Introduction
Oral squamous cell carcinoma (OSCC) is the most
prevalent type of head and neck cancer and it accounts for nearly
90% of all oral cancer cases (1).
Despite improvement in OSCC treatment over the past years, the
5-year survival rate of OSCC patients has not significantly
improved (2). Therefore,
identifying effective biomarkers and therapeutic targets is
essential to acknowledge the molecular mechanisms underlying the
progression of OSCC.
Long non-coding RNAs (lncRNAs), a class of
regulatory transcripts greater than 200 nucleotides without
protein-coding function, play a critical role in tumorigenesis
through multiple mechanisms, including interaction with microRNAs
(miRNAs) or proteins (3,4). miRNAs can post-transcriptionally
regulate gene expression via binding to the 3′-untranslated region
(UTR) of mRNAs (5). Both
dysregulated lncRNAs and miRNAs were reported to be closely related
with tumor cellular processes, including proliferation,
differentiation, invasion and apoptosis (6,7). For
example, NEAT1 was found to be upregulated in several types of
cancers (8–11). NEAT1 contributed to cell growth and
metastasis and acted as a competing endogenous RNA (ceRNA) for
miR-377-3p in lung cancer (12).
NEAT1 epigenetically suppressed miR-129-5p expression by promoting
the miR-129 related CpG island methylation (13). In addition, several other miRNAs,
including miR-335, miR-107 and miR-101, were demonstrated to
interact with NEAT1 and involved in NEAT1 regulated tumor-related
biological processes (14–16). However, the expression and function
of NEAT1 in the development of OSCC remain unclear.
Among the dysregulated miRNAs, miR-365 was reported
to be decreased in colon cancer, inhibited cell cycle progression
and induced apoptosis (17).
NKX2-1, a direct target of miR-365, attenuated the suppressive
function of miR-365 on cell proliferation in lung cancer (18). In gastric cancer, activation of Akt
decreased the expression of miR-365, consequently promoting cell
growth by increasing the expression of cyclin D1 and CDC25A
(19).
In the present study, we determined that the
expression of NEAT1 was increased in OSCC and associated with tumor
progression. Knockdown of NEAT1 inhibited cell proliferation and
invasion and induced cell cycle arrest at the G0/G1 phase.
Furthermore, we found that NEAT1 could act as a ceRNA of miR-365
and therefore regulate its target gene RGS20.
Materials and methods
Cell lines and clinical samples
OSCC cell lines (SCC-9, SCC-25, HN4, Tca-8113 and
Cal-27) were obtained from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China) and cultured in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.), and 100 µg/ml
penicillin/streptomycin (BioLight, Shanghai, China). A human normal
oral keratinocyte cell line (hNOK) was used as a control. All cells
were incubated at 37°C in a humidified atmosphere with 5%
CO2.
Thirty OSCC tissues and their adjacent non-tumor
tissues were obtained from patients at the Department of
Stomatology, General Hospital of Benxi Iron and Steel Co., Ltd.
(Benxi, China), between 2010 and 2012. The present study was
approved by the Research Ethics Committee of the General Hospital
of Benxi Iron and Steel Group Co., Ltd., and written informed
consents from patients were signed before surgery. None of the
patients had a prior history of cancern or had received
radiochemotherapy before surgery. All tissues were immediately
snap-frozen in liquid nitrogen and stored at −80°C until use. The
clinicopathological characteristics of patients were summarized in
Table I.
 | Table I.The expression levels of NEAT1 and
miR-365 in subgroups of OSCC cases. |
Table I.
The expression levels of NEAT1 and
miR-365 in subgroups of OSCC cases.
|
Characteristics | Cases, n=30 | NEAT1 levels | P-value | miR-365 levels | P-value |
|---|
| Age (years) |
|
| 0.657a |
| 0.966a |
|
<55 | 12 | 3.057±1.094 |
| 0.616±0.328 |
|
|
≥55 | 18 | 3.297±1.268 |
| 0.627±0.396 |
|
| Sex |
|
| 0.094a |
| 0.933a |
|
Female | 13 | 3.432±1.231 |
| 0.593±0.301 |
|
|
Male | 17 | 2.680±1.067 |
| 0.645±0.413 |
|
| LNM status |
|
|
0.009a |
| 0.244a |
|
Negative | 20 | 2.562±0.769 |
| 0.669±0.353 |
|
|
Positive | 10 | 3.892±1.394 |
| 0.529±0.385 |
|
| TNM stage |
|
|
0.018a |
|
0.039a |
| I,
II | 16 | 2.507±0.778 |
| 0.743±0.355 |
|
| III,
IV | 14 | 3.576±1.328 |
| 0.486±0.334 |
|
| Smoking |
|
| 0.148a |
| 0.258a |
|
Never | 9 | 2.445±0.809 |
| 0.761±0.419 |
|
|
Quit | 21 | 3.246±1.250 |
| 0.563±0.331 |
|
|
Differentiation |
|
| 0.603a |
| 0.835a |
|
Well | 16 | 2.893±1.241 |
| 0.605±0.364 |
|
|
Moderate/poor | 14 | 3.135±1.144 |
| 0.643±0.377 |
|
| Location |
|
| 0.443b |
| 0.929b |
|
Tongue | 19 | 2.987±1.361 |
| 0.644±0.425 |
|
|
Cheek | 8 | 3.234±0.837 |
| 0.574±0.212 |
|
|
Gingiva | 3 | 2.517±0.767 |
| 0.621±0.357 |
|
Quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). To assess NEAT1 and
RGS20 expression, a High Capacity cDNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City,
CA, USA) and SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.,
Dalian, China) were used for reverse transcription (RT) and qPCR,
respectively. For the expression of miR-365, the TaqMan MicroRNA
Reverse Transcription kit and the TaqMan Universal Master Mix II
(both from Applied Biosystems; Thermo Fisher Scientific, Inc.) were
used for RT and qPCR, respectively. The results for NEAT1 and RGS20
were normalized to the expression of ATCB and miR-365 to the
expression of U6. The relative expression level of each gene was
calculated and normalized using the 2−ΔΔCt method.
