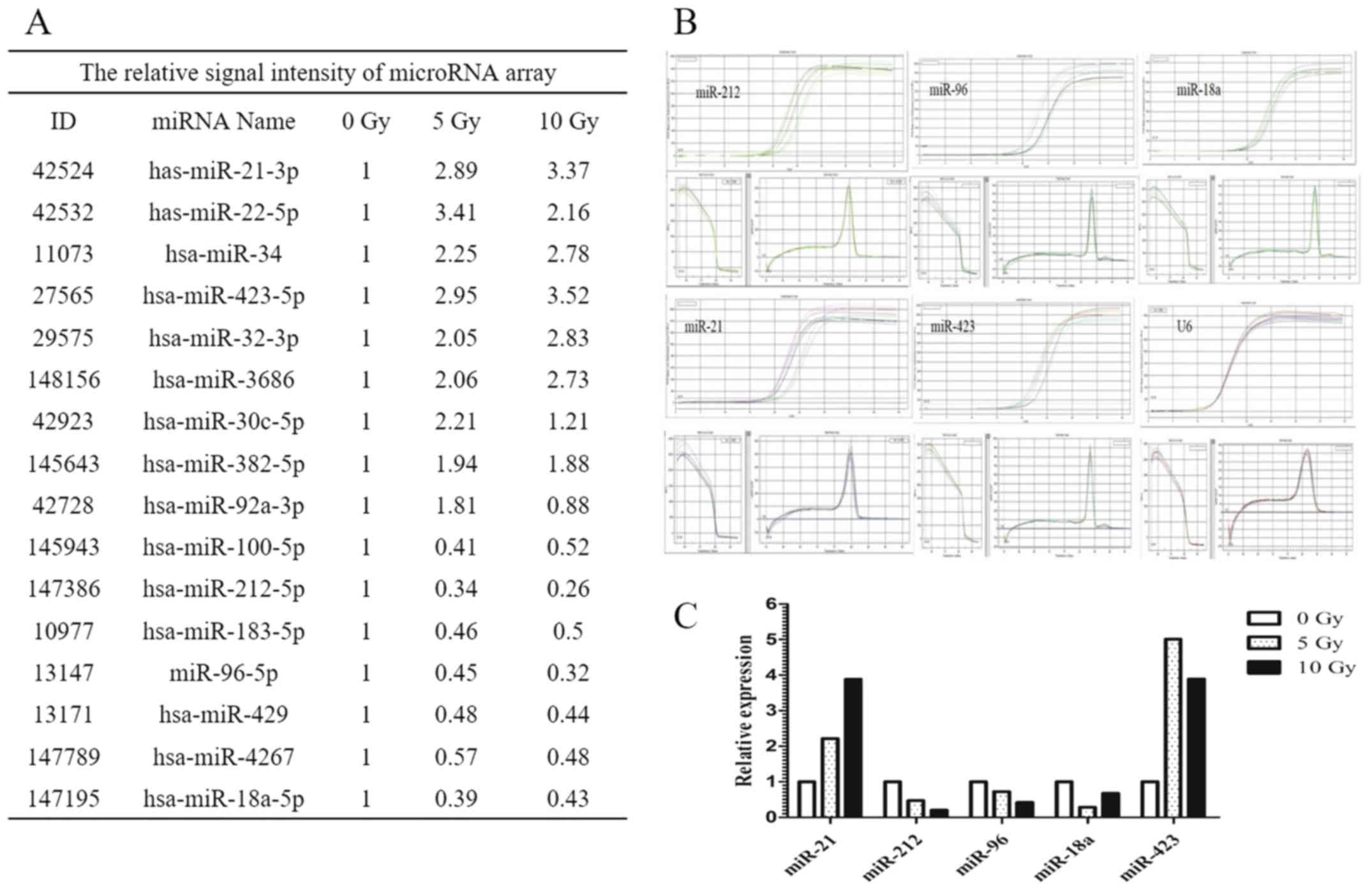
assay. The signal intensity of miRNAs relative to that of
unirradiated U251 cells as controls is shown in Fig. 1A. Of the 16 miRNAs showing
alterations in expression, five were further examined by real-time
PCR, that is, miR-212, miR-96, miR-18a, miR-21, and miR-423
(Fig. 1B). The relative expression
levels of these five miRNAs relative to that of U6 as the
endogenous control are shown in Fig.
