Introduction
Gastric cancer is the fifth most common cancer
worldwide and a leading cause of cancer-related deaths (1,2). More
than 60% of gastric cancer cases occur in China, Japan and Korea,
and in 2015 there were 670,000 newly diagnosed cases of gastric
cancer and 498,000 deaths attributed to gastric cancer in China
(3). Although surgery is
potentially curative for early gastric cancer, the prognosis of
advanced adenocarcinoma remains poor despite improvements in
chemotherapy (4). Identification of
the molecular factors involved in gastric cancer, including
epigenetic modifications, will help us to understand the disease
process and facilitate early diagnosis.
Loss of intercellular tight junctions is a hallmark
of malignant transformation of gastric epithelium. Disassembly of
tight junctions is thought to increase the metastatic potential of
tumor cells by causing epithelial cell polarity loss and
epithelial-mesenchymal transition (5). As transmembrane proteins, the claudin
family comprises 27 members and plays a critical role in the
formation, integrity and function of tight junctions (6). Various members of the claudin family
have been implicated in gastric cancer. For example, increased
expression of claudin-1, claudin-4, claudin-6, claudin-7 and
claudin-9 and decreased expression of claudin-18 have been reported
to be associated with migration, invasion, proliferation and/or
prognosis of gastric cancer (7–11).
There is also evidence that claudin-3 plays a role in gastric
cancer, although the findings have not been entirely consistent
with previous studies. On the one hand, it has been reported that
tissues of gastric intestinal metaplasia and dysplasia show
enhanced expression of claudin-3 compared with normal gastric
mucosa (12,13), while on the other hand, Jung et
al found that the expression of claudin-3 was significantly
lower in cases of advanced gastric cancer (T3 or T4 stage)
(14). Furthermore, an
immunohistochemistry (IHC) study suggested that downregulation of
claudin-3 was associated with increased proliferative potential in
early gastric cancer (15), while
loss of claudin-3 expression at the invasive front was associated
with an enhanced grade of malignancy of gastric cancer in
vivo (16).
DNA methylation in the promoter regions of genes is
a frequent epigenetic mechanism that is involved in a variety of
cellular processes and can be modified during tumorigenesis and
cancer progression (17). There is
accumulating evidence that alterations in DNA methylation
contribute to the deregulation of claudin expression in cancer. For
example, colorectal cancer tissue showed DNA hypomethylation of the
claudin-1 gene as well as reduced membrane expression and increased
cytoplasmic expression of claudin-1, as compared with adjacent
non-neoplastic mucosa (18).
Ovarian cancer cells that exhibit high expression of claudin-3
showed increased DNA methylation and enhanced histone H3
acetylation of the promoter region (19). Interestingly, downregulation of
claudin-3 expression was associated with hypermethylation of its
promoter in hepatocellular cancer cell lines and was an independent
predictor of poorer survival in patients with hepatocellular
carcinoma (20). Epigenetic
modifications of claudin genes have also been reported in gastric
cancer. Kwon et al showed that claudin-4 upregulation in
gastric tissues and gastric cancer cells was correlated with DNA
hypomethylation and histone modifications (7). Furthermore, Agarwal et al
demonstrated that hypermethylation of the claudin-11 promoter was
associated with downregulation of claudin-11 in gastric cancer
tissues and with increased invasive potential of gastric cancer
cells (21). However, the role of
claudin-3 in gastric cancer remains unknown.
We hypothesized that changes in claudin-3 promoter
methylation and claudin-3 expression are associated with gastric
cancer prognosis. Therefore, the aims of the present study were to
establish the relationship between claudin-3 promoter methylation
and claudin-3 mRNA and protein expression in gastric adenocarcinoma
and to determine the associations of these factors with
clinicopathological characteristics.
Materials and methods
Study participants
One hundred and twenty-two patients with advanced
gastric adenocarcinoma were enrolled between January 2012 and
December 2014 at the First Affiliated Hospital of Fujian Medical
University, Fuzhou, Fujian, China. The inclusion criteria were as
follows: i) pathologic diagnosis of advanced gastric
adenocarcinoma; ii) disease stage group IIB to IV; iii) lymph node
metastasis; and iv) complete follow-up and clinical pathologic data
available. The exclusion criteria were: i) preoperative
chemotherapy; ii) death due to postoperative complications; or iii)
loss during follow-up. Pathologic diagnosis and cancer staging were
performed independently by two pathologists according to the
classification criteria of the World Health Organization. The study
protocol was approved by Fujian Medical University Ethics
Committee, and informed written consent was obtained from each
participant before enrollment in the study.
The following clinicopathological information was
collected: age, sex, Lauren classification subtype (intestinal,
diffuse or mixed), tumor size, depth of invasion (T3 or T4), number
of metastatic lymph nodes and TNM stage. For each patient,
formalin-fixed and paraffin-embedded tissue samples representative
of normal tissue, intestinal metaplasia, primary tumor and lymph
node metastasis were obtained from the archival tissue bank of our
institution's pathology laboratory. Claudin-3 protein expression
and promoter methylation status were examined in samples from all
patients (see below). Claudin-3 mRNA expression was determined for
a subset of participants whose tissue samples had been stored
appropriately (i.e. the tissue samples had been immediately snap
frozen in liquid nitrogen after surgical excision and then stored
at 80°C).
Tissue microarray construction and IHC
experiment
Three representative staining areas from each core
were selected and imaged (10× and 40× objectives) for IHC scoring.
Claudin-3 immunoreactivity was assessed based on a combined score
of the extent and intensity of staining. Scores 0–3 were assigned
according to the percentage of positive tumor cells (0, 0%; 1,
<25%; 2, 25–50%; 3, >50%) and the intensity of staining in
the tumor (0, 0; 1, 1+; 2, 2+; 3, 3+). The two scores were
multiplied to provide an overall score of 0–9; 0–3 was defined as
weak expression, 4–9 was defined as high expression (22).
RNA isolation and real-time qPCR
(23)
Total RNA was extracted using TRIzol reagent (Thermo
Fisher Scientific, Inc. Waltham, MA, USA), and reverse
transcription was performed using cDNA Reverse Transcription Kit
(Applied Biosystems, Thermo Fisher Scientific, Inc., Waltham, MA,
USA), in accordance with the manufacturer's instructions. Real-time
reverse-transcription polymerase chain reaction (RT-PCR) was
conducted using SYBR Premix Ex Taq (Takara Bio, Beijing, China) and
the Applied Biosystems 7500 Fast Real-Time PCR System (Thermo
Fisher Scientific, Inc. Waltham, MA, USA). Briefly, 1 µg (20 µl) of
total cDNA solution was added to 9 µl of Fast-SYBR mixture (with
ROX), which contained 2.5 mmol/l MgCl2 and 0.5 µmol/l
primers (as shown in Table I). A
sample of non-reverse-transcribed RNA and a non-template control
were used as negative controls. Melting curve analysis and
electrophoresis were applied to confirm the specificity of the
amplified products. All experiments were performed in triplicate,
and the results were normalized to the expression of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as
the internal control. The primer sequences are listed in Table I.
 | Table I.Primers used for methylation-specific
PCR, bisulfite-sequencing PCR and real-time PCR. |
Table I.
