Introduction
Esophageal cancer, is the fourth leading cause of
cancer-related deaths, causing ~220.000 deaths worldwide per year,
with an annual mortality rate almost matching its incidence
(1). Early diagnosis of esophageal
cancer has been proved to be difficult, along with the unavailable
therapeutic responses (2). At the
early stages of esophageal cancer, certain changes in cell-cell and
cell-matrix interactions lead to abnormal cell behavior and result
in the invasive and malignant cell transformation. In the late
stages of tumor development, metastasis is the predominant
complication rendering esophageal cancer difficult to control,
which has been identified as the main cause of high mortality
(3). Therefore, it is urgently
required to elucidate the underlying mechanism of disease
progression and metastasis.
Normal cells constantly monitor the cellular
microenvironment. They are equipped with an inherent molecular
defense to detect changes and then block several oncogenic
signaling cascades in the internal and external cell environment
(4). During embryonic development,
epithelial cells escape the structural constraints imposed by
tissue architecture and adopt a phenotype more amenable to cell
movement, which has been known as epithelial-mesenchymal transition
(EMT). In addition, the progression of carcinomas to invasive and
metastatic disease may involve localized occurrences of EMT
(5). Research has demonstrated that
the Notch signaling pathway participated in the invasion and
metastasis of esophageal carcinoma through EMT.
The Notch pathway plays an important role in normal
embryonic development, in maintaining the characteristics of cancer
stem cells and in cell metastasis (6). DLL4 belongs to the Notch protein
family, along with DLL1, DLL3, Jagged 1 (JAG1) and JAG2. In recent
years, Notch receptor ligands have been frequently reported to be
involved in tumor proliferation and metastasis. Specifically, DLL1
promoted the metastasis of melanoma by regulating the adhesion
molecules (7) and Jag1 played an
important role in bone metastasis of breast cancer (8). Breast cancer cells with high Jag1
expression directly affected osteoblasts and osteoclasts,
increasing the secretion of IL6 and TGF-β and finally promoting the
proliferation and metastasis of cancer cells (8,9).
Similarly, Jag2 promoted cancer metastasis by regulating the Notch
signaling pathway in several types of cancer, such as pancreatic,
bladder, breast and lung cancer (10–13).
Conversely, the role of DLL4 in normal vascular development was
first established by Reinacher-Schick et al (18).
Previous studies, have mainly focused on the
effectiveness of DLL4 in regulating angiogenesis. However, the role
that DLL4 plays in tumor metastasis has not been fully explored. In
2014, Huang et al (14)
reported that the overexpression of DLL4 promoted the migration and
invasion of renal cell carcinoma by activating Notch signaling
cascades, which promoted further studies of DLL4 in regulating
tumor metastasis. Similarly, evidence indicated that DLL4 was
highly expressed in breast cancer patients and furthermore, DLL4
expression was also positively related to lymphatic metastasis
(15). Additionally, by detecting
DLL4 plasma levels, total dll4+ breast cancer
cell and vasculature activity can be reliably estimated, which
indicated that DLL4 could be regarded as a novel target in tumor
therapeutic management (16).
In the present study, in order to elucidate the
mechanism which DLL4 is involved in cell invasion and metastasis in
esophagus cancer, initially, the dll4 gene was stably
silenced in the Eca109 cells, then the expression of DLL4 in
esophagus cancer and adjacent non-cancerous tissues was detected
and further analysis of its correlation in vitro. Notably,
we observed that downregulated DLL4 suppressed the expression of
p-Akt, and subsequently, led to the induction of migration and
invasion, as well as apoptosis of esophagus cancer cells. Stable
silencing of dll4 also reduced esophageal tumor growth and
extended survival in xenograft animal models by affecting the
phosphorylation of Akt. These findings associated DLL4 with
metastasis in esophagus cancer and also provided a promising target
for treating late stage patients with esophagus cancer.