1C. Of the five miRNAs analyzed, miR-212 was identified as a
negatively associated radiation-induced miRNA that was
downregulated in human glioma cells after radiation exposure. This
was confirmed in SHG-44 glioma cells by real-time PCR (data not
shown).
miR-212 promotes cell colony formation
in U251, U-118MG and SHG-44 cell lines exposed to γ-radiation
To determine whether miR-212 affects the
radiosensitivity of glioma cells, a colony formation assay was
performed in the U251, U-118MG (data not shown) and SHG-44 (data
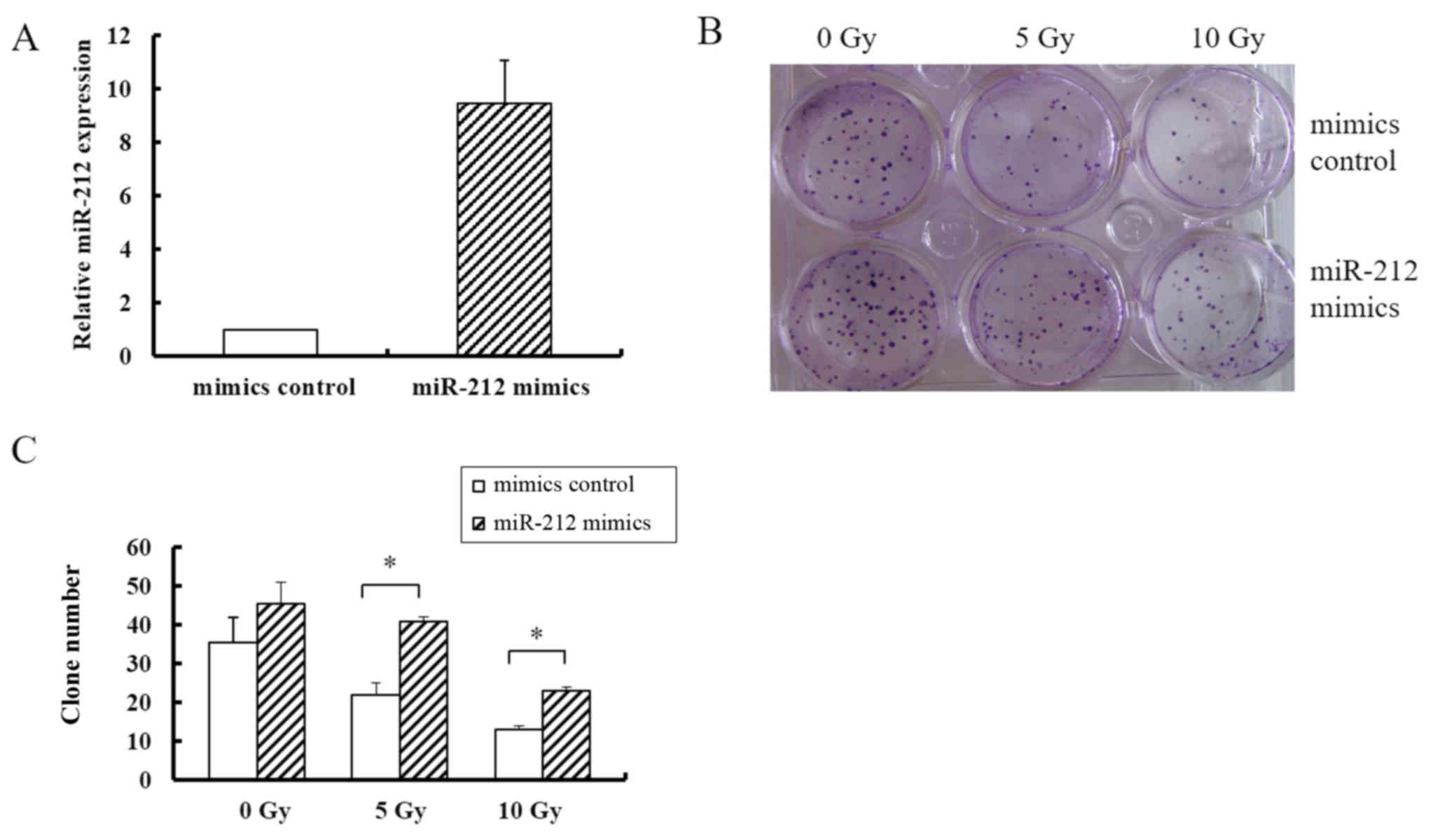
not shown) cell lines. miR-212 was overexpressed by transfecting
cells with pcDNA3/EGFP-miR-212 mimics, which resulted in a 9-fold
higher expression level of miR-212 than that in cells transfected
with the pcDNA3/EGFP-mimics control (Fig. 2A). As shown in Fig. 2B, overexpression of miR-212
increased the colony formation rate of U251 cells by ~2-fold
compared with that in the controls in the groups receiving 5, and
10 Gy radiation; but in groups without radiation exposure, miR-212
transfection did not significantly affect colony formation. These
data indicated that miR-212 increased colony formation ability in
response to radiation, suggesting that it induces radioresistance.