Primers used for methylation-specific
PCR, bisulfite-sequencing PCR and real-time PCR.
|
| Primer | Size (bp) | Tm (°C) |
|---|
| MSP | M-claudin-3-MF:
5′-TTTTTAGGTTTTGGAGAGCGC-3′ | 125 | 58 |
|
| M-claudin-3-MR:
5′-ATAACTTTATAAACGAACGACGACG-3′ |
|
|
|
| M-claudin-3-UMF:
5′-TGTTTTTAGGTTTTGGAGAGTGTG-3′ | 133 | 59 |
|
| M-claudin-3-UMR:
5′-CTACCTATAACTTTATAAACAAACAACAACA-3′ |
|
|
| qPCR | Claudin-3-F:
5′-GCCACCAAGGTCGTCTACTC-3′ | 102 | 60 |
|
| Claudin-3-R:
5′-CCCTGCGTCTGTCCCTTAGA-3′ |
|
|
|
| GAPDH-F:
5′-TGCACCACCAACTGCTTAGC-3′ | 87 | 58 |
|
| GAPDH-R:
5′-GGCATGGACTGTGGTCATGAG-3′ |
|
|
| BSP | M-CLDN3-F:
5′-TYGGTGAAGGTGGGAGGTAG-3′ | 287 | 59 |
|
| M-CLDN3-R:
5′-AAACCRCTAAACCTAACRAAAACTAC-3′ |
|
|
Bisulfite modification and methylation
analysis
Genomic DNA was extracted from paraffin-embedded
tissues of normal tissues, intestinal metaplasia, primary tumors
and metastatic lymph nodes using the DNeasy Blood and Tissue kit
(Qiagen, Germantown, MD, USA) in accordance with the manufacturer's
instructions, and bisulfite modification was performed using the
Epi-Tect Bisulfite kit (Qiagen). The status of claudin-3 promoter
methylation was determined using methylation-specific PCR
(MSP).
Two primer pairs were designed according to the
location of the claudin-3 CpG islands (Sangon, Shanghai, China).
The claudin-3 primer sets used for MSP were designated as
hypermethylated (M), or hypomethylated (U). Sensitivity was
determined using a series dilutions of methylated DNA in
unmethylated DNA: MSP with the claudin-3 primers was able to
reliably detect the 1% methylated standards. Briefly, MSP
amplifications of claudin-3 were performed in a total volume of 25
µl, containing 2 µl DNA template, 1X PCR buffer, 200 µmol/l dNTP,
20 pmol of each primer, 1 U HotStarTaq DNA polymerase (Qiagen) and
2.0 mmol/l MgCl2. The reaction conditions were as
follows: 95°C for 4 min; 95°C for 30 sec, 25 cycles, 58°C for 30
sec and 72°C for 30 sec; and 72°C for 5 min. Electrophoresis on a
2% agarose gel was performed for confirmation. The methylation
status of the CpG islands of the claudin-3 promoter was determined
according to the electrophoresis bands: 125 bp represented
methylated and 133 bp represented unmethylated.
Primers for bisulfite sequencing PCR (BSP) were
designed as shown in Table I.
Amplification of bisulfite-treated DNA was performed in a total
volume of 50 µl, containing 3 µl DNA template, 1X PCR buffer, 200
µM dNTP, 20 pmol of each primer, 4 U Taq DNA polymerase
(Qiagen) and 3.0 mmol/l MgCl2. The PCR conditions were
as follows: 98°C for 4 min; 40 cycles of (94°C for 45 sec, 66°C for
45 sec and 72°C for 1 min); 72°C for 8 min. PCR was confirmed by
electrophoresis on a 2% agarose gel. Purification of PCR products
was performed using QIAquick PCR Purification Kit (Qiagen).
Purified PCR products were cloned to the pUC18T vector and
transformed into competent SK9307 cells. SK9307 cells carrying the
vectors were selected on agar plates containing
ampicillin/Xgal/IPTG, and white colonies were selected and grown in
LB medium. Plasmids containing the target DNA were extracted using
the QIAprep Spin Miniprepkit (Qiagen) and subjected to standard
sequencing analysis with an ABI Prism 3130XL DNA sequencer (Applied
Biosystems; Thermo Fisher Scientific Inc. Waltham, MA, USA).
Statistical analysis
All statistical analyses were performed using SPSS
18.0 (SPSS, Inc., Chicago, IL, USA). Comparisons of quantitative
data were made using analysis of variance (ANOVA) followed by the
Tukey's post hoc test. Comparisons of promoter methylation level
and claudin-3 expression level among the different tissues (normal
gastric tissue, intestinal metaplasia tissue, primary tumor and
metastastic lymph node) were made by nonparametric rank-based
tests. The nonparametric Spearman's rank correlation test was used
to determine the correlation of claudin-3 mRNA (mean ± SD),
promoter methylation (hypermethylation, partial methylation and
hypomethylation) and protein expression levels (low and high
expression) with the clinical parameters (Tables II and III). The relationship between claudin-3
mRNA expression, promoter methylation levels and claudin-3 protein
expression (IHC score) was determined by calculation of Spearman's
rank correlation coefficient (rs). Survival analysis was
carried out using the Kaplan-Meier method, and statistical
comparisons were made using the log-rank test. Univariate and
multivariate analyses of prognostic factors were carried out using
a Cox regression model. Hazard ratios (HRs) and corresponding 95%
confidence intervals (95% CIs) were used to evaluate the
associations between risk factors and overall survival. A two sided
P-value <0.05 was considered as statistically significant.
 | Table II.Association of claudin-3 protein
expression and promoter methylation with various clinical
characteristics of the 122 patients with gastric cancer. |
Table II.
Association of claudin-3 protein
expression and promoter methylation with various clinical
characteristics of the 122 patients with gastric cancer.
|
|
| Claudin-3 protein
expression |
| Claudin-3 promoter
methylation status |
|
|---|
|
|
|
|
|
|
|
|---|
|
| n | Weak (0–3) | Strong (4–9) | P-value | U | M+U | M | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
Male | 84 | 32 | 52 | 0.217 | 41 | 23 | 20 | 0.323 |
|
Female | 38 | 19 | 19 |
| 16 | 8 | 14 |
|
| Age (years) |
|
|
|
|
|
|
|
|
|
≤60 | 61 | 24 | 37 | 0.582 | 24 | 18 | 19 | 0.259 |
|
>60 | 61 | 27 | 34 |
| 33 | 13 | 15 |
|
| Lauren subtype |
|
|
|
|
|
|
|
|
|
Intestinal | 48 | 20 | 28 | 0.078 | 42 | 6 | 0 |
|
|
Mixed | 30 | 20 | 10 |
| 8 | 12 | 10 | <0.001 |
|
Diffuse | 44 | 36 | 8 |
| 7 | 13 | 24 |
|
| Depth of
invasion |
|
|
|
|
|
|
|
|
| T3 | 19 | 5 | 14 | 0.136 | 13 | 4 | 2 | 0.196 |
| T4 | 103 | 46 | 57 |
| 44 | 27 | 32 |
|
| Lymph nodes |
|
|
|
|
|
|
|
|
|
1–3 | 13 | 5 | 8 | 0.146 | 9 | 3 | 1 | 0.163 |
|
4–6 | 34 | 12 | 22 |
| 19 | 7 | 8 |
|
| ≥7 | 75 | 34 | 41 |
| 29 | 21 | 25 |
|
| Tumor size
(cm) |
|
|
|
|
|
|
|
|
| ≤5 | 89 | 29 | 60 | 0.596 | 47 | 21 | 21 | 0.075 |
|
>5 | 33 | 22 | 11 |
| 10 | 10 | 13 |
|
| TNM stage |
|
|
|
|
|
|
|
|
|
IIB | 8 | 3 | 5 | 0.012 | 5 | 1 | 2 | <0.001 |
|
IIIA-IIIB | 85 | 56 | 29 |
| 15 | 23 | 47 |
|
|
IIIC-IV | 29 | 19 | 10 |
| 5 | 7 | 17 |
|
 | Table III.Association of claudin-3 mRNA
expression, protein expression and promoter methylation with
various clinical characteristics of the 50 patients with gastric
cancer. |
Table III.