Materials and methods
Cell culture
293T cells and Eca109 cells were purchased from the
Shanghai Institute of Cell Biology (Shanghai, China). 293T and
Eca109 cells were separately maintained in Dulbecco's modified
Eagle's medium (DMEM) and RPMI-1640 (Invitrogen, Carlsbad, CA, USA)
with 10% fetal bovine serum (FBS) and 100 U/ml
penicillin-streptomycin solution at 37°C in a humidified atmosphere
of 5% CO2.
dll4 shRNA construction and lentiviral
production
Three target sequences for dll4 mRNA were
chosen according to the RNAi Consortium (TRC) shRNA Library. The
dll4 shRNA single-strand oligonucleotides are listed as
follows: shRNA1 forward,
5′-CCGGCTCTCCAACTGCCCTTCAATTCTCGAGAATTGAAGGGCAGTTGGAGAGTTTTTG-3′
and reverse,
5′-AATTCAAAAACTCTCCAACTGCCCTTCAATTCTCGAGAATTGAAGGGCAGTTGGAGAG-3′;
shRNA2 forward,
5′-CCGGGTGTCGGATATCAGCGATATGCTCGAGCATATCGCTGATATCCGACACTTTTTG-3′
and reverse,
5′-AATTCAAAAAGTGTCGGATATCAGCGATATGCTCGAGCATATCGCTGATATCCGACAC-3′;
shRNA3 forward,
5′-CCGGACCACACATTGGACTATAATCCTCGAGGATTATAGTCCAATGTGTGGTTTTTTG-3′
and reverse,
5′-AATTCAAAAAACCACACATTGGACTATAATCCTCGAGGATTATAGTCCAATGTGTGGT-3′.
Additionally, shRNA without homology to dll4 gene and other
genes was selected as the negative control (shScramble), and its
sequence was as follows: shScramble forward,
5′-CCGGACTCATCTATTCGTCCACTCCCTCGAGGGAGTGGACGAATAGATGAGTTTTTTG-3′
and reverse,
5′-AATTCAAAAAACTCATCTATTCGTCCACTCCCTCGAGGGAGTGGACGAATAGATGAGT-3′.
Two single-stranded, complementary oligonucleotides
containing the target sequences were chemically synthesized by
Sangon Biotech (Shanghai, China) and annealed. The double-stranded
oligonucleotides were then inserted into pLKO lentiviral vector
between the AgeI and EcoRI restriction sites [New
England Biolabs (NEB), Ipswich, MA, USA]. pLKO lentivirus vector
contains CMV promoter-driven enhanced green fluorescent protein
(eGFP) reporter gene and puromycin selectable marker. After
ligation, plasmid was transformed into E. coli DH5α
competent cells (maintained by our own laboratory) for plasmid
amplification. The plasmids from positive colonies were identified
by XhoI (NEB) digestion and confirmed by DNA sequencing
(Boshang Biotech, Hangzhou, China).
Recombinant lentiviruses were produced by
co-transfecting with three combinant lentiviral vectors, Δ8.91 and
pVSVG (10:10:1, mass ratio) in 293T cells and the transfection
reagent X-tremeGENE™ HP DNA (Roche Diagnostics, Indianapolis, IN,
USA) was used to facilitate the transfection. The culture
supernatants containing the lentiviral particles were harvested at
24, 48 and 72 h after transfection, mixed and centrifuged at 10,000
× g at 4°C for 10–15 min to discard cell debris. Eca109 cells were
subcultured at a density of 1×106 cells/6 cm dish. The
cells were divided into 5 groups: the control group (infected with
vehicle plasmids), the vector group (infected with shScramble
lentiviral vector) and three dll4-shRNA groups (infected
with 3 different shRNA sequences designed for dll4).
Subsequently, the Eca109 cells were infected with the same titer
virus for 3 days continuously, followed by screening with 2 µg/ml
puromycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The
cells were maintained and allowed to grow for 7–9 days, changed to
fresh medium without puromycin, and then passaged for the following
assays.