In addition, the colony number did somewhat decrease in response to
an increasing radiation dose despite miR-212 overexpression,
indicating that miR-212 did not fully abrogate the effect of
radiation.
miR-212 attenuates radiation-induced
apoptosis and affects the expression levels of apoptosis-related
proteins
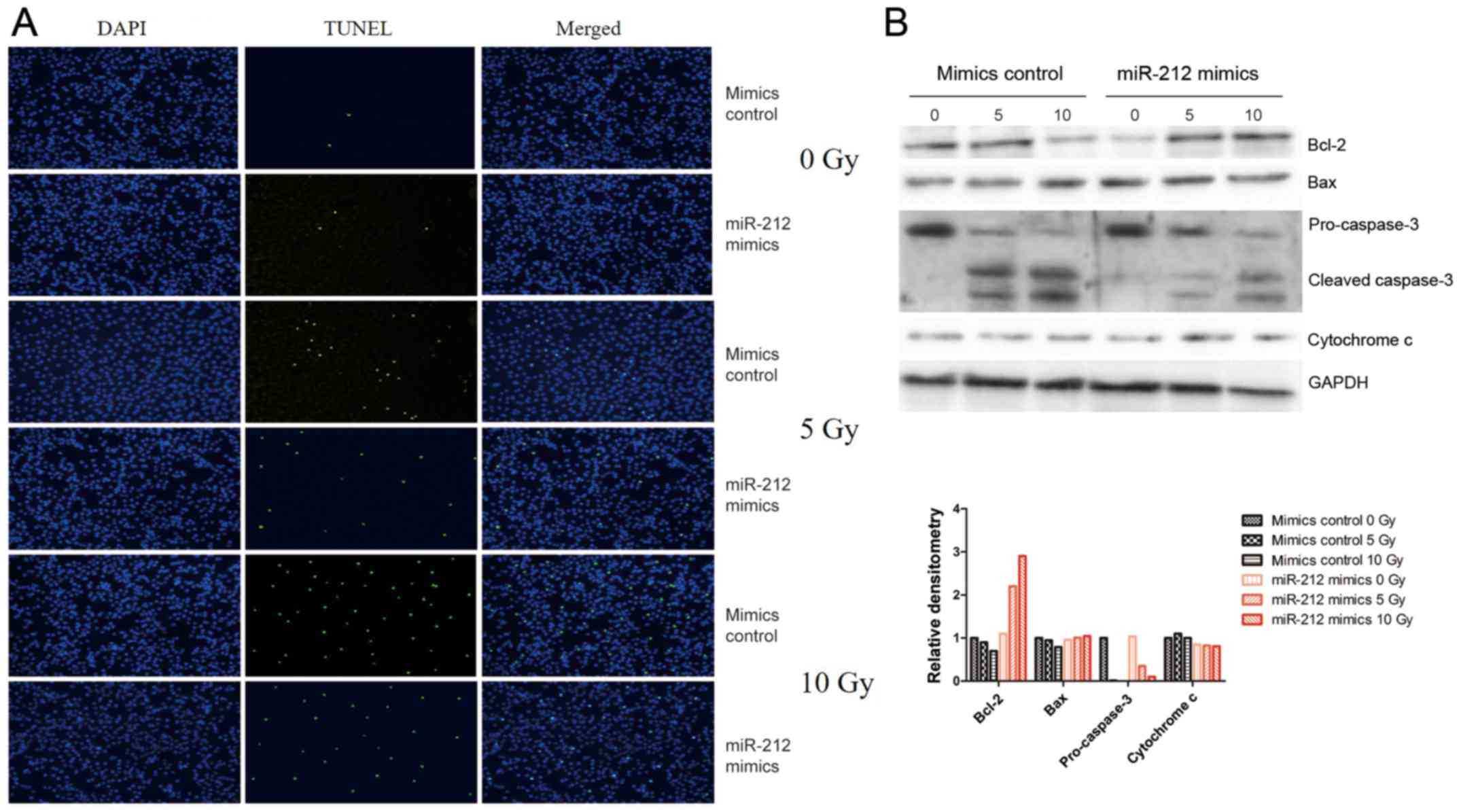
Apoptosis is a common response to irradiation in
most cells. In the present study, the potential role of miR-212 in
radiation-induced apoptosis was tested by fluorescence microscopy
assay and confirmed by western blot analysis of apoptosis-related
proteins such as Bcl-2, Bax, caspase-3, and cytochrome c,
with GAPDH as the endogenous control. As shown in Fig. 3A, overexpression of miR-212 (miR-212
mimics) attenuated radiation-induced apoptosis compared with that
in cells transfected with the mimic control, whereas overexpression
of miR-212 had no effect on unirradiated U251 cells. Consistent
with this result, western blot analysis indicated that miR-212
overexpression attenuated the radiation-induced downregulation of
Bcl-2 and upregulation of cleaved-caspase-3, but there was no
significance change in Bax or cytochrome c (Fig. 3B). These results suggest that
miR-212 plays a role in radiation-induced apoptosis.