Plasmidand oligonucleotide
The small interfering RNAs (siRNAs) specifically
targeting NEAT1 (si-NEAT1 sense, 5′-GAGGGAUGAGGGUGAAGAA-3′ and
antisense, 5′-UUCUUCACCCUCAUCCCUC-3′) and the negative control
siRNA (si-NC) were obtained from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For in vivo analysis, cells were
transfected with 6 µg of sh-NEAT1 or the empty lentiviral vector,
cultured with DMEM containing 20% FBS for 36 h. Lentiviral
particles were harvested and used for infection. The target
sequence of sh-NEAT1 was as follows: 5′-GCCATCAGCTTTGAATAAATT-3′.
The human NEAT1 gene was ligated into the pGCMV/MCS/RFP/Neo vector
(Shanghai Genepharma Co., Ltd., Shanghai, China) and stable cell
lines were generated by selection with Geneticin® (G418;
Invitrogen; Thermo Fisher Scientific, Inc.). The packaged
lentiviruses were named sh-NEAT1 and the empty lentiviral vector
(sh-Ctrl) was used as a control. The miR-365 mimic, miR-365
inhibitor, mimic negative control (mim-NC), inhibitor negative
control (inh-NC) sequences were obtained from Shanghai Genepharma
Co., Ltd. Cells were transfected using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). To restore RGS20
expression, the Cal-27 cells were transfected with a pcDNA3.1-RGS20
plasmid (pcRGS20), which contained the coding sequences but lacked
the 3′-UTR of RGS20. Cells transfected with the empty vector were
used as a control and named pcDNA.
Cell proliferation analysis
Cells (1.5×103/well) were plated in
96-well culture plates and cell viability was assessed every 24 h
after transfection. MTT [5 mg/ml in phosphate-buffered saline
(PBS); Sigma-Aldrich, St. Louis, MO, USA] was added to each well
and the plates were incubated at 37°C. After 4 h, 150 µl dimethyl
sulfoxide (DMSO) was added to each well. The absorbance was
measured at 490 nm on a microplate reader (Molecular Devices, LLC,
Sunnyvale, CA, USA).
Cell cycle and apoptosis analysis
At 48 h post-transfection, cells were harvested,
washed with PBS, and fixed with 70% ethanol. Then the fixed cells
were washed with PBS, centrifuged at 1,500 × g for 5 min and
subsequently treated with RNase A (0.1 mg/ml) and propidium iodide
(PI; 0.05 mg/ml) at 37°C for 30 min. The stained cells were
analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose,
CA, USA). For the apoptosis assay, 48 h post-transfection, the
cells were collected by trypsinization and washed twice with
serum-containing medium. The cells were collected and resuspended
in 1X Annexin V Binding buffer (Annexin V-FITC Apoptosis Detection
kit; BD Pharmingen; BD Biosciences) at a concentration of
1×106 cells/ml. Then, 5 µl of FITC Annexin V and 5 µl PI
(BD Pharmingen; BD Biosciences) were added to 100 µl of the cell
suspension. After incubation for 10 min at room temperature in the
dark, 400 µl of binding buffer was added. Apoptosis was analyzed by
flow cytometry (FACSCalibur; BD Biosciences) and the data were
analyzed using CellQuest software (BD Biosciences).
Cell invasion assay
Cell invasion abilities were detected using
Transwell chambers precoated with Matrigel (BD Biosciences). DMEM
with 10% FBS was added to the lower chamber. OSCC cells were
transfected, incubated, and then starved in serum-free DMEM
overnight. Subsequently, they were resuspended (1×105
cells) in serum-free medium, which was added to the upper chamber.
Twenty-four hours later, the cells that had invaded to the lower
surface of the membrane were fixed, stained and counted under an
inverted microscope (Olympus, Tokyo, Japan) by counting five random
fields.
Luciferase activity assay
The fragment from NEAT1 containing the predicted
miR-365 binding site was amplified by PCR and cloned into a pmirGLO
Dual-Luciferase Target Vector (Promega Corp., Madison, WI, USA) to
form the NEAT1-wild-type reporter vector (NEAT1-WT). The mutant was
generated by mutating the miR-365 seed region binding site and
named NEAT1-MUT. Cells were co-transfected with either wild-type
fragments or mutant fragments and miR-365 mimic or mim-NC using
Lipofectamine 2000. A luciferase reporter assay was performed using
the Dual-Luciferase Reporter Assay system (Promega Corp.).
The 3′-UTR of RGS20 containing the putative binding
sites for miR-365 was amplified by PCR and cloned into the
pGL3-luciferase reporter plasmid (Promega Corp.). Mutations in the
miR-365-binding site of RGS20 3′-UTR were generated by the
QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA,
USA). Cells were co-transfected with miR-365 mimic (or mim-NC) and
the reported vector with the wild-type (WT) or mutant (MUT) 3′-UTR
of RGS20. Luciferase activity was assessed after incubation for 48
h at 37°C.
Western blot analysis
Cells were lysed in RIPA lysis buffer (Beyotime
Institute of Biotechnology, Shanghai, China) and the protein
concentration was measured using a BCA protein assay kit (Thermo
Fisher Scientific, Inc., Rockford, IL, USA). Equal amounts of
protein were isolated using SDS-PAGE and then transferred to a
polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Bedford,
MA, USA). The membranes were blocked in 5% non-fat milk/TBST and
incubated with primary antibodies. The primary antibodies RGS20
(1:500; cat. no. ab191500), cyclin D1 (1:1,000; cat. no. ab134175),
E-cadherin (1:2,000; cat. no. ab15148), N-cadherin (1:2,000; cat.
no. ab18203), vimentin (1:2,000; cat. no. ab24525) and β-actin
(1:2,000; cat. no. ab8227) were purchased from Abcam (Cambridge,
MA, USA). Subsequently, the membranes were incubated with goat
anti-rabbit secondary antibodies (1:2,000; cat. no. ab150077) and
the proteins were detected with ECL reagents (Pierce; Thermo Fisher
Scientific, Rockford, IL, USA).