Western blotting
After being washed with cold PBS for 3 times, the
cells were lysed in RIPA buffer which composed of 0.6 M Tris-HCl
(pH 6.8), 10% SDS and protease inhibitor cocktail. Cell lysates
were collected and centrifuged at 10,000 × g for 15 min at 4°C. The
supernatants were transferred, mixed with 2X sampling buffer and
boiled for 10 min. Following the BCA assay, protein was separated
by SDS-polyacrylamide gel electrophoresis, and then transferred to
a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories,
Hercules, CA, USA). The membrane was blocked with 5% non-fat milk
(Nestlé S.A, Vevey, Vaud, Switzerland) in TBS-T (10 mM Tris-Cl, pH
7.5, 100 mM NaCl and 1% Tween-20) at room temperature for 1 h.
Subsequently, the membranes were incubated with primary antibodies
such as DLL4 (1:1,000; cat. no. ab7280; Abcam, Cambridge, MA, USA),
AKT (1:1,000; cat. no. 9272S; Cell Signaling Technology, Inc.
Danvers, MA, USA), p-AKT (1:1,000; cat. no. 9271S; Cell Signaling
Technology), E-cadherin (1:500; cat. no. 743712; BD Biosciences,
Franklin Lakes, NJ, USA) and β-actin (1:1,000; cat. no. 4767; Cell
Signaling Technology) at 4°C overnight. The membrane was washed by
TBS-T three times (3×10 min), and then incubated with HRP-labeled
secondary antibody (1:2,000; cat. no. 7074s; Cell Signaling
Technology) for 1 h at room temperature. After being washed again
for three times (3×10 min), the blots were subjected to
chemiluminescence using an ECL kit (cat. no. 6883s; Cell Signaling
Technology) and signals were recorded under the C-DiGit Western
Blot Scanner (LI-COR Biosciences, Lincoln, NE, USA).
DAPI staining
The cells were plated in a 6-well-dish with
coverslip to perform the DAPI staining. Twenty-four hours later,
the cells were washed by 1X PBS for 3 times and stained with DAPI
(1 µg/ml, dissolved with 1X PBS) for 10 sec, then fixed with 4%
paraformaldehyde. The coverslips were washed by 1X PBS for another
3 times, and then were observed under a fluorescence
microscope.
Annexin V-FITC/PI double-staining
The Annexin V-FITC/PI double-staining was used to
detect cell apoptosis. The Eca109 cells treated with shScramble or
dll4-shRNA1 for 48 h were collected, washed twice with PBS,
and then suspended with 1X binding buffer to adjust the cell
concentration to 1×108/ml. Samples were treated
according to the Annexin V/PI apoptosis detection kit and analyzed
by flow cytometry. Cells (10,000) were analyzed each time. The
experiment was repeated three times, and representative data from
one experiment were presented.
Scratch wound healing assay
The migration ability of the Eca109 cells was
assessed by scratch assay as follows: when cells grew to 100%
confluency on the 6-well plate, straight scratches were made by a
sterile 10 µl pipette tip. The medium was discarded and cells were
washed once with PBS to remove the floating cells. For minimizing
the interference of cell proliferation, 10% FBS complete medium was
replaced by RPMI-1640 without FBS. After 24-h incubation, the plate
was removed from the incubator and images were captured under the
microscope (Olympus IX71 inverted system microscope; Olympus Corp.,
Tokyo, Japan). The average wound closure rate was calculated using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA). All the experiments were repeated three times.
Matrigel invasion assay
A 24-well plate with inner chamber (Corning Inc.,
Corning, NY, USA) was used for the invasion assay. The 8-µm pore
membranes were pre-coated with Matrigel (BD Biosciences), which was
mixed with RPMI-1640 without FBS at a dilution of 1:8. After
incubation and hydration with RPMI-1640 for 0.5 h, the Eca109 cells
of the control group and the dll4 shRNA group at the density
of 2×104/chamber were inoculated in the inner chamber
with RPMI-1640 with 1% FBS and 0.1% bovine serum albumin (BSA).