miR-212 directly targets BRCA1 and
negatively regulates its expression
To explore the mechanism underlying the inhibition
of radiation-induced apoptosis by miR-212 in human glioma cells,
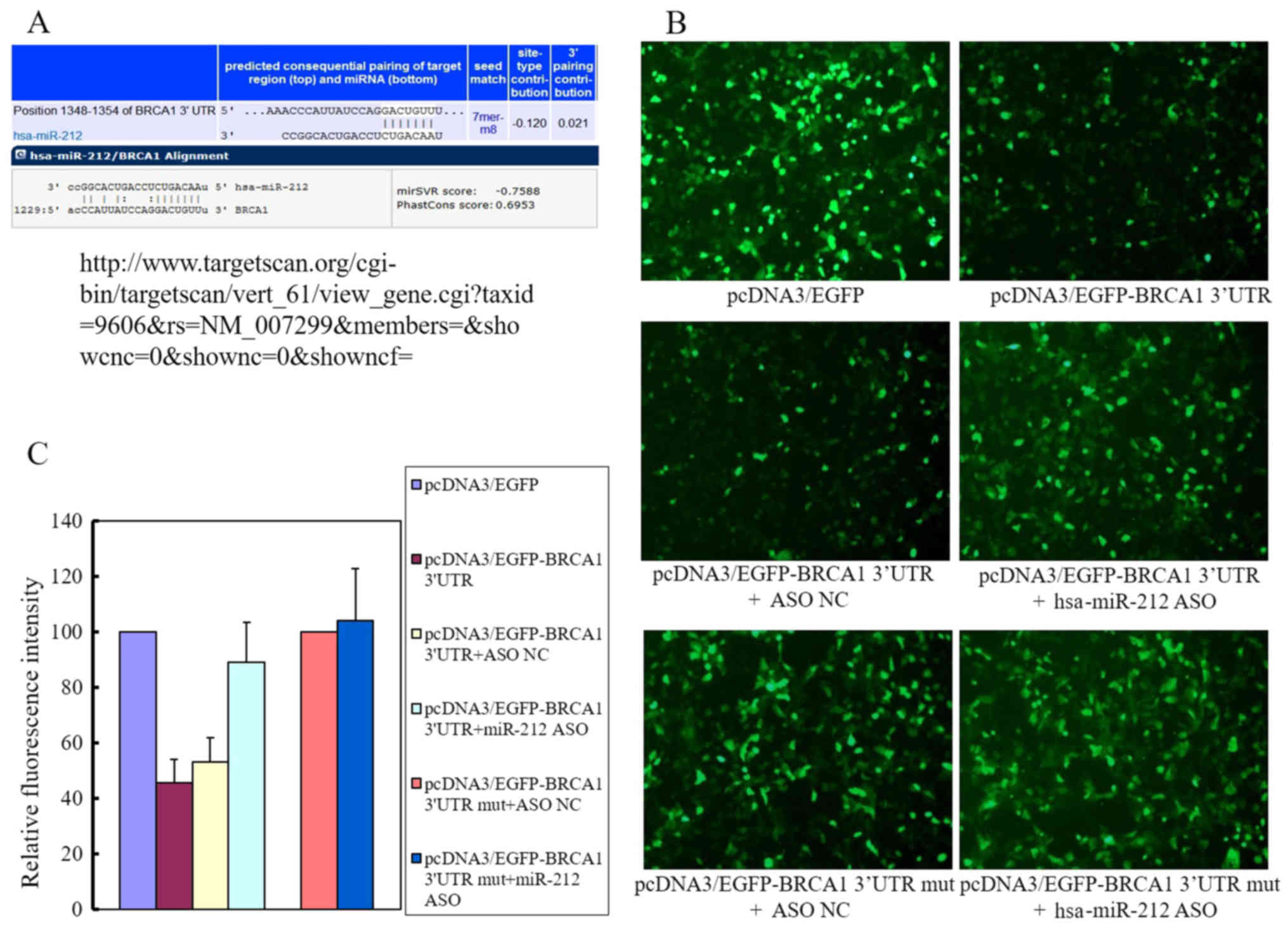
three algorithms were used to predict the target genes of miR-212,
namely, microRNA, TargetScan, and PicTar. Among the predicted
genes, we selected BRCA1 as our candidate target. The
binding sites for miR-212 on the 3′-UTR of BRCA1 are shown
in Fig. 4A. The results of the EGFP
fluorescence reporter assay (Fig.
4B) showed that endogenous miR-212 suppressed the upregulation
of BRCA1 induced by pcDNA3/EGFP-BRCA1 transfection by ~55%
(Fig. 4C) compared with that in
cells transfected with pcDNA3/EGFP. Co-transfection with
pcDNA3/EGFP-BRCA1 and pcDNA3/EGFP-hsa-miR-212 ASO partly reversed
this effect, mainly because the pcDNA3/EGFP-hsa-miR-212 ASO
eliminated the endogenous miR-212 expression. The fluorescence
intensity was restored to ~90% of the levels in cells transfected
with pcDNA3/EGFP. The mutant BRCA1 containing vector
pcDNA3/EGFP-BRCA1 3′-UTR mut transfected alone or co-transfected
with pcDNA3/EGFP-hsa-miR-212 had no significant effect on
fluorescence intensity compared with that in cells transfected with
pcDNA3/EGFP. Taken together, these results indicated that miR-212
interacted with the 3′-UTR of BRCA1.