In vivo tumor growth assay
All animal procedures were in line with the
guidelines of the Laboratory Animal Centre and were approved by the
Ethics Committee of the General Hospital of Benxi Iron and Steel
Co., Ltd. Ten female athymic BALB/c nude mice (4–5 weeks) were
used. A total of 200 µl of PBS containing 2×107 Cal-27
cells expressing sh-NEAT1 or sh-NC were injected subcutaneously to
the flanks of each mouse (n=5 for each group). The tumor size was
measured every 7 days and was calculated using the following
formula: 0.5 × length × width2. Four weeks later, the
mice were sacrificed and the tumors were harvested.
Statistical analysis
SPSS 16.0 software (IBM, Armonk, NY, USA) and
GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA,
USA) were used for statistical analysis. The paired samples t-test
was used to compare gene expression levels between OSCC and
non-tumor controls. The overall survival of patients was analyzed
by the Kaplan-Meier method. One-way ANOVA or Student's t-test was
used for comparisons between the groups. P<0.05 was considered
to indicate a statistically significant result.
Results
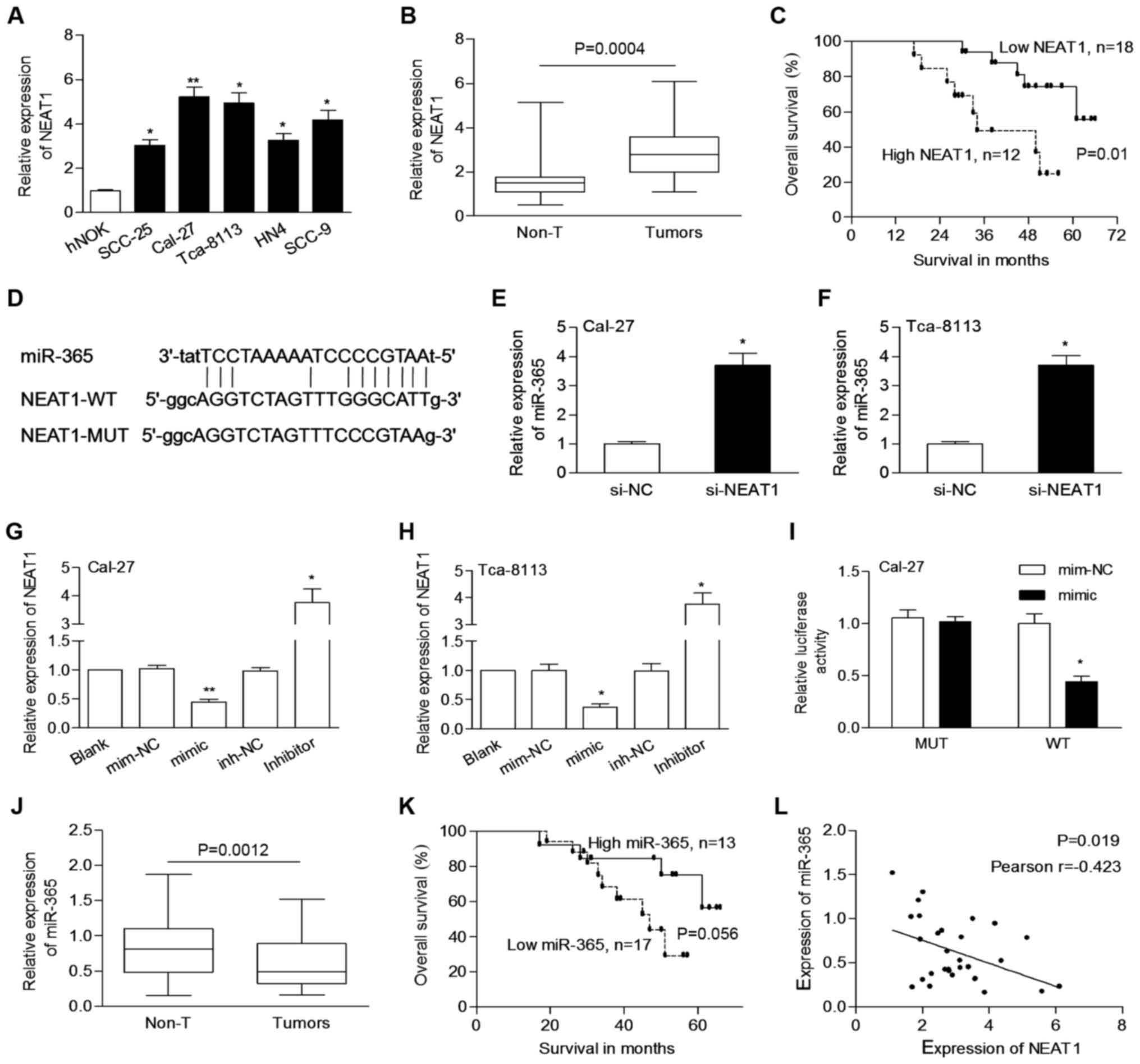
NEAT1 is overexpressed in OSCC cells
and tissues
In order to know the relevance of NEAT1 in OSCC
development, we assessed the endogenous levels of NEAT1 in OSCC
cells. As shown in Fig. 1A, the
expression of NEAT1 was significantly increased in OSCC cells
compared to hNOK cells. Then, a qPCR assay was performed to
evaluate the expression of NEAT1 in clinical samples. The
expression of NEAT1 in OSCC tissues was significantly higher than
that in matched non-tumor tissues (3.006±1.182 vs. 1.712±0.971,
P=0.0004; Fig. 1B).