Subsequently, 500 µl RPMI-1640 containing 10% FBS was placed in the
lower chamber. After incubation at 37°C for 36 h, cells on the
upper surface of the filters were removed using a cotton swab. The
chamber was kept at room temperature for 30 min and then immersed
with 0.5% crystal violet containing 1% methanol for another 30 min.
The crystal violet was washed with PBS for three times. Cells on
the lower chamber were counted under a microscope in four randomly
selected fields. All of the experiments were repeated three
times.
Xenograft model
Mice were treated according to the NIH Guide for the
Care and Use of Laboratory Animals (17) and the animal study was approved by
the Ethics Committee of Ningbo University. The mice were housed in
a temperature-controlled room with proper dark-light cycles, fed
with a regular diet and maintained under the care of the Laboratory
Animal Unit of Ningbo University, China. The mice were acclimated
for 1 week before the beginning of the experiment. The Eca109
control, Eac109 vector as well as Eca109 with dll4-silenced
cells grown in logarithmic phase were trypsinized, resuspended and
harvested. After being stained with trypan blue, the cells were
counted under a hemocytometer to assess their viability. Viable
cell concentration was adjusted with culture medium to
1×107 cells/ml. The nude mice were randomly divided into
3 groups (n=10 of each), corresponding to the 3 different cell
lines. Each nude mouse was inoculated with 0.5 ml of cell
suspension. The needle was stopped internally for 5 sec, rotated
and pulled out to avoid leakage of the cell suspension. The
activity, diet and mental state of nude mice were observed daily.
The tumor volume was evaluated by a caliper every 5 days. At the
30th day of the experiment, the mice were intraperitoneally
injected with 200 µl of D-luciferin (15 mg/ml), anaesthetized with
isoflurane by a gas manifold at air flow rate of 2%, and then
placed under the animal imaging system to record bioluminescent
signals after 10–15 min of anesthesia. At the end of the
experiment, the mice were sacrificed by cervical vertebra
dislocation. The tumors and pulmonary nodules were stripped and
counted, and then tumors were frozen under liquid nitrogen.
Immunohistochemistry (IHC) and western blot analysis were performed
to detect the expression of proliferating cell nuclear antigen
(PCNA) and DLL4, respectively.
IHC assay
IHC assay was performed to detect the protein
expression in xenografts. Anti-rabbit polyclonal immunoglobulin G
antibody against PCNA (1:200; cat. no. ab18197; Abcam) was used as
the primary antibody. Immunohistochemical kit (SP-9001) was
obtained from Beijing Zhongshan Golden Bridge Biotechnology, Co.,
Ltd., (Beijing, China). For each sample, one score was given
according to the percent of positive cells as follows: no positive
cells, 0; <5% of the cells, 1 point; 5–35% of the cells, 2
points; 36–70% of the cells, 3 points; >70% of the cells, 4
points. To achieve objectivity, the intensity of positive staining
was also used in a 4-scoring system as follows: 0 (negative
staining), 1 (weak staining exhibited as light yellow), 2 (moderate
staining exhibited as yellow brown), and 3 (strong staining
exhibited as brown). A final score was then calculated by
multiplying the above two scores. If the final score was equal or
>4, the tumor was considered as high expression, otherwise, it
was considered as low expression.
Statistical analysis
All statistical analyses were performed using the
SPSS 13.0 statistical software package (SPSS, Inc., Chicago, IL,
USA). The data evaluated were normal distribution and variance
homogeneity, and were expressed as the means ± SD. Survival curves
were plotted by the Kaplan-Meier method and compared by the
log-rank test. The significance of various variables for survival
was analyzed using the Cox proportional hazard model in the
multivariate analysis. In vitro data statistical analysis
was performed using two-tailed Student's t-test or one-way ANOVA
followed by the Tukey's test by GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 in all cases was
considered to indicate a statistically significant difference.