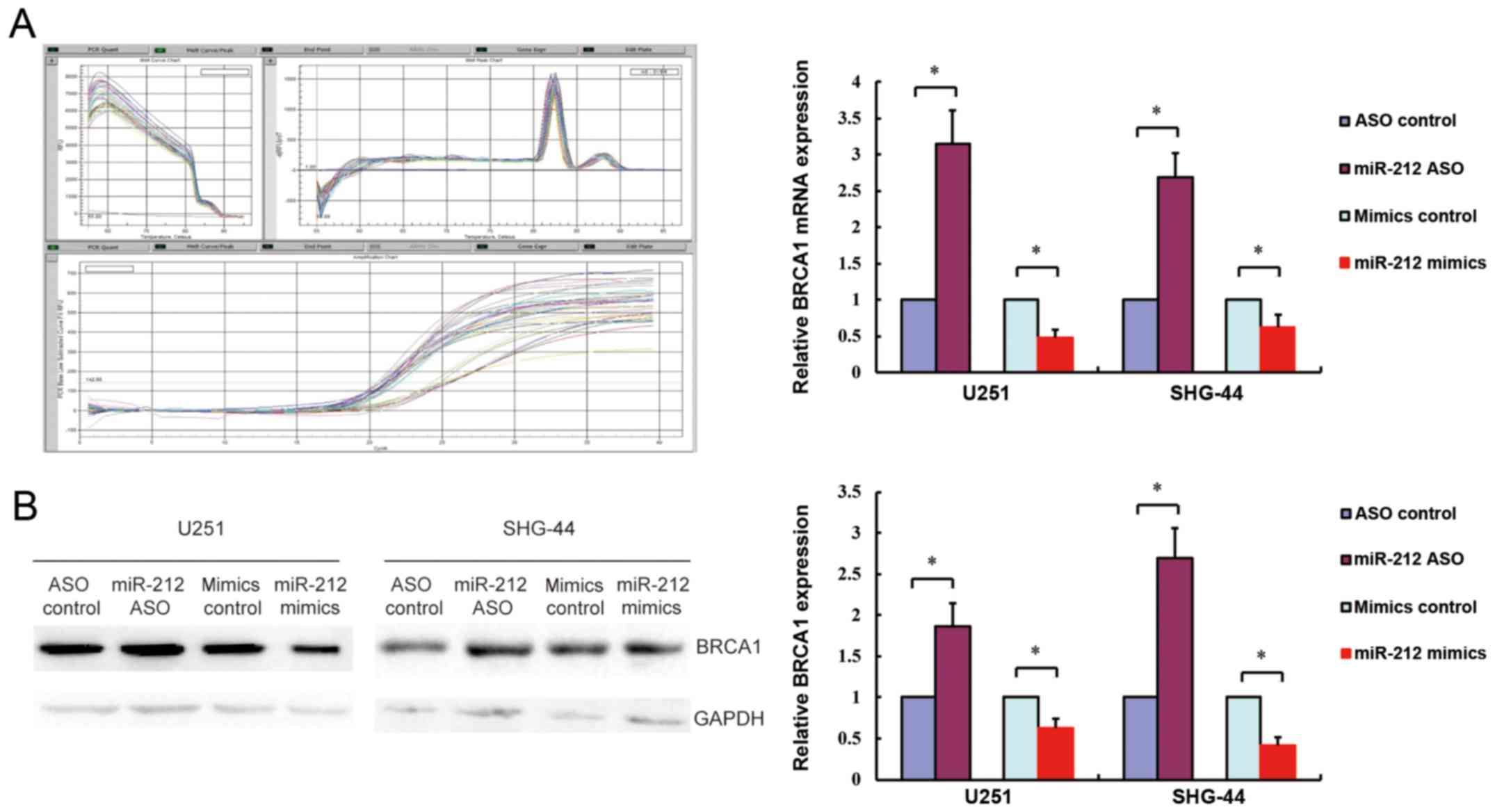
To confirm the association of hsa-miR-212 with
BRCA1, BRCA1 gene and protein expression levels were
measured by real-time PCR and western blotting in response to
miR-212 upregulation or downregulation in SHG-44 and U251 cells. As
shown in Fig. 5, overexpression of
miR-212 (miR-212 mimics) downregulated BRCA1 gene and protein
expression, whereas inhibition of miR-212 (miR-212 ASO) had the
opposite effect. Consistent with the fluorescence microscopy
results, these findings confirmed that miR-212 negatively regulated
BRCA1 expression by interacting with its 3′-UTR.