Elevated NEAT1 is correlated with
aggressive tumor phenotypes and poor prognosis
OSCC cases were classified into different subgoups
such assex (male vs. female) and TNM stage (I/II vs. III/IV). We
determined that the expression levels of NEAT1 were significantly
increased in cases with lymph node metastasis (P=0.009) and higher
clinical stage (P=0.018; Table I),
respectively. The median level of NEAT1 in tumors was used as a
cut-off value to divide cases into two groups. Patients with high
NEAT1 expression had poor survival when compared to patients with
low NEAT1 expression (P=0.01; Fig.
1C).
NEAT1 negatively regulates miR-365 in
OSCC
The exact function and underlying mechanism of NEAT1
in OSCC warranted further investigation. Using starBase 2.0 and
RegRNA2.0, miR-365 was determined to potentially bind to NEAT1
(Fig. 1D), implying a possible
interaction between miR-365 and NEAT1. Cal-27 and Tca-8113 cells,
which express a relatively high level of NEAT1, were used for
further analysis. We determined that the levels of miR-365 were
significantly increased by si-NEAT1 transfection in both Cal-27 and
Tca-8113 cells (Fig. 1E and F). The
expression of NEAT1 was downregulated by miR-365 mimic, while it
was upregulated by the miR-365 inhibitor (Fig. 1G and H). Co-transfection of miR-365
and NEAT1-WT significantly decreased the luciferase activity
(Fig. 1I).
miR-365 was significantly downregulated in OSCC
tissues compared to non-tumor tissues (0.622±0.364 vs. 0.819±0.428,
P=0.0012; Fig. 1J). miR-365 was
significantly downregulated in tumors of advanced stage (P=0.039,
Table I). Cases were grouped in a
low or high group according to the median level of miR-365.
Patients with low expression of miR-365 appeared to have poor
prognosis but without statistical significance (P=0.056; Fig. 1K). In addition, NEAT1 expression was
negatively correlated with the expression of miR-365 in tumors
(P=0.019, Pearson r=−0.423; Fig.
1L). These findings indicated that an interaction between NEAT1
and miR-365 may be involved in the development of OSCC.
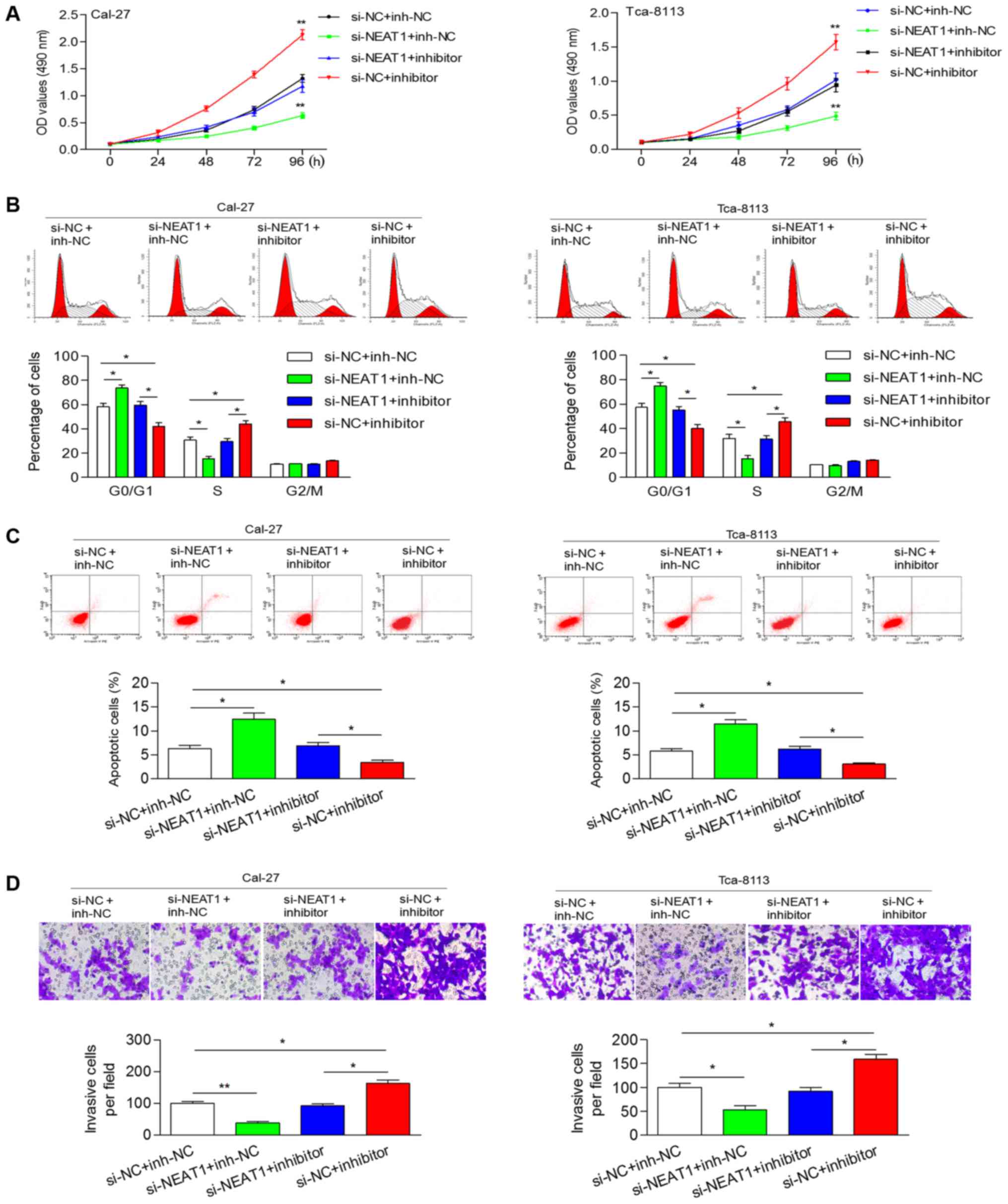
Inhibition of miR-365 attenuates the
NEAT1 knockdown-induced inhibition of cellular processes
To explore the effect of NEAT1 knockdown and miR-365
inhibition on cellular processes, we transfected OSCC cells with
si-NEAT1 (or si-NC) and miR-365 inhibitor (or inh-NC). Knockdown of
NEAT1 significantly reduced cell proliferation and abrogated the
miR-365 inhibitor-induced increase of the cell proliferation rate
(Fig. 2A). Flow cytometric analysis
revealed that si-NEAT1 induced an increase in the percentage of
cells at the G0/G1 phase and a reduction in the percentage of cells
at the S phase, while the miR-365 inhibitor had an opposite effect
on cell cycle distribution (Fig.