Results
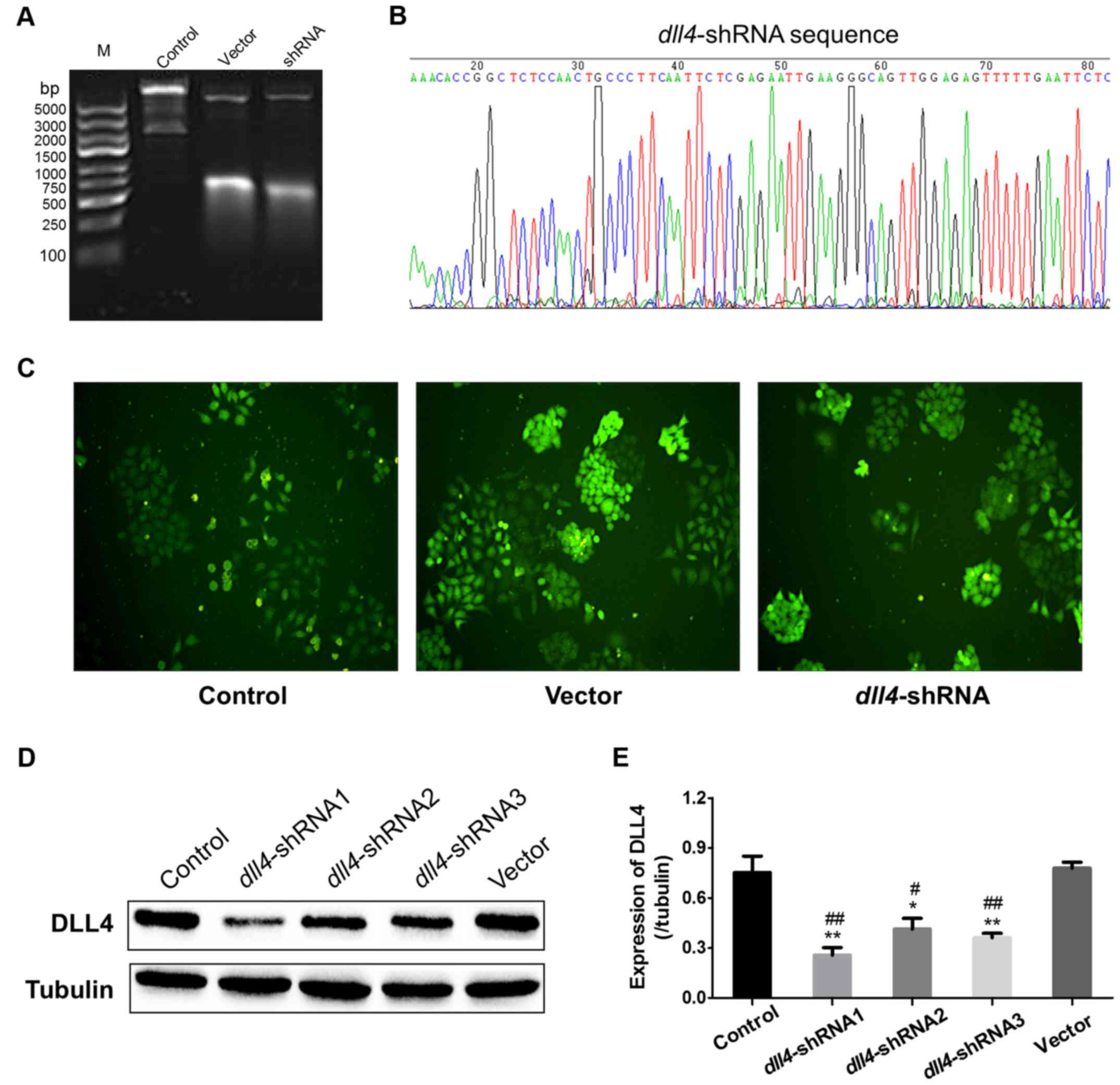
Construction and identification of
stable knockout DLL4 cells
The primary plasmids of pLKO-EGFR-TRC as well as the
recombinant plasmids were digested by XhoI at 37°C for 2 h,
ensuing with an examination by 0.8% agarose gel electrophoresis.