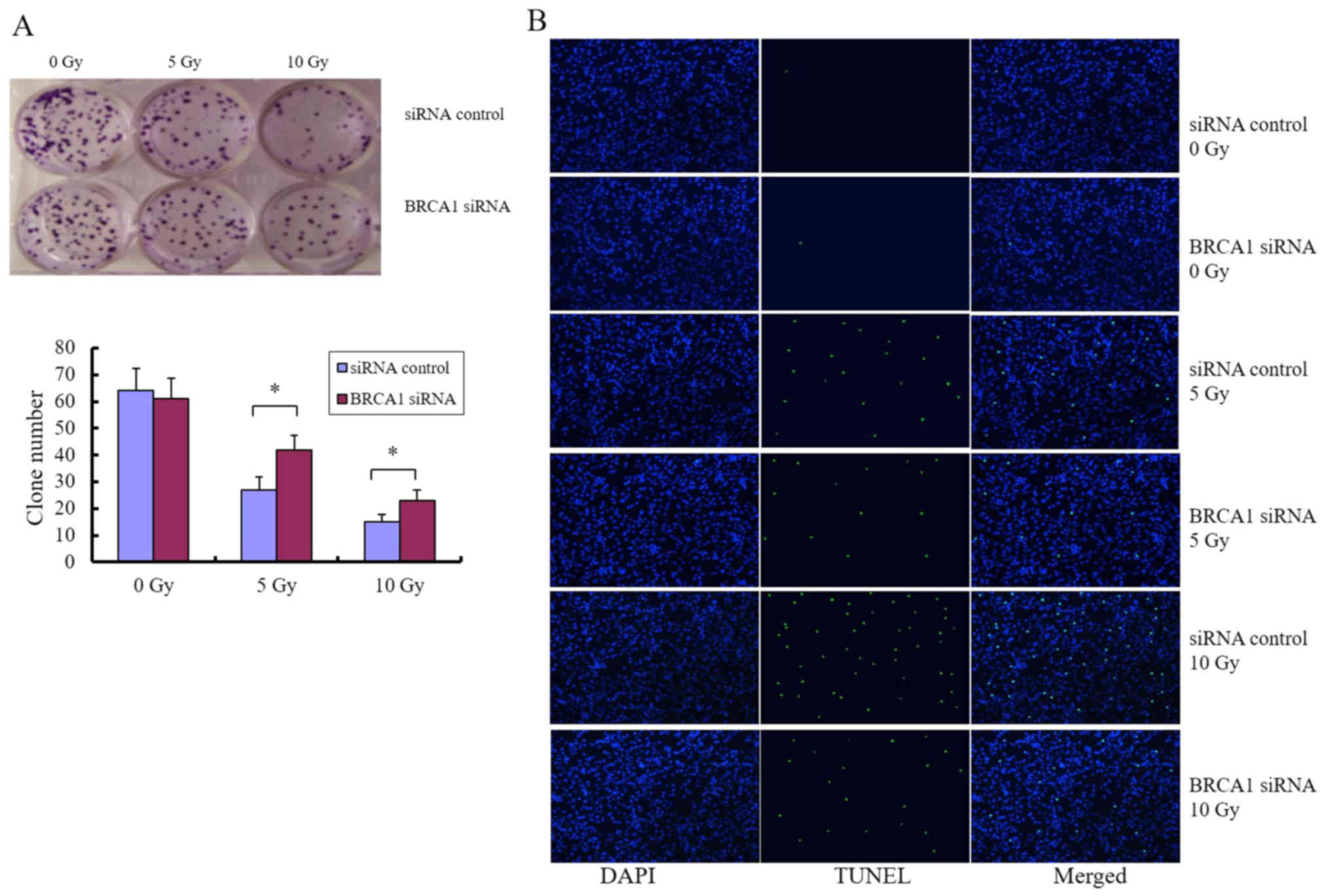
Inhibition of BRCA1 by siRNA
phenocopies the roles of miR-212
We next sought to determine whether the
miRNA-mediated downregulation of BRCA1 can be applied as a
molecular treatment strategy for patients with glioma, particularly
as a strategy to modulate radiosensitivity. Cells were treated with
siRNA targeting BRCA1, and colony formation assay and
fluorescence microscopy analysis of apoptosis were performed after
exposure to different doses of radiation. Knockdown of BRCA1
enhanced the colony forming ability of U251 cells after radiation
(Fig. 6A) and attenuated
radiation-induced apoptosis (Fig.
6B), indicating that BRCA1 is positively associated with
radiosensitivity in glioma cells. U-118MG and SHG-44 cell lines
were used to confirm these observations (data not shown).
Overall, these data indicated that miR-212 is
involved in the response to radiation and functions in the
regulation of BRCA1 expression, which is important for
tumorigenesis and response to treatment.