2B). Knockdown of NEAT1 induced cell apoptosis and abolished
the miR-365 inhibitor-induced decrease of apoptotic cells (Fig. 2C). In addition, knockdown of NEAT1
could inhibit the invasive ability of cells, which was promoted by
the miR-365 inhibitor (Fig. 2D).
These results revealed that NEAT1 contributed to cell proliferation
and invasion by negatively-mediated miR-365.
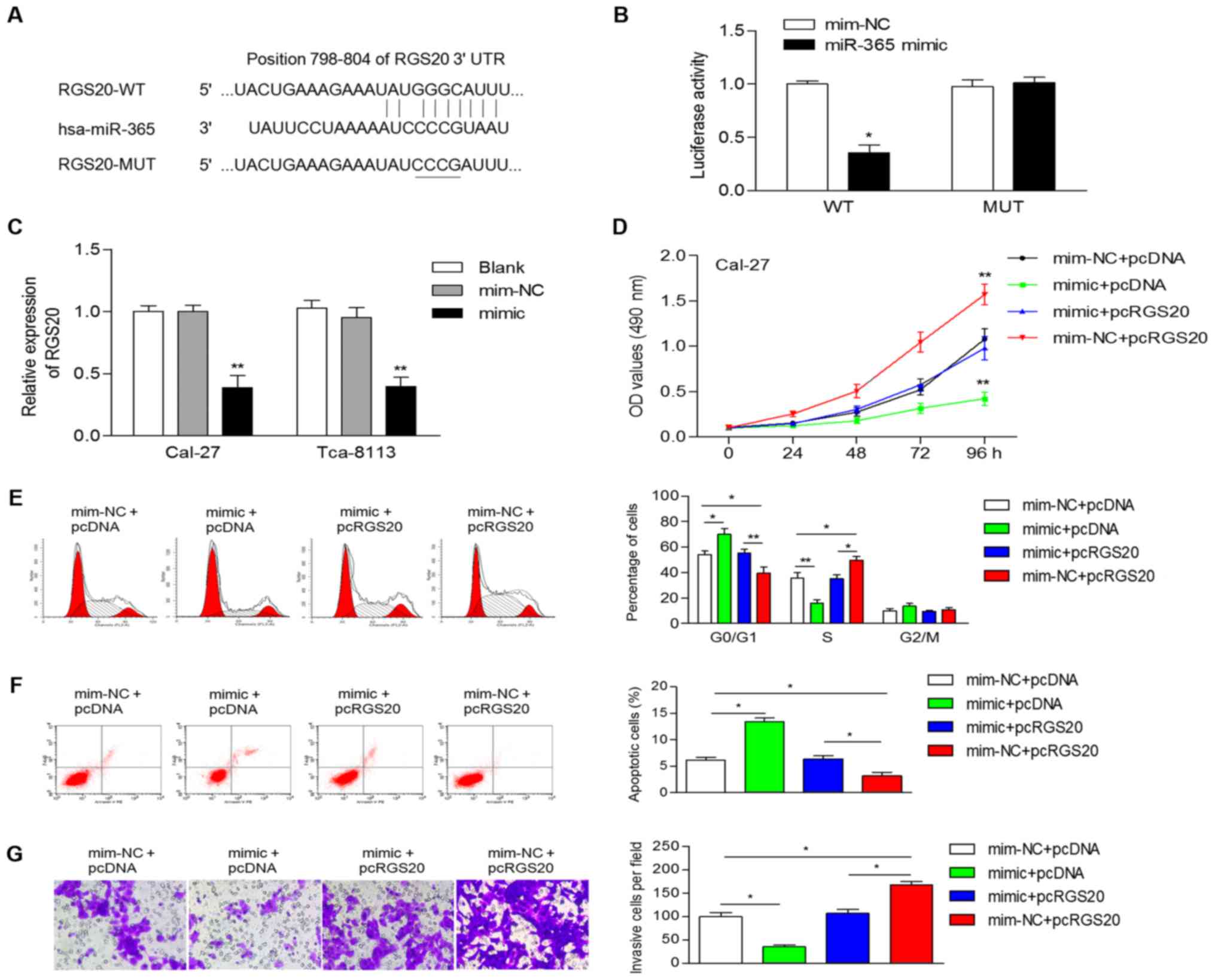
RGS20 is a target of miR-365
Numerous studies have revealed that lncRNAs could
competitively suppress miRNAs by acting as ceRNAs, and ultimately
regulate the expression of protein-coding genes. Thus, we searched
for candidate genes of miR-365 using TargetScan, microRNA, miRDB
and TargetMiner. Among the predicted targets, in the present study
we focused on RGS20, considering the involvement of RGS20 in human
cancers. In addition, by analyzing datasets from Oncomine, we
determined that the mRNA levels of RGS20 were significantly
upregulated in tongue squamous cell carcinoma (data not shown). All
four bioinformatics tools revealed that the 3′-UTR of RGS20 mRNA
has a potential binding site of miR-365 (Fig. 3A). Furthermore, a luciferase
reporter assay revealed that the miR-365 mimic significantly
reduced the luciferase activity of the wild-type 3′-UTR of RGS20
(Fig. 3B). In addition,
overexpression of miR-365 significantly decreased the mRNA
expression of RGS20 in OSCC cells (Fig.