Association of claudin-3 mRNA
expression, protein expression and promoter methylation with
various clinical characteristics of the 50 patients with gastric
cancer.
|
| Protein
expression | Methylation
status | mRNA
expressiona |
|---|
|
|
|
|
|
|---|
|
| n | Low (≤3) | High (>3) | P-value | U | M+U | M | P-value | Mean ± SD | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
|
|
Male | 31 | 16 | 15 | 0.425 | 10 | 7 | 14 | 0.837 | 2.39±1.283 | 0.957 |
|
Female | 19 | 12 | 7 |
| 7 | 3 | 9 |
| 2.37±1.012 |
|
| Age (years) |
|
|
|
|
|
|
|
|
|
|
|
≤60 | 26 | 14 | 12 | 0.783 | 9 | 5 | 12 | 0.989 | 2.54±1.140 | 0.326 |
|
>60 | 34 | 24 | 10 |
| 18 | 5 | 11 |
| 2.21±1.215 |
|
| Lauren subtype |
|
|
|
|
|
|
|
|
|
|
|
Intestinal | 8 | 1 | 7 | 0.011 | 8 | 0 | 0 | 0.001 | 4.00±0.756 | 0.005 |
|
Mixed | 14 | 7 | 7 |
| 4 | 4 | 6 |
| 2.36±1.336 |
|
|
Diffuse | 28 | 20 | 8 |
| 5 | 6 | 17 |
| 1.93±0.716 |
|
| Depth of
invasion |
|
|
|
|
|
|
|
|
|
|
| T3 | 4 | 1 | 3 | 0.193 | 2 | 1 | 1 | 0.671 | 3.25±0.50 | 0.124 |
| T4 | 46 | 27 | 19 |
| 15 | 9 | 22 |
| 2.30±1.190 |
|
| Lymph nodes |
|
|
|
|
|
|
|
|
|
|
|
4–6 | 9 | 4 | 5 | 0.473 | 3 | 0 | 6 | 0.108 | 2.44±1.163 | 0.454 |
| ≥7 | 41 | 24 | 17 |
| 15 | 9 | 17 |
| 2.11±1.269 |
|
| Tumor size
(cm) |
|
|
|
|
|
|
|
|
|
|
| ≤5 | 34 | 14 | 20 | 0.002 | 14 | 6 | 14 | 0.295 | 2.56±1.160 | 0.118 |
|
>5 | 16 | 14 | 2 |
| 3 | 4 | 9 |
| 2.00±1.155 |
|
| TNM stage |
|
|
|
|
|
|
|
|
|
|
|
IIIA-IIIB | 31 | 14 | 17 | 0.049 | 15 | 8 | 8 | 0.001 | 2.77±1.146 | 0.002 |
|
IIIC-IV | 19 | 14 | 5 |
| 2 | 2 | 15 |
| 1.74±0.933 |
|
Results
Study participants
A total of 122 patients (84 males and 38 females)
with a median age of 61 years (ranging from 33 to 84 years) at
diagnosis were enrolled. The clinicopathological characteristics of
the study participants are presented in Table II.
Claudin-3 protein expression
Representative images of claudin-3 protein
expression in the different types of gastric tissue are presented
in Fig. 1A. The IHC scores are
shown in Fig. 1B. Strong staining
in both the cytoplasm and cell membrane was observed in the
majority of tissues of intestinal metaplasia (112/122, 91.8%) and
primary tumors (71/122, 58.2%) but in the minority of tissues of
metastatic lymph node (37/122, 30.3%) and normal gastric epithelium
(17/122, 13.9%) (Fig. 1C). The
expression level of claudin-3 differed significantly for different
TNM stages (P=0.012). The expression was stronger for stage III
than for more advanced stages (Table
II). Also, the expression seemed to be stronger in the
intestinal type than in the mixed or diffuse types of gastric
cancer, although no statistical significance was obtained (P=0.078;
Table II). No significant
association between claudin-3 protein expression and other
clinicopathological features such as age, sex, tumor size, depth of
invasion and number of metastatic lymph nodes was observed
(Table II).
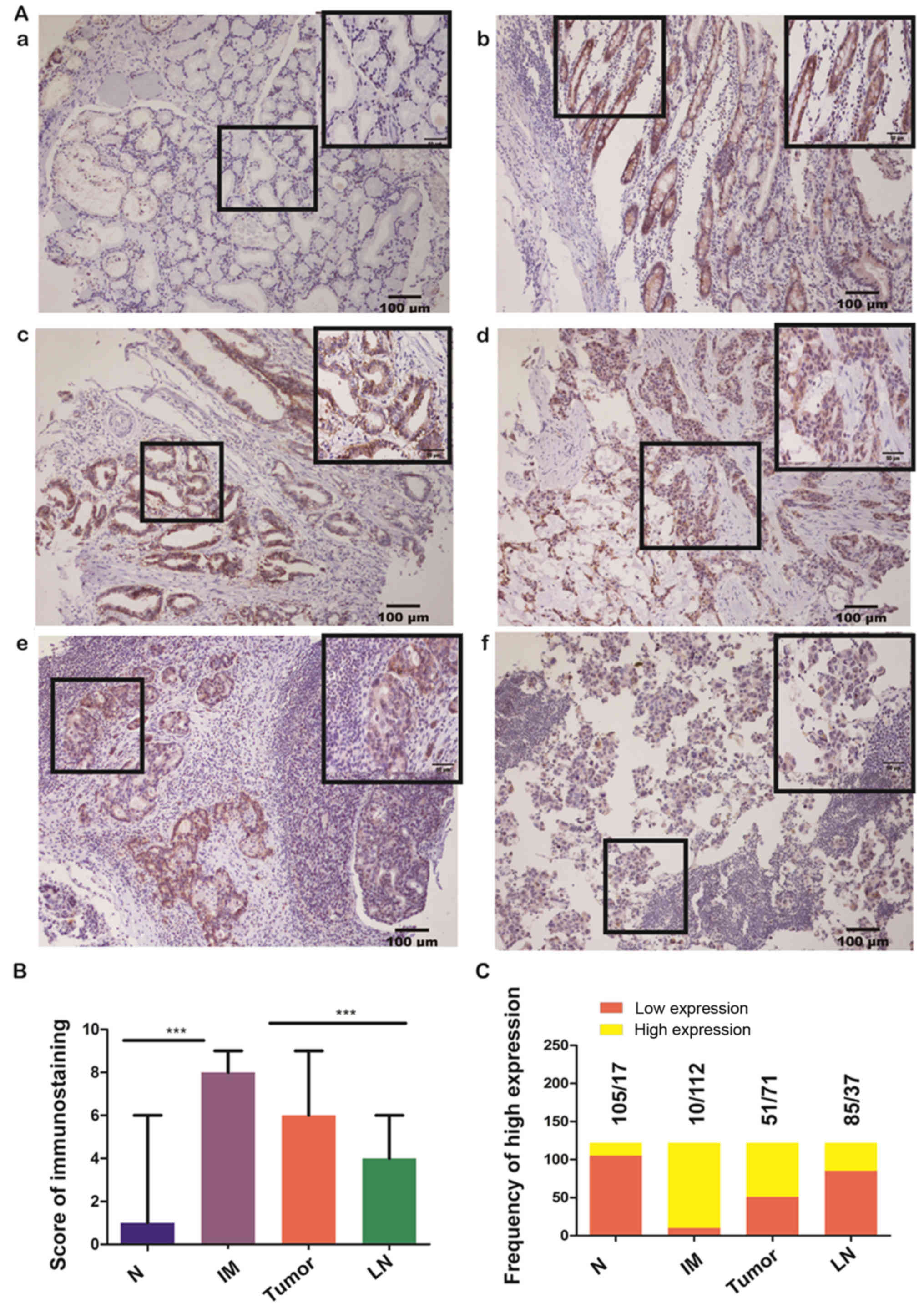 | Figure 1.Claudin-3 protein expression assessed
using immunohistochemistry. (A) Images illustrating
immunohistochemical (IHC)staining of claudin-3 protein in gastric
tissue samples representative of: (a) normal gastric epithelium
(IHC staining score=1, weak expression); (b) intestinal metaplasia
(IHC staining score=9, strong expression); (c) primary tumor,
intestinal type (IHC staining score=7, strong expression); (d)
primary tumor, diffuse type (IHC staining score=4, moderate
expression); (e) lymph node metastasis (IHC staining score=3, weak
expression). The images are representative of data from 122
participants with advanced gastric cancer. Claudin-3 was stained
yellow or brown. The inset in the top right of each figure is a
magnified view of the region highlighted in the box to its left.