Discussion
miRNAs have been investigated extensively in recent
years and have been found to function as post-transcriptional
regulators of many genes involved in cancer initiation,
development, metastasis and the response to radiation (23). miRNAs have therefore been regarded
as attractive targets in the development of more powerful therapies
(24). Since the ability of miRNAs
to act as negative gene regulators allows them to modulate
signaling pathways that regulate multiple cellular processes,
although numerous miRNAs are involved, only a few can be targeted
therapeutically. Concerning radiotherapy, miRNAs have the potential
to be used as either radiosensitizers or radioprotectors.
Previous studies (16,19,21,25)
have revealed many functions of miR-212, such as tumor-promoting
properties in NSCLC (16) and
proliferation-inhibition properties in gastric cancer (19). To the best of our knowledge, the
present study indicated for the first time that miR-212 is
associated with the response to radiation by targeting
BRCA1. Gain-of-function experiments showed that miR-212
overexpression increased colony formation in U251 cells after
radiation exposure by approximately 2-fold compared with that in
the control groups. The present data indicated that miR-212
attenuated radiation-induced apoptosis and affected the expression
of apoptosis-related proteins.
Furthermore, we used bioinformatic tools to predict
target genes, and BRCA1 was chosen as a candidate target.
The EGFP fluorescence reporter assay, a direct method for target
validation, verified that BRCA1 is a direct target gene of
miR-212 (Fig. 4B and C). miR-212
was shown to be a negative regulator of BRCA1 expression, as
overexpression of miR-212 downregulated BRCA1 expression at
the gene (Fig. 5A) and protein
levels (Fig. 5B), consistent with
previous reports that a large number of miRNAs regulate gene
expression negatively.
We showed that BRCA1 downregulation by
miR-212 can be replaced by siRNA, as knockdown of BRCA1
attenuated radiation-induced apoptosis. This indicated that
BRCA1 may have a positive correlation with radiosensitivity
in glioma cells, in contrast to previous reports in ovarian
(26) and other tumors (27). However, BRCA1 has been shown
to act as a tumor suppressor, and to be involved in the response to
radiation and cisplatin. The role of BRCA1 in DNA damage
(6,28) and tumor development has been
investigated extensively; however, the precise mechanisms
underlying the effect of BRCA1 in radiosensitivity or
chemosensitivity considering tumor heterogeneity remain elusive.
The BRCA1 status should be more carefully considered in the
planning of anti-glioma treatment (29–31).
Our findings revealed that the miR-212/BRCA1
axis was involved in gliomas cell radiosensitivity, which indicates
that miR-212-mediated modulation of BRCA1 gene expression
plays a potential role in glioma radiotherapy. Further experiments
are necessary to better define the role of the association between
miR-212 and BRCA1 in radioresistance.
Acknowledgements
This research was supported by the National Natural
Science Foundation of China (no. 81402541) for data analysis, CAMS
Innovation Fund for Medical Sciences (CIFMS, 2017-I2M-1-016) and
the Fundamental Research Funds for the Chinese Academy of Medical
Sciences (CAMS) (no. 2016ZX310078) for design of the study and
collection, interpretation of data and in writing the manuscript.
We gratefully acknowledge the invaluable contribution of Naling
Song, Aimin Meng and Jinjian Liu.
Glossary
Abbreviations
Abbreviations:
|
BRCA1
|
breast cancer susceptibility gene
1
|
|
miRNA
|
microRNA
|
|
3′-UTR
|
3′-untranslated region
|
|
FBS
|
fetal bovine serum
|
|
ASO
|
antisense oligonucleotide
|
|
RT
|
reverse transcription
|
|
TUNEL
|
TdT-mediated dUTP nick-end
labeling
|
|
TdT
|
terminal deoxynucleotidyl
transferase
|
References
|
1
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos
MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, et al: BRCA1
inhibition of estrogen receptor signaling in transfected cells.
Science. 284:1354–1356. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Narod SA and Foulkes WD: BRCA1 and BRCA2:
1994 and beyond. Nat Rev Cancer. 4:665–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu Y: BRCA1, hormone, and tissue-specific
tumor suppression. Int J Biol Sci. 5:20–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caestecker KW and Van de Walle GR: The
role of BRCA1 in DNA double-strand repair: Past and present. Exp
Cell Res. 319:575–587. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huszno J, Budryk M, Kołosza Z and Nowara
E: The influence of BRCA1/BRCA2 mutations on toxicity related to
chemotherapy and radiotherapy in early breast cancer patients.