The primary plasmids of DNA fragment were identified to be 7,872,
2,155 and 190 bp, while fragment of the recombinant plasmids after
digestion should be changed to 7,872, 190, 303 and 42 bp, since the
agarose concentration was at low concentration of 0.8% for
distinguishing the large fragments, the small fragments basically
run out of the agarose, therefore, it was difficult to detect both
the large and small fragments in one gel, however, we could still
judge that the recombinant plasmid was successfully constructed
according to the typical bands at large size (Fig. 1A). After plasmids sequencing to
identify the insertion sequence (Fig.
1B), the lentivirus was packaged by the recombinant plasmids
coupled with VSV-G and Δ8.91. Using lentivirus infection and
screening by puromycin, the cell lines of control, vector and
dll4-shRNA were successfully constructed and observed under
a fluorescence microscope (Fig.
1C). Finally, the expression of DLL4 was detected by western
blotting and the results indicated that dll4-shRNA1 could
lead to an ~85% downregulation of DLL4 in the Eca109 cells
(Fig. 1D and E). Therefore,
dll4-shRNA1 was chosen for the stable silencing of
dll4 in Eca109 cells.
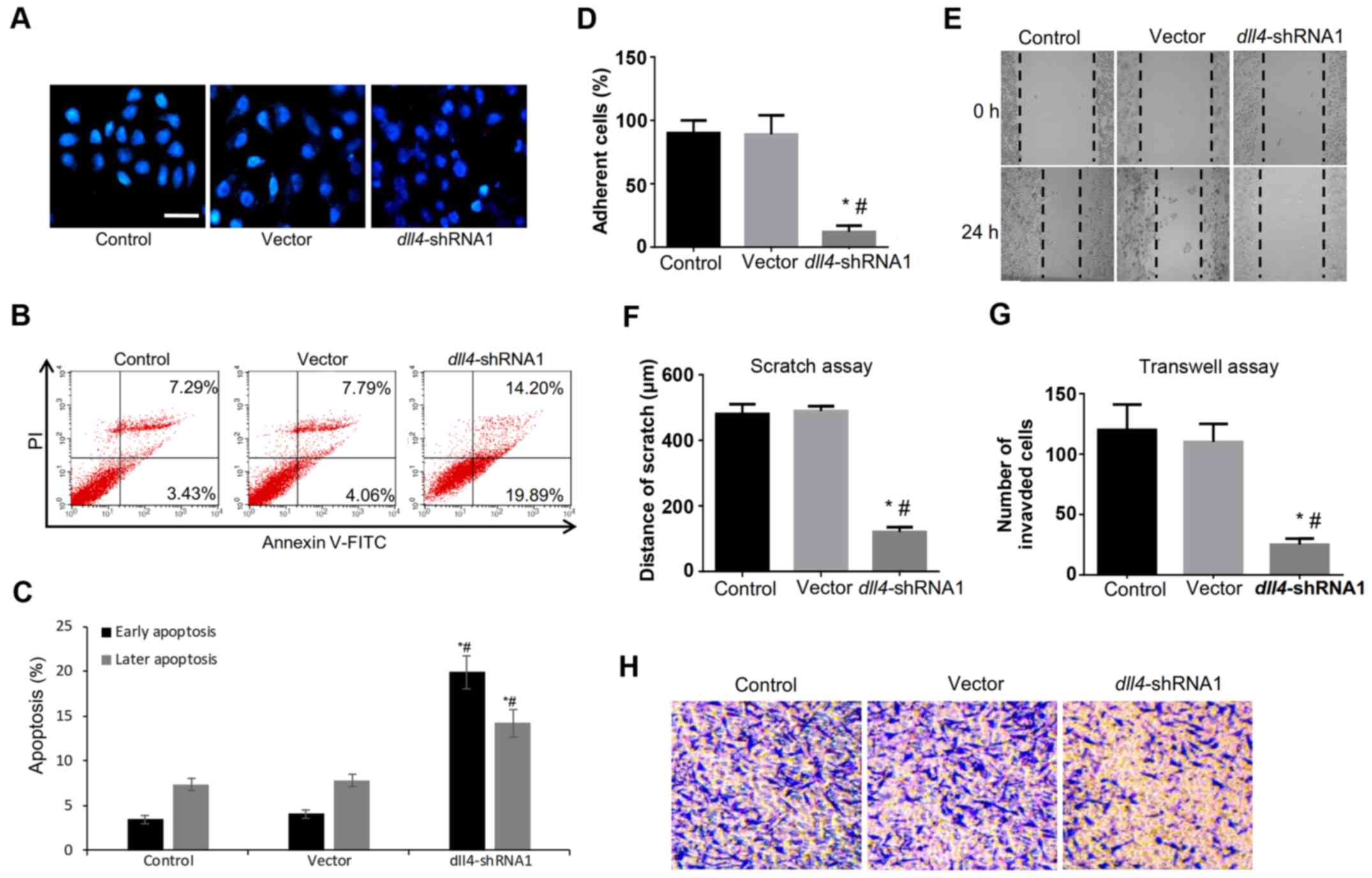
The role of DLL4 in regulating cell
apoptosis, migration and invasion
In cancer, cell invasion allows cancer cells to
acquire the ability to enter lymphatic and/or blood vessels for
dissemination into the circulation, followed by invasion to distant
organs for metastatic growth. To examine the role of DLL4 in
esophagus carcinogenesis, we examined the effects of dll4
knockdown on the proliferation, migration and invasion of the
Eca109 cell line.
After DAPI and Annexin V-FITC/PI staining, a trend
of apoptosis induction was observed in the dll4-shRNA group
(Fig. 2A and B). There was a
significant difference in cell apoptosis rate between the
dll4-shRNA group and the control or vector group (34.09 vs.
10.72 or 11.85%) (Fig. 2C). Cell
adherent assay also indicated the abolished ability of cell
adhesion to fibronectin after the downregulation of DLL4 (Fig. 2D). Furthermore, the scratch assay
(Fig. 2E) and the Transwell assay
demonstrated that migration (Fig.
2F) and invasion (Fig. 2G and
H) of the Eca109 cells significantly decreased after the
dll4 silencing. These results indicated that DLL4 may play a
vital role in regulating the apoptosis and invasion ability of
esophagus cancer cells.