(B) IHC staining score for each tissue type was calculated based on
the intensity and area of the staining for claudin-3 (see Materials
and methods for details). Data are presented as the mean ± standard
deviation (n=122 participants). ***P<0.001 (nonparametric test).
(C) Frequency of weak expression (IHC score 0–3) and strong
expression (IHC score 4–9) of claudin-3 in each tissue type (n=122
participants). N, normal gastric epithelium; IM, intestinal
metaplasia; Tumor, primary tumor; LN, lymph node metastasis. |
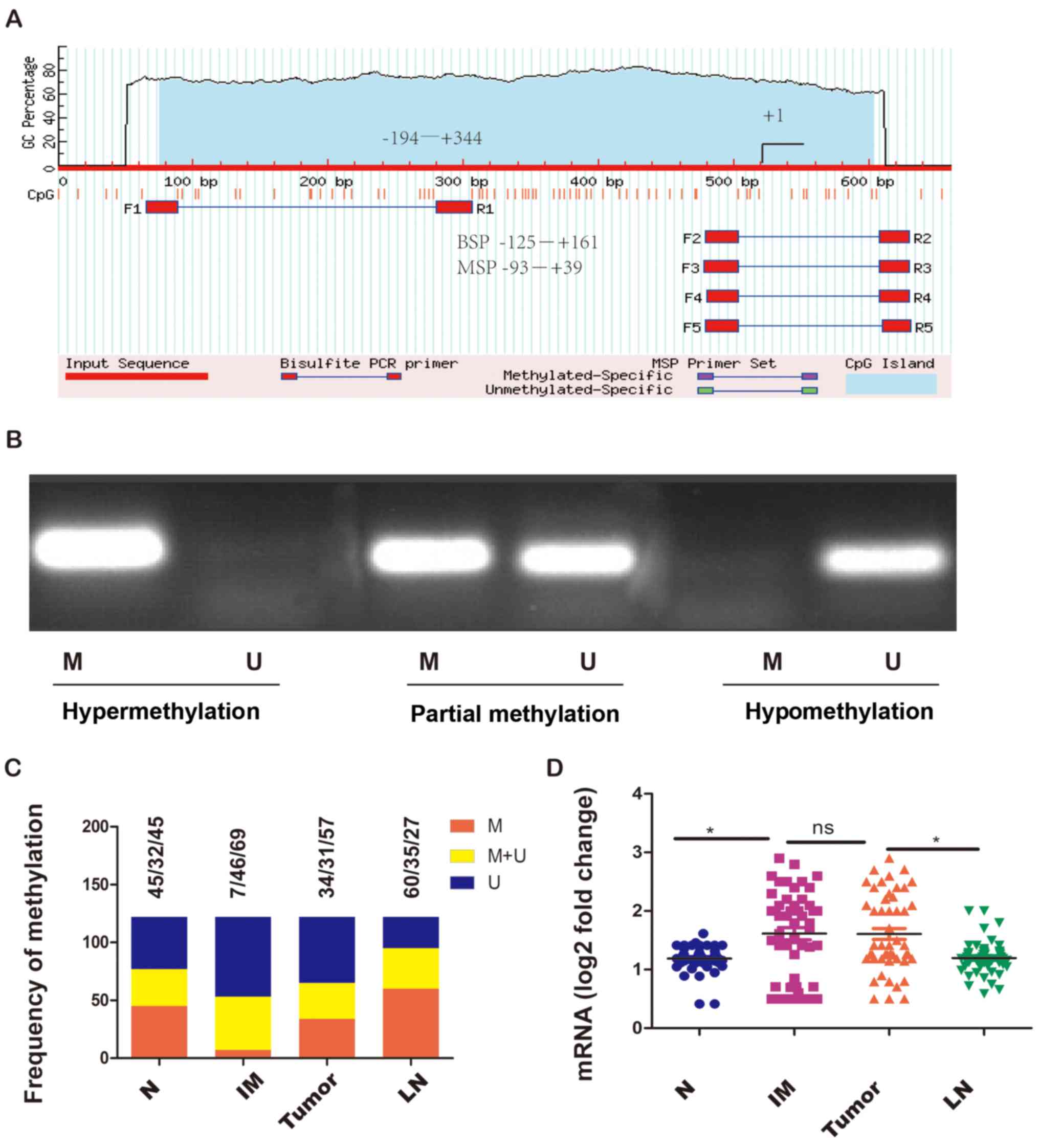
Claudin-3 promoter methylation
The methylation status of the claudin-3 gene
promoter was first detected by MSP (Fig. 2A and B). The hypermethylation rate
for normal gastric epithelium, intestinal metaplasia, primary
tumors and metastatic lymph node were 36.9% (45/122), 5.7% (7/122),
27.9% (34/122) and 49.2% (60/122), respectively (Fig. 2C). There were significant
differences in the methylation status of the claudin-3 promoter
among the different Lauren subtypes and different TNM stages:
Gastric adenocarcinoma of the intestinal subtype and in low stage
(IIB-IIIB) had a lower frequency of hypermethylation (Table II). No significant associations
between the methylation status of the claudin-3 promoter and other
clinicopathological features such as age, sex, tumor size, depth of
invasion and number of metastatic lymph nodes were observed
(Table II).
Claudin-3 mRNA expression levels were detected in
tissue samples from 50 of the 122 participants using real-time
qPCR. Elevated levels of claudin-3 mRNA were observed in intestinal
metaplasia and primary tumors compared with that observed in normal
gastric epithelium and metastatic lymph node (Fig. 2D), which was consistent with the
protein expression data described above. The data from these 50
participants also revealed significant subgroup differences in
claudin-3 mRNA expression, claudin-3 protein expression and
claudin-3 promoter methylation between Lauren subtypes and between
TNM stages. Gastric cancer tissue of the intestinal subtype
exhibited higher mRNA and protein expression of claudin-3 and a
lower frequency of promoter hypermethylation than diffuse and mixed
subtypes (Table III). Similarly,
elevated mRNA and protein expression of claudin-3 and a lower
incidence of promoter hypermethylation were observed in stage
IIIA-IIIB than in stage IIIC-IV (Table III). There were no significant
associations between claudin-3 mRNA expression and other
clinicopathological features such as age, sex, tumor size, depth of
invasion and number of metastatic lymph nodes (Table III).