3C).
RGS20 is a functional target of
miR-365
To ascertain whether miR-365 performs its
suppressive function through downregulation of RGS20, Cal-27 cells
were co-transfected with pcRGS20 or the 3′-UTR (or pcDNA) and with
miR-365 mimic (or mim-NC). Cell proliferation was stimulated by
overexpression of RGS20 and inhibited by the miR-365 mimic
(Fig. 3D). Cell cycle analysis
revealed that pcRGS20 transfected cells displayed a higher
frequency of cells at the S phase and a lower frequency of cells at
the G1 phase and ectopic expression of RGS20 reversed the
miR-365-induced accumulation of G0/G1 phase cells (Fig. 3E). The apoptosis of Cal-27 cells was
increased by the miR-365 mimic, while it was promoted decreased by
the overexpression of RGS20 (Fig.
3F). Overexpression of RGS20 increased cell invasion (Fig. 3G), which was similar to the effect
of the miR-365 inhibitor. RGS20 overexpression also significantly
attenuated miR-365-induced inhibition on cellular invasion
(Fig. 3G). These data indicated
that miR-365 performs its tumor-suppressive function by regulating
RGS20.
Dysregulation of the
NEAT1/miR-365/RGS20 axis is involved in epithelial-mesenchymal
transition (EMT) and tumor growth
Western blot analysis was performed to evaluate the
effect of NEAT1/miR-365/RGS20 on the protein expression of cell
cycle- and EMT-related markers. As shown in Fig. 4A, the protein expression of RGS20
was decreased by knockdown of NEAT1, while it was increased by the
miR-365 inhibitor. Inhibition of NEAT1 abolished the miR-365
inhibitor-induced upregulation of RGS20 (Fig. 4A). Downregulation of NEAT1 led to an
increase of E-cadherin and a reduction of cyclin D1, N-cadherin and
vimentin, a phenomenon that could be reversed by the miR-365
inhibitor (Fig. 4A). Subsequently,
the protein level of RGS20 could be inhibited by the miR-365 mimic
(Fig. 4B), which was consistent
with previous data shown in Fig.
3C. Restoration of RGS20 promoted the protein expression of
cyclin D1, N-cadherin and vimentin, but suppressed the protein
expression of E-cadherin (Fig.
4B).
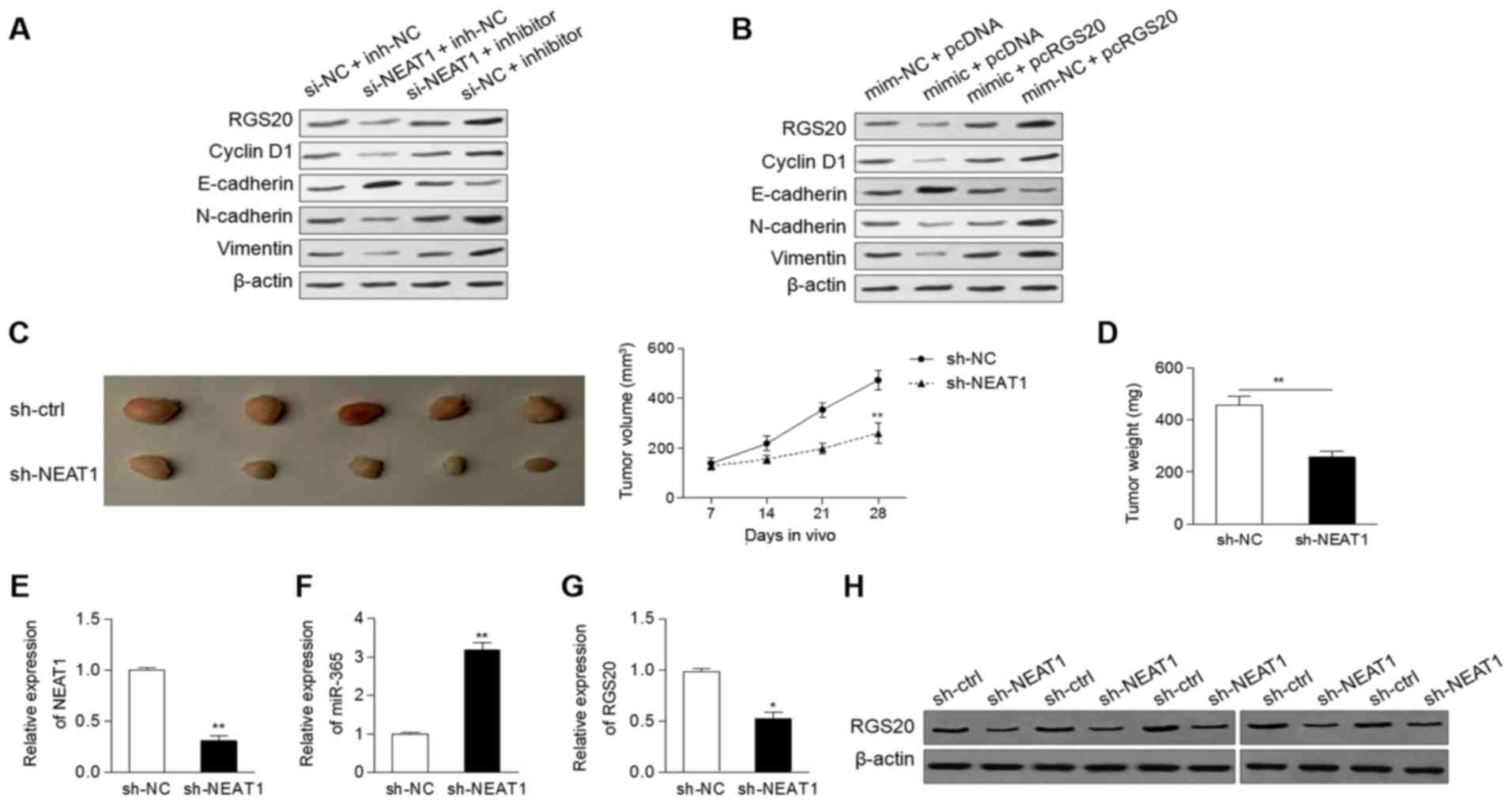 | Figure 4.Dysregulationof the
NEAT1/miR-365/RGS20 axis is involved in EMT and tumor growth. (A
and B) The protein levels of RGS20, cyclin D1, E-cadherin,
N-cadherin and vimentin in Cal-27 cells following (A) knockdown of
NEAT1 and/or inhibition of miR-365, as well as in (B) cells
transfected with mimic (or mim-NC) and pcRGS20 (or pcDNA). (C and
D) Knockdown of NEAT1 led to a marked reduction of (C) the tumor
volume and (D) tumor weight. The expression of (E) NEAT1, (F)
miR-365 and (G) RGS20 was determined in mouse tumors. (H) The
protein levels of RGS20 of mouse tumors were determined by western
blot analysis. *P<0.05, **P<0.01. EMT, epithelial-mesenchymal
transition; mim-NC, mimic negative control; si-NC, negative control
siRNA; inh-NC, inhibitor negative control; si-NEAT1, specifically
targeting NEAT1. |
To ascertain whether knockdown of NEAT1 inhibits
tumor growth in vivo, Cal-27 cells (expressing sh-NEAT1or
sh-ctrl) were injected into the flanks of nude mice. The results
indicated that the tumor volumes and weights formed by the sh-NEAT1
cells were markedly lower than those formed by the sh-ctrl cells
(Fig. 4C and D). In addition, the
tumors of sh-NEAT1-treated mice had a significantly low level of
NEAT1, increased expression of miR-365, and downregulation of RGS20
(Fig. 4E-G). Furthermore, the
protein levels of RGS20 were decreased in mouse tumors transfected
with sh-NEAT1 compared to the control group (Fig. 4H).
Discussion
Numerous studies have shown that lncRNAs function as