Oncology. 85:278–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang
C, Liu X, Wang X, Li H, Kang C, et al: MiR-124 governs glioma
growth and angiogenesis and enhances chemosensitivity by targeting
R-Ras and N-Ras. Neuro Oncol. 16:1341–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stupp R, Mason WP, Van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups, ; National Cancer Institute of
Canada Clinical Trials Group, : Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lim LPLN, Lau NC, Garrett-Engele P,
Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson
JM: Microarray analysis shows that some microRNAs downregulate
large numbers of target mRNAs. Nature. 433:769–773. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsai NP, Lin YL and Wei LN: MicroRNA
mir-346 targets the 5′-untranslated region of receptor-interacting
protein 140 (RIP140) mRNA and up-regulates its protein expression.
Biochem J. 424:411–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang HW, Wang F, Wei Q, Zhao YF, Liu M, Li
X and Tang H: miR-20a promotes migration and invasion by regulating
TNKS2 in human cervical cancer cells. FEBS Lett. 586:897–904. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Díaz-López A, Moreno-Bueno G and Cano A:
Role of microRNA in epithelial to mesenchymal transition and
metastasis and clinical perspectives. Cancer Manag Res. 6:205–216.
2014.PubMed/NCBI
|
|
15
|
Zhao L, Bode AM, Cao Y and Dong Z:
Regulatory mechanisms and clinical perspectives of miRNA in tumor
radiosensitivity. Carcinogenesis. 33:2220–2227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zhang D, Chen C, Ruan Z, Li Y and
Huang Y: MicroRNA-212 displays tumor-promoting properties in
non-small cell lung cancer cells and targets the hedgehog pathway
receptor PTCH1. Mol Biol Cell. 23:1423–1434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumor progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang X, Zeng J, Wang L, Fang M, Wang Q,
Zhao M, Xu X, Liu Z, Li W, Liu S, et al: Histone demethylase
retinoblastoma binding protein 2 is overexpressed in hepatocellular
carcinoma and negatively regulated by hsa-miR-212. PLoS One.
8:e697842013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiping Z, Ming F, Lixiang W, Xiuming L,
Yuqun S, Han Y, Zhifang L, Yundong S, Shili L, Chunyan C, et al:
MicroRNA-212 inhibits proliferation of gastric cancer by directly
repressing retinoblastoma binding protein 2. J Cell Biochem.
114:2666–2672. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu L, Wang F, Xu XF, Mo WH, Xia YJ, Wan R,
Wang XP and Guo CY: Down-regulation of miR-212 expression by DNA
hypermethylation in human gastric cancer cells. Med Oncol. 28 Suppl
1:S189–S196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng X, Wu J, Pan C, Wang H, Ying X, Zhou
Y, Yu H, Zuo Y, Pan Z, Liu RY, et al: Genetic and epigenetic
down-regulation of microRNA-212 promotes colorectal tumor
metastasis via dysregulation of MnSOD. Gastroenterology.
145:426–436.e1-e6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA Profiling of Prostate Cancer. J Cancer.
4:350–357. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gandellini P, Rancati T, Valdagni R and
Zaffaroni N: miRNAs in tumor radiation response: Bystanders or
participants? Trends Mol Med. 20:529–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sakaguchi M, Hisamori S, Oshima N, Sato F,
Shimono Y and Sakai Y: miR-137 regulates the tumorigenicity of
colon cancer stem cells through the inhibition of DCLK1. Mol Cancer
Res. 14:354–362. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang X, Chen X, Chen L, Ma Y, Zhou L, Qi
Q, Liu Y, Zhang S, Luo J and Zhou X: Upregulation of the
miR-212/132 cluster suppresses proliferation of human lung cancer
cells. Oncol Rep. 33:705–712. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou C, Smith JL and Liu J: Role of BRCA1
in cellular resistance to paclitaxel and ionizing radiation in an
ovarian cancer cell line carrying a defective BRCA1. Oncogene.
22:2396–2404. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kan C and Zhang J: BRCA1 mutation: A
predictive marker for radiation therapy? Int J Radiat Oncol Biol
Phys. 93:281–293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Saha J and Davis AJ: Unsolved mystery: The
role of BRCA1 in DNA end-joining. J Radiat Res (Tokyo). 57 Suppl
1:i18–i24. 2016. View Article : Google Scholar
|
|
29
|
Bencokova Z, Pauron L, Devic C, Joubert A,
Gastaldo J, Massart C, Balosso J and Foray N: Molecular and
cellular response of the most extensively used rodent glioma models
to radiation and/or cisplatin. J Neurooncol. 86:13–21. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boukerroucha M, Josse C, Segers K,
El-Guendi S, Frères P, Jerusalem G and Bours V: BRCA1 germline
mutation and glioblastoma development: Report of cases. BMC Cancer.
15:1812015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chai KM, Wang CY, Liaw HJ, Fang KM, Yang
CS and Tzeng SF: Downregulation of BRCA1-BRCA2-containing complex
subunit 3 sensitizes glioma cells to temozolomide. Oncotarget.
5:10901–10915. 2014. View Article : Google Scholar : PubMed/NCBI
|