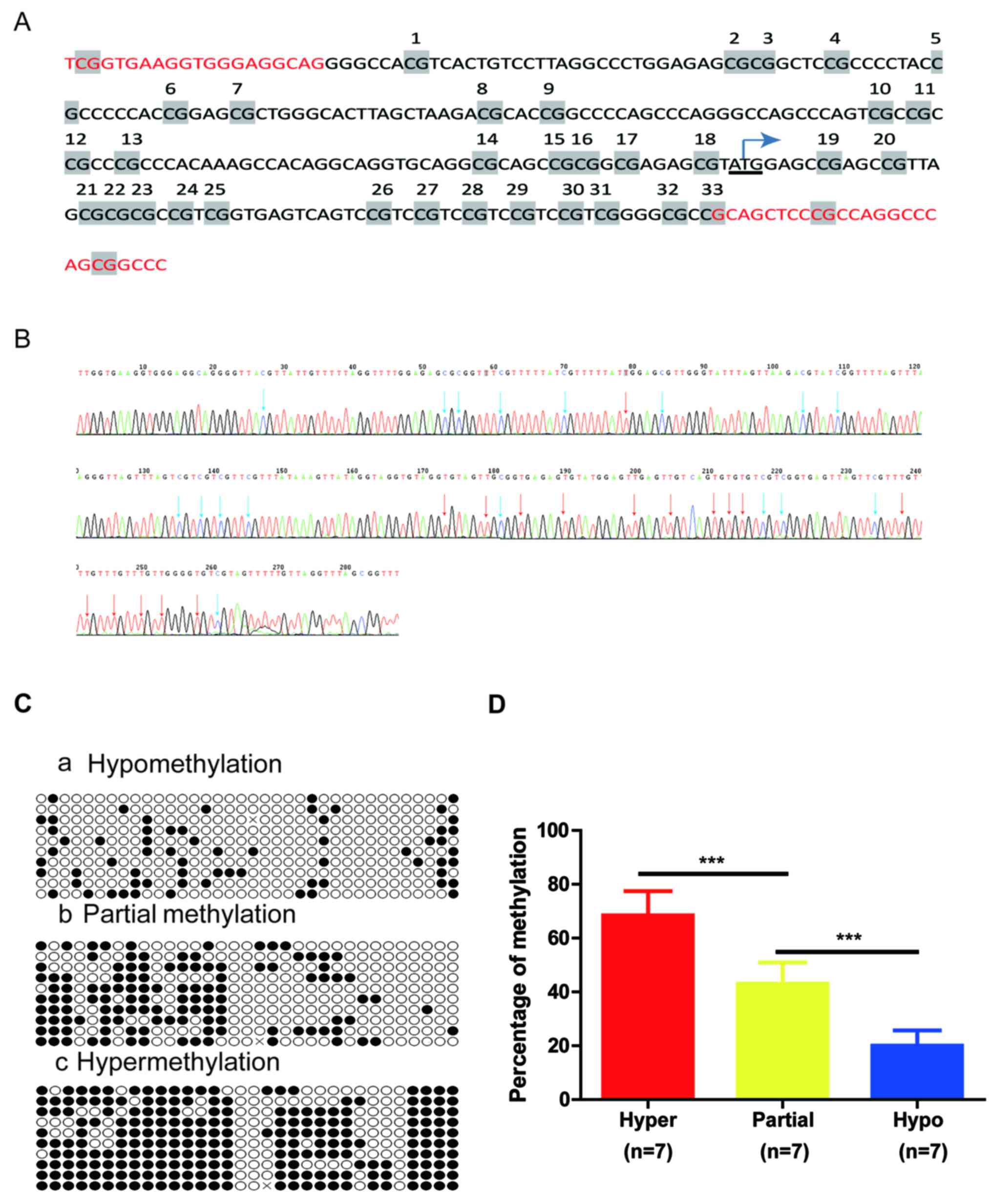
To validate the claudin-3 promoter methylation
status determined by methylation-specific PCR, 21 samples from
primary tumors with different methylation levels (hypomethylation,
partial methylation and hypermethylation) were further examined by
BSP using the primers indicated in Table I. The 33 CpG sites spanned from
nucleotide −107 to + 242 (NCBI accession: NC_000007) (Fig. 3A and B). Sequence analysis was made
using Quantification Tool for Methylation Analysis (http://quma.cdb.riken.jp/). In Fig. 3C, each CpG site is represented as a
circle and each row represents one cloned PCR product (open circle,
unmethylated; filled circle, methylated). The percentage CpG
methylation per sequence was quantified as the percentage of DNA
methylation relative to the total (%MET). As shown in Fig. 3D, the percentage methylation ranged
from 57.6 to 81.8% (68.5±5.5%, n=7) in cases with hypermethylation,
30.3 to 54.5% (43.0±7.5%, n=7) in cases with partial methylation
and 9.1 to 27.2% (20.0±5.5%, n=7) in cases with hypomethylation,
with significant differences among groups (P<0.001). This was
consistent with the MSP results.
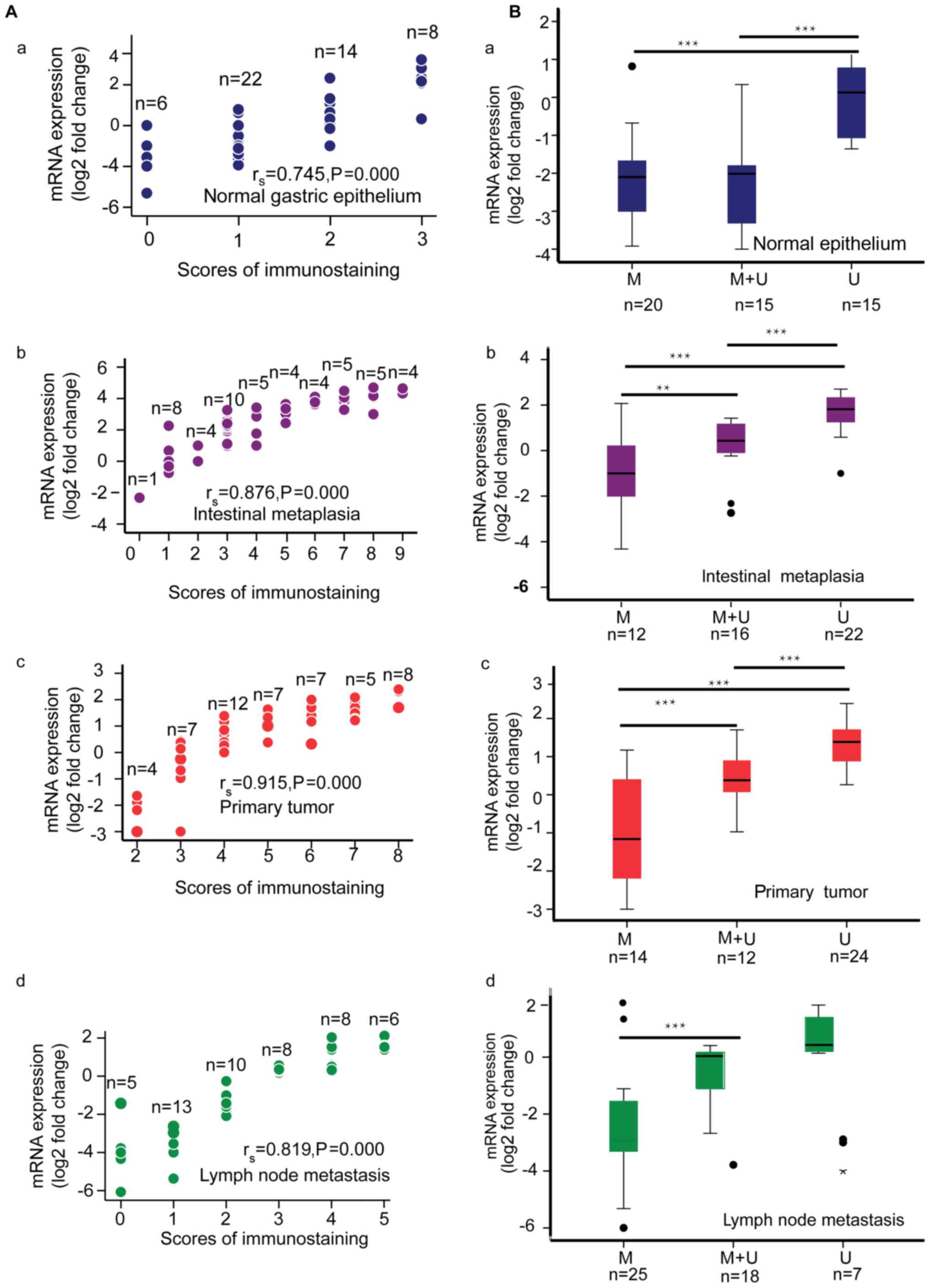
Scatterplots illustrating the correlation between
claudin-3 mRNA expression and claudin-3 protein expression (IHC
score) are shown in Fig. 4A.