oncogenes or tumor-suppressor genes to regulate carcinogenesis, and
that they can be used as diagnostic or prognostic markers (20). In the present study, the expression
of NEAT1 was markedly increased in OSCC cells and tissues, and
upregulation of NEAT1 was correlated with advanced stage and
unfavorable prognosis of OSCC patients. Similarly, high expression
of NEAT1 was associated with metastasis and vaso-invasion in
hepatocellular carcinoma (21).
High NEAT1 was closely related to larger tumor size and
independently associated with risk of death in glioma (22), as well as clinical pathologic grade
in bladder cancer (23). Our
results revealed the oncogenic role of NEAT1 in the development of
OSCC. Certainly, further analysis based on a larger number of cases
would provide more knowledge on the clinical relevance of NEAT1 in
OSCC.
Functionally, knockdown of NEAT1 exerted a
tumor-suppressive effect by inhibiting cell proliferation, cell
cycle progression, and invasion in vitro and tumorigenesis
in vivo, which was consistent with previous studies
(10,23–25).
NEAT1 could negatively regulate the expression of miR-365. miR-365
inhibition abrogated the inhibitory effect of NEAT1 knockdown.
Several other miRNAs, including miR-377, miR-335, miR-107, miR-98
and miR-506, were identified to interact with NEAT1 in different
types of cancers (12,14,26–28),
suggesting that NEAT1 plays an oncogenic role in different types of
cancer through the regulation of different miRNAs.
Our findings revealed that miR-365 suppressed cell
proliferation and invasion and expanded on the knowledge of miR-365
as a tumor suppressor in OSCC. The inhibitory effect of miR-365 on
tumorigenesis has also been reported in several studies (17–19,29).
However, miR-365 displayed the opposite effect in cutaneous tumors
by facilitating tumor growth (30).
Thus, miR-365 exerts a tumor-suppressive or oncogenic function
depending on its target genes. In the present study, RGS20 was
identified as a direct target of miR-365 and overexpression of
RGS20 impaired the miR-365-induced inhibition of cell growth and
invasion. In addition, cyclin D1, CDC25A, WNT5A and ADAM10 were
identified as targets of miR-365 and were correlated with
miR-365-mediated cell growth and metastasis (19,29,31).
RGS20 was first reported to be overexpressed in
metastatic melanomas (32). High
expression of RGS20 indicated the progression and poor survival of
triple-negative breast cancer (33). RGS20 facilitated cell aggregation,
invasion and the expression of vimentin, but decreased the
expression of E-cadherin (34). By
gain-of-function approaches, we revealed similar results. RGS20
increased cell viability, motility and protein expression of cyclin
D1, N-cadherin but decreased the protein level of E-cadherin,
suggesting the oncogenic function of RGS20 in OSCC. The protein
level of RGS20 was regulated by NEAT1/miR-365, suggesting that
NEAT1 acted as a ceRNA of miR-365 and enhanced the expression of
RGS20. Moreover, cell cycle- and EMT-related indicators were
regulated by the NEAT1/miR-365/RGS20 pathway, supporting the
regulatory effect of the NEAT1/miR-365/RGS20 axis on cell growth
and metastasis in vitro. A previous study also demonstrated
that NEAT1 is a regulator of EMT-related proteins in gastric cancer
(35). Our findings revealed that
RGS20, a direct target of miR-365, could mediate the biological
effects that NEAT1 exerted.
In conclusion, we determined that upregulated NEAT1
was correlated with an aggressive tumor phenotype and an adverse
prognosis in OSCC. NEAT1 promoted OSCC cell proliferation, cell
cycle progression and invasion through the miR-365/RGS20 axis.