Claudin-3 mRNA and protein expression were significantly positively
correlated for normal gastric epithelium (rs=0.745,
P<0.001), intestinal metaplasia (rs=0.876,
P<0.001), primary gastric adenocarcinoma (rs 0.915,
P<0.001) and metastatic lymph node (rs=0.819,
P<0.001). Boxplots comparing claudin-3 mRNA expression between
hypermethylated, partially methylated and hypomethylated claudin-3
promoter groups are presented in Fig.
4B. For normal gastric epithelium, promoter hypermethylation or
partial methylation was associated with lower claudin-3 mRNA
expression. For intestinal metaplasia and primary tumors, claudin-3
mRNA expression progressively decreased from hypomethylated to
partially methylated to hypermethylatedgroup. For metastatic lymph
node, claudin-3 mRNA expression was lower in the hypermethylated
group than in the other two groups.
Univariate and multivariate analyses
of factors associated with overall survival
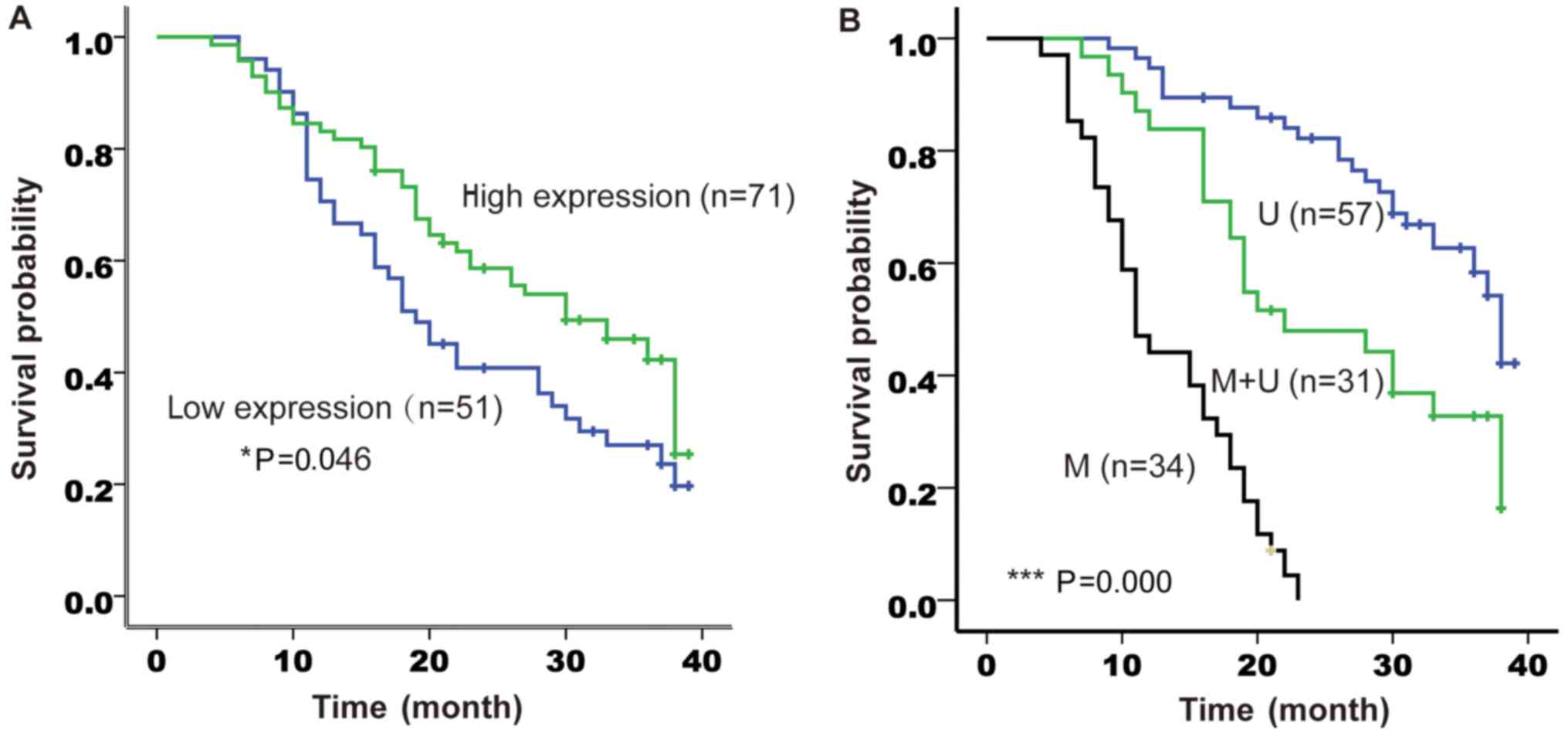
The 122 participants were followed up for a median
of 19 months (range, 4–41 months). A total of 114 patients (93.4%)
succumbed to gastric cancer during the follow-up period. The median
survival time was 30 months (range, 21.0–39.0 months) in the high
claudin-3 expression group and 19 months (range, 14.7–23.3 months)
in the low claudin-3 expression group (P<0.05, log-rank test;
Fig. 5A). The median survival time
was 38 months (range, 35.7–40.3 months) in the hypomethylated
promoter group, 22 months (range, 13.2–30.8 months) in the
partially methylated promoter group and 11 months (range, 8.7–13.3
months) in the methylated promoter group (P<0.001, log-rank
test; Fig. 5B). Univariate
regression analysis demonstrated that Lauren classification, tumor
size, TNM stage, number of metastatic lymph nodes and the status of
claudin-3 promoter methylation were significantly associated with
overall survival (Table IV).
Multivariate analysis suggested that only the status of claudin-3
promoter methylation (HR, 5.67; 95% CI, 2.27–14.17) was an
independent predictor of overall survival (Table IV). However, claudin-3 protein
expression level was not associated with overall survival in both
the univariate and multivariate regression analyses (Table IV).
 | Table IV.Univariate and multivariate
regression analyses of factors associated with overall survival in
patients with gastric cancer. |
Table IV.
Univariate and multivariate
regression analyses of factors associated with overall survival in
patients with gastric cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) |
|
|
|
|
|
<60 | 1 |
|
|
|
|
≥60 | 1.021
(0.657–1.587) | 0.927 |
|
|
| Sex |
|
|
|
|
|
Female | 1 |
|
|
|
|
Male | 1.080
(0.802–2.055) | 0.299 |
|
|
| Tumor size
(cm) |
|
|
|
|
| ≤5 | 1 |
| 1 |
|
|
>5 | 1.654
(1.027–2.664) | 0.038 | 1.102
(0.626–1.080) | 0.704 |
| Depth of
invasion |
|
|
|
|
|
T2-T3 | 1 |
|
|
|
| T4 | 1.631
(0.839–3.172) | 0.149 |
|
|
| Lauren subtype |
|
|
|
|
|
Intestinal | 1 |
| 1 |
|
|
Mixed | 2.331
(1.232–4.407) | 0.009 | 1.066
(0.416–2.731) | 0.894 |
|
Diffuse | 5.585
(3.178–9.815) | <0.001 | 2.376
(0.931–6.064) | 0.070 |
| Lymph nodes
involved |
|
|
|
|
|
1–6 | 1 |
| 1 |
|
| ≥7 | 2.388
(1.031–5.531) | 0.040 | 2.245
(0.927–5.435) | 0.073 |
| TNM stage |
|
|
|
|
|
IIB-IIIB | 1 |
| 1 |
|
|
IIIC-IV | 3.905
(2.399–6.358) | <0.001 | 1.536
(0.879–2.684) | 0.132 |
| Promoter
methylation |
|
|
|
|
| M | 1 |
|
|
|
|
M+U | 2.340
(1.304–4.202) | 0.004 | 1.553
(0.647–3.728) | 0.325 |
| U | 10.89
(5.868–20.23) | <0.001 | 5.674
(2.271–14.17) | <0.001 |
| Claudin-3
expression |
|
|
|
|
|
Low | 1 |
|
|
|
|
High | 0.644
(0.413–1.005) | 0.053 |
|
|
Discussion
Gastric adenocarcinoma is a severe disease that is
progressively driven by genetic and epigenetic changes, and DNA
methylation contributes to the pathogenesis and progression of
gastric adenocarcinoma (24). In
the present study, claudin-3 protein and mRNA expression and
promoter methylation profile were compared between normal gastric
epithelium, intestinal metaplasia, primary tumors and metastatic
lymph nodes. Intestinal metaplasia increases the risk of the
intestinal-type of gastric cancer, and previous studies have
demonstrated a higher expression of claudin-3 both in intestinal
metaplasia and intestinal-type gastric adenocarcinoma (12), although the underlying regulatory
mechanisms remain unclear. Here we found that normal gastric
epithelium showed promoter hypermethylation and lower claudin-3
expression compared with the paired samples of intestinal
metaplasia or primary tumors. Furthermore, claudin-3 mRNA
expression was strongly positively correlated with claudin-3
protein expression and was negatively associated with promoter
methylation status. This suggests that hypomethylation of the
claudin-3 promoter in intestinal metaplasia or primary tumor may be
involved in the genesis of intestinal-type gastric adenocarcinoma
through regulation of the transcription of the claudin-3 gene.
These findings are consistent with previous reports that claudin-3
is more highly expressed in tissues of intestinal metaplasia than
in tissues of normal gastric mucosa (12,13).
Notably, an inverse correlation between promoter methylation level
and claudin expression level in gastric cancer has also been
observed for claudin-4 (7) and
claudin-11 (21). Furthermore, the
association of claudin-3 promoter hypermethylation with
downregulation of claudin-3 expression has been reported previously
in hepatocellular cancer cells (20).
An important finding of our study was that claudin-3
expression and promoter methylation differed between the various
Lauren subtypes of gastric adenocarcinoma. Notably, diffuse-type
adenocarcinoma showed lower expression of claudin-3 and
hypermethylation of its promoter as compared with the
intestinal-type. This implies that claudin-3 expression and
promoter methylation status are related to tumor heterogeneity in
gastric adenocarcinoma. Diffuse-type adenocarcinoma is more
invasive and has greater metastatic potential than intestinal-type
adenocarcinoma (25), which raises
the possibility that promoter hypermethylation and decreased
expression of claudin-3 may be a marker of poorly differentiated
phenotype and higher metastatic potential of gastric cancer.
Indeed, it was noted that promoter hypermethylation and reduced
claudin-3 expression were also positively associated with higher
TNM stage. Our data are in agreement with several previous
investigations. For example, downregulation of claudin-3 in gastric
cancer has been found to be associated with increased proliferative
potential, higher grade of malignancy and advanced stage (14–16).
Furthermore, similar to our findings, hypermethylation of the
claudin-11 promoter and downregulation of claudin-11 were observed
to be related to an increased invasive potential of gastric cancer
cells (21). Another observation in
our study was that metastatic lymph node tissue showed decreased
claudin-3 expression and increased promoter methylation than
primary tumor tissues. This suggests that gastric cancer cells
showing enhanced claudin-3 promoter methylation and reduced
claudin-3 expression may have a greater potential to metastasize to
the lymph nodes.
A major finding of this study was that claudin-3
promoter methylation and reduced claudin-3 expression were both
significantly associated with shorter survival. Indeed, promoter
methylation status was an independent predictor of survival in
multivariate analysis. This finding suggests that enhanced
claudin-3 promoter methylation and reduced claudin-3 expression may
be associated with a poorer prognosis. Our observations in patients
with gastric cancer are consistent with those of a previous study
in hepatocellular carcinoma, which showed that downregulation of
claudin-3 expression was an independent predictor of poorer
survival (20). Currently, there
are few defined biomarkers with prognostic and diagnostic value for
gastric tumors. Clinically, HER-2 amplification or p16
hypermethylation are valuable for the prediction of therapeutic
response, and CDH1 gene methylation pattern can be detected in
peritoneal fluid or serum to predict tumor recurrence and
metastasis (26). In our
preliminary study, claudin-3 promoter methylation in primary tumors
proved to be an alternative biomarker to predict the prognosis of
advanced gastric cancer. In addition, the reversible methylation
pattern of the claudin-3 promoter has the potential to be used as a
non-invasive predictable biomarker similar to the CDH1 gene and may
be a future therapeutic target for advanced gastric
adenocarcinoma.
Our MSP results also showed the presence of promoter
partial methylation, which has been suggested to play a role during
carcinogenesis (27). According to
the correlation analysis, partial methylation of the claudin-3
promoter was generally associated with claudin-3 mRNA expression
levels. Thus, partial methylation of DNA promoter regions may also
induce gene transcription.
However, the study has some limitations. Firstly,
there are tissue-specific intra and inter-genic differences in
claudin-3 promoter methylation and expression, thus it is
impossible to definitively associate claudin-3 methylation and
expression profiles with the initiation or progression of gastric
adenocarcinoma (28). However, our
results suggest that claudin-3 expression level and promoter
methylation status may be relevant to tumor initiation and
progression. Secondly, heterogeneity at both the protein level and
epigenetic level is an important feature of a tumor, and the sample
size in our study was likely too small to accurately determine
differential expression of claudin-3 among the various tissue
types. Therefore, additional experiments in a larger series are
needed. Thirdly, the possible roles of other members of the claudin
family were not investigated. Although the claudin family members
share similar structure and function, claudin-3 may not be
representative of the other family members in terms of differential
expression between tissue types in gastric cancer. Further analysis
of other claudin family members may reveal novel and interesting
findings. Fourthly, BSP was too laborious to further confirm all
the methylation statuses in the different tissue types. Sun and
colleagues have developed a partial methylation pipeline for
identifying partial methylation patterns in different samples by
next generation sequencing (29),
and such a technique could be used for further tests of the
claudin-3 partial methylation status.