These data provide new insights into the regulatory function of
NEAT1/miR-365/RGS20 in the development of oral malignancy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
GH conceived and designed the experiments. GH and
XLW performed the experiments. XH analyzed the data. GH and XH
wrote the manuscript. All authors contributed toward data analysis,
drafting and critically revising the paper, gave final approval of
the version to be published.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the General Hospital of Benxi Iron and Steel
Co., Ltd., and written informed consents from patients were signed
before surgery. All animal procedures were in line with the
guidelines of the Laboratory Animal Centre and were approved by the
Ethics Committee of the General Hospital of Benxi Iron and Steel
Co., Ltd.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoffman HT, Porter K, Karnell LH, Cooper
JS, Weber RS, Langer CJ, Ang KK, Gay G, Stewart A and Robinson RA:
Laryngeal cancer in the United States: Changes in demographics,
patterns of care, and survival. Laryngoscope. 116:1–13. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bayoumi AS, Sayed A, Broskova Z, Teoh JP,
Wilson J, Su H, Tang YL and Kim IM: Crosstalk between long
noncoding RNAs and microRNAs in health and disease. Int J Mol Sci.
17:3562016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Tian H, Yang J and Gong Z: Long
noncoding RNAs regulate cell growth, proliferation, and apoptosis.
DNA Cell Biol. 35:459–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015.PubMed/NCBI
|
|
9
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z and Xu Y: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y
and Li D: Long non-coding RNA NEAT1 promotes non-small cell lung
cancer progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016.PubMed/NCBI
|
|
13
|
Lo PK, Zhang Y, Wolfson B, Gernapudi R,
Yao Y, Duru N and Zhou Q: Dysregulation of the BRCA1/long
non-coding RNA NEAT1 signaling axis contributes to breast
tumorigenesis. Oncotarget. 7:65067–65089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao J, Zhang Y, Yang J, He S, Li M, Yan S,
Chen Y, Qu C and Xu L: NEAT1 regulates pancreatic cancer cell
growth, invasion and migration though mircroRNA-335-5p/c-met axis.
Am J Cancer Res. 6:2361–2374. 2016.PubMed/NCBI
|
|
15
|
Yang X, Xiao Z, Du X, Huang L and Du G:
Silencing of the long non-coding RNA NEAT1 suppresses glioma
stem-like properties through modulation of the miR-107/CDK6
pathway. Oncol Rep. 37:555–562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen
Z and Xu X: The long non-coding RNA NEAT1 interacted with miR-101
modulates breast cancer growth by targeting EZH2. Arch Biochem
Biophys. 615:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y,
Du X and Han W: microRNA-365, down-regulated in colon cancer,
inhibits cell cycle progression and promotes apoptosis of colon
cancer cells by probably targeting cyclin D1 and Bcl-2.
Carcinogenesis. 33:220–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang SM, Lee HJ and Cho JY: MicroRNA-365
regulates NKX2-1, a key mediator of lung cancer. Cancer Lett.
335:487–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo SL, Ye H, Teng Y, Wang YL, Yang G, Li
XB, Zhang C and Yang X, Yang ZZ and Yang X: Akt-p53-miR-365-cyclin
D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN
deficiency. Nat Commun. 4:25442013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong
M, Dang Y, Feng Z and Chen G: Clinical implication of long
non-coding RNA NEAT1 expression in hepatocellular carcinoma
patients. Int J Clin Exp Pathol. 8:5395–5402. 2015.PubMed/NCBI
|
|
22
|
He C, Jiang B, Ma J and Li Q: Aberrant
NEAT1 expression is associated with clinical outcome in high grade
glioma patients. APMIS. 124:169–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong W, Zheng J, Liu X, Ma J, Liu Y and
Xue Y: Knockdown of NEAT1 restrained the malignant progression of
glioma stem cells by activating microRNA let-7e. Oncotarget.
7:62208–62223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jen J, Tang YA, Lu YH, Lin CC, Lai WW and
Wang YC: Oct4 transcriptionally regulates the expression of long
non-coding RNAs NEAT1 and MALAT1 to promote lung cancer
progression. Mol Cancer. 16:1042017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang P, Wu X, Wang X, Huang W and Feng Q:
NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin
sensitivity in lung cancer cells. Oncotarget. 7:43337–43351.
2016.PubMed/NCBI
|
|
28
|
Huang B, Liu C, Wu Q, Zhang J, Min Q,
Sheng T, Wang X and Zou Y: Long non-coding RNA NEAT1 facilitates
pancreatic cancer progression through negative modulation of
miR-506-3p. Biochem Biophys Res Commun. 482:828–834. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Xu C, Wang Y and Zhang X:
MicroRNA-365 inhibits ovarian cancer progression by targeting
Wnt5a. Am J Cancer Res. 7:1096–1106. 2017.PubMed/NCBI
|
|
30
|
Zhou M, Zhou L, Zheng L, Guo L, Wang Y,
Liu H, Ou C and Ding Z: miR-365 promotes cutaneous squamous cell
carcinoma (CSCC) through targeting nuclear factor I/B (NFIB). PLoS
One. 9:e1006202014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Zhang W, Liu S, Liu K, Ji B and
Wang Y: miR-365 targets ADAM10 and suppresses the cell growth and
metastasis of hepatocellular carcinoma. Oncol Rep. 37:1857–1864.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Riker AI, Enkemann SA, Fodstad O, Liu S,
Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al: The
gene expression profiles of primary and metastatic melanoma yields
a transition point of tumor progression and metastasis. BMC Med
Genomics. 1:132008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Q, Jin W, Cai Y, Yang F, Chen E, Ye D,
Wang Q and Guan X: Regulator of G protein signaling 20 correlates
with clinicopathological features and prognosis in triple-negative
breast cancer. Biochem Biophys Res Commun. 485:693–697. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang L, Lee MM, Leung MM and Wong YH:
Regulator of G protein signaling 20 enhances cancer cell
aggregation, migration, invasion and adhesion. Cell Signal.
28:1663–1672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu JW, Kong Y and Sun X: Long noncoding
RNA NEAT1 is an unfavorable prognostic factor and regulates
migration and invasion in gastric cancer. J Cancer Res Clin Oncol.
142:1571–1579. 2016. View Article : Google Scholar : PubMed/NCBI
|