In conclusion, our results demonstrated dynamic
change in the claudin-3 promoter methylation status and expression
profile among different loci in patients with gastric
adenocarcinoma. Although we need to extend our findings to a larger
series, our results suggest that promoter hypomethylation and
higher claudin-3 expression contribute to the initiation of
intestinal-type gastric adenocarcinoma. On the contrary, promoter
hypermethylation and reduced expression of claudin-3 may contribute
to the progression of diffuse-type gastric adenocarcinoma, which is
associated with poor prognosis.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant
from the Natural Science Foundation of China (no. 81101893), the
Construction Plan of High Level Hospital in Fujian Province and
Young and Middle-Aged Scientists Research of the First Affiliated
Hospital of Fujian Medical College (no. JGG201306).
Availabilty of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZS and ZZZ conceived and designed the study. ZZZ,
YWX, CCQ and CYP performed the experiments. ZZZ wrote the paper.
ZS, CCQ and CLY revised and edited the manuscript. All authors read
and approved the manuscript and agreed to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The study protocol was approved by and informed
written consent was obtained from each participant before
enrollment in the study.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LN
|
lymph node
|
|
IHC
|
immunohistochemistry
|
|
H&E
|
hematoxylin and eosin
|
|
RT-PCR
|
real-time reverse-transcription
polymerase chain reaction
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
ANOVA
|
analysis of variance
|
|
BSP
|
bisulfite sequencing PCR
|
|
MSP
|
methylation-specific PCR
|
References
|
1
|
Catalano V, Labianca R, Beretta GD, Gatta
G, de Braud F and Van Cutsem E: Gastric cancer. Gastric Rev Oncol
Hematol. 71:127–164. 2009. View Article : Google Scholar
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maruyama K: The most important prognostic
factors for gastric cancer patients: A study using univariate and
multivariate analyses. Scand J Gastroenterol. 22:63–68. 1987.
View Article : Google Scholar
|
|
5
|
Huang L, Wu RL and Xu AM:
Epithelial-mesenchymal transition in gastric cancer. Am J Transl
Res. 7:2141–2158. 2015.PubMed/NCBI
|
|
6
|
Turksen K and Troy TC: Junctions gone bad:
Claudins and loss of the barrier in cancer. Biochim Biophys Acta.
1816:73–79. 2011.PubMed/NCBI
|
|
7
|
Kwon MJ, Kim SH, Jeong HM, Jung HS, Kim
SS, Lee JE, Gye MC, Erkin OC, Koh SS, Choi YL, et al: Claudin-4
overexpression is associated with epigenetic derepression in
gastric carcinoma. Lab Invest. 91:1652–1667. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zavala-Zendejas VE, Torres-Martinez AC,
Salas-Morales B, Fortoul TI, Montano LF and Rendon-Huerta EP:
Claudin-6, 7, or 9 overexpression in the human gastric
adenocarcinoma cell line AGS increases its invasiveness, migration,
and proliferation rate. Cancer Invest. 29:1–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sanada Y, Oue N, Mitani Y, Yoshida K,
Nakayama H and Yasui W: Down-regulation of the claudin-18 gene,
identified through serial analysis of gene expression data
analysis, in gastric cancer with an intestinal phenotype. J Pathol.
208:633–642. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu YL, Zhang S, Wang GR and Chen YP:
Expression transformation of claudin-1 in the process of gastric
adenocarcinoma invasion. World J Gastroenterol. 14:4943–4948. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang J, Zhang L, He C, Qu Y, Li J, Zhang
J, Du T, Chen X, Yu Y and Liu B: Claudin-1 enhances tumor
proliferation and metastasis by regulating cell anoikis in gastric
cancer. Oncotarget. 6:1652–1665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Satake S, Semba S, Matsuda Y, Usami Y,
Chiba H, Sawada N, Kasuga M and Yokozaki H: Cdx2 transcription
factor regulates claudin-3 and claudin-4 expression during
intestinal differentiation of gastric carcinoma. Pathol Int.
58:156–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang H and Yang X: The expression patterns
of tight junction protein claudin-1, −3, and −4 in human gastric
neoplasms and adjacent non-neoplastic tissues. Int J Clin Exp
Pathol. 8:881–887. 2015.PubMed/NCBI
|
|
14
|
Jung H, Jun KH, Jung JH, Chin HM and Park
WB: The expression of claudin-1, claudin-2, claudin-3, and
claudin-4 in gastric cancer tissue. J Surg Res. 167:e185–e191.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okugawa T, Oshima T, Chen X, Hori K,
Tomita T, Fukui H, Watari J, Matsumoto T and Miwa H:
Down-regulation of claudin-3 is associated with proliferative
potential in early gastric cancers. Dig Dis Sci. 57:1562–1567.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsuda Y, Semba S, Ueda J, Fuku T, Hasuo
T, Chiba H, Sawada N, Kuroda Y and Yokozaki H: Gastric and
intestinal claudin expression at the invasive front of gastric
carcinoma. Cancer Sci. 98:1014–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Auclair G and Weber M: Mechanisms of DNA
methylation and demethylation in mammals. Biochimie. 94:2202–2211.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hahn-Strömberg V, Askari S, Ahmad A,
Befekadu R and Nilsson TK: Expression of claudin 1, claudin 4, and
claudin 7 in colorectal cancer and its relation with CLDN DNA
methylation patterns. Tumour Biol. 39:10104283176975692017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Honda H, Pazin MJ, D'Souza T, Ji H and
Morin PJ: Regulation of the CLDN3 gene in ovarian cancer cells.
Cancer Biol Ther. 6:1733–1742. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang L, Yang YD, Fu L, Xu W, Liu D, Liang
Q, Zhang X, Xu L, Guan XY, Wu B, et al: CLDN3 inhibits cancer
aggressiveness via Wnt-EMT signaling and is a potential prognostic
biomarker for hepatocellular carcinoma. Oncotarget. 5:7663–7676.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agarwal R, Mori Y, Cheng Y, Jin Z, Olaru
AV, Hamilton JP, David S, Selaru FM, Yang J, Abraham JM, et al:
Silencing of claudin-11 is associated with increased invasiveness
of gastric cancer cells. PLoS One. 4:e80022009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu S, Singh K, Mangray S, Tavares R, Noble
L, Resnick MB and Yakirevich E: Claudin expression in high-grade
invasive ductal carcinoma of the breast: Correlation with the
molecular subtype. Mod Pathol. 26:485–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
VanGuilder HD, Vrana KE and Freeman WM:
Twenty-five years of quantitative PCR for gene expression analysis.
Biotechniques. 44:619–626. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sonohara F, Inokawa Y, Hayashi M, Kodera Y
and Nomoto S: Epigenetic modulation associated with carcinogenesis
and prognosis of human gastric cancer. Oncol Lett. 13:3363–3368.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng H, Takahashi H, Murai Y, Cui Z,
Nomoto K, Miwa S, Tsuneyama K and Takano Y: Pathobiological
characteristics of intestinal and diffuse-type gastric carcinoma in
Japan: An immunostaining study on the tissue microarray. J Clin
Pathol. 60:273–277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pinheiro Ddo R, Ferreira WA, Barros MB,
Araujo MD, Rodrigues-Antunes S and Borges Bdo N: Perspectives on
new biomarkers in gastric cancer: Diagnostic and prognostic
applications. World J Gastroenterol. 20:11574–11585. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ehrlich M and Lacey M: DNA hypomethylation
and hemimethylation in cancer. Adv Exp Med Biol. 754:31–56. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mroz EA and Rocco JW: The challenges of
tumor genetic diversity. Cancer. 123:917–927. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun S and Li P: HMPL: A pipeline for
identifying hemimethylation patterns by comparing two samples.
Cancer Inform. 14 Suppl 2:S235–S245. 2015.
|