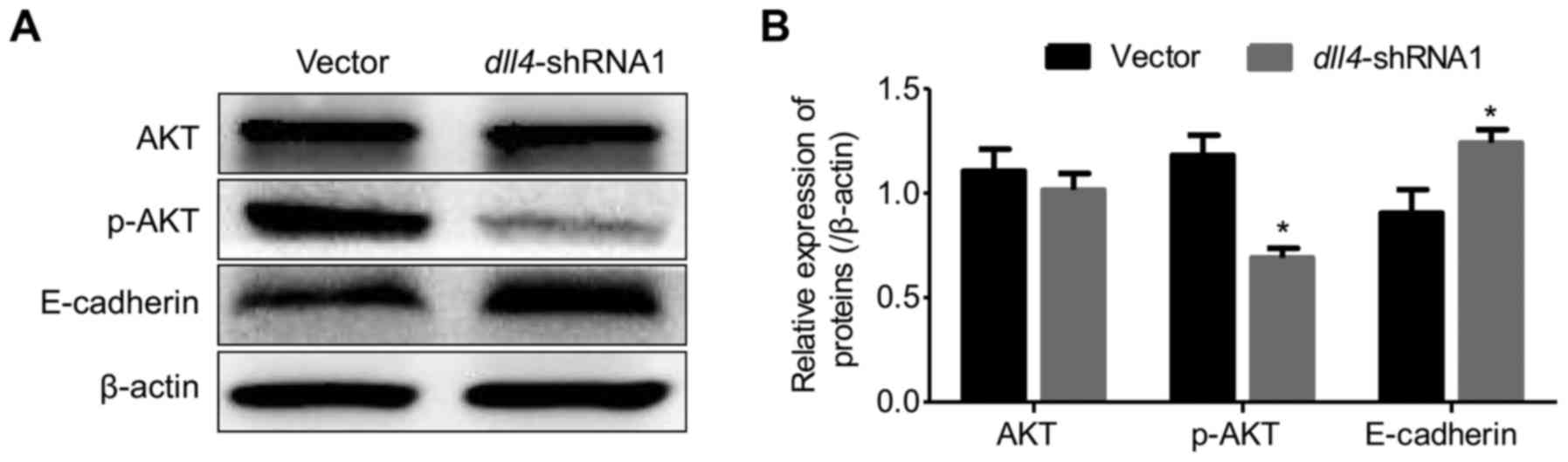
Knockdown of dll4 attenuates the
phosphorylation of Akt and downregulates the expression of
E-cadherin
Given that DLL4 positively correlated with enhanced
esophagus cancer cell migration and invasion, we examined whether
p-Akt or E-cadherin is an underlying mechanism. Dysregulation of
E-cadherin has been considered to be the major critical molecule in
epithelial-mesenchymal transition (EMT)-mediated cancer cell
metastasis. In the present study, we explored the role of DLL4 in
the regulation of p-Akt and E-cadherin expression in the Eca109
cell. The expression of Akt and p-Akt was detected by western
blotting. The results indicated that p-Akt was significantly
decreased after the dll4-gene silencing (Fig. 3). In contrast, as a key molecule
that regulates cell migration, the expression of E-cadherin
significantly increased in the dll4-shRNA1 group, which
indicated that E-cadherin may be responsible for regulating cell
adhesion in esophagus cancer (Fig.
3).
Downregulation of DLL4 abolishes the
growth and metastasis of Eca109 cells
To examine the effect of DLL4 on tumor growth, we
subcutaneously injected Eca109 cells transfected with vector or
dll4 shRNA into nude mice, and then monitored tumor growth
for 60 days. As displayed in Fig.
4A, the tumor formation rate was 100%. Palpable tumors size was
assessed during the experiment in the scrambled control sides.
Downregulation of DLL4 had significant effect on tumor size
(Fig. 4B) and growth curve
(Fig. 4C). Compared with the vector
(1,222.73±95 mm3) and control group (1,009.5±95
mm3), the dll4-shRNA group had significantly
smaller tumor volumes at day 60 (P<0.05). Stripped xenografts
were then subjected to pathological sections and stained with
hematoxylin and eosin (H&E) (Fig.
4D) and PCNA was stained to evaluate the proliferation index of
cancer cells. As displayed in Fig.
4F, the PCNA index was significantly lower in the
dll4-shRNA group than in the vector and control group
(P<0.05), which was also in consistense with the observation
results from the tumor growth curve. Furthermore, to determine
whether dll4 knockdown would alter the expression of DLL4
and p-Akt in vivo, Akt and DLL4 expression were detected by
western blotting. Densitometric analysis of western blotting
revealed that the expression of both DLL4 and p-Akt was
significantly lower in the dll4 knocked-down cells
(P<0.05; Fig. 4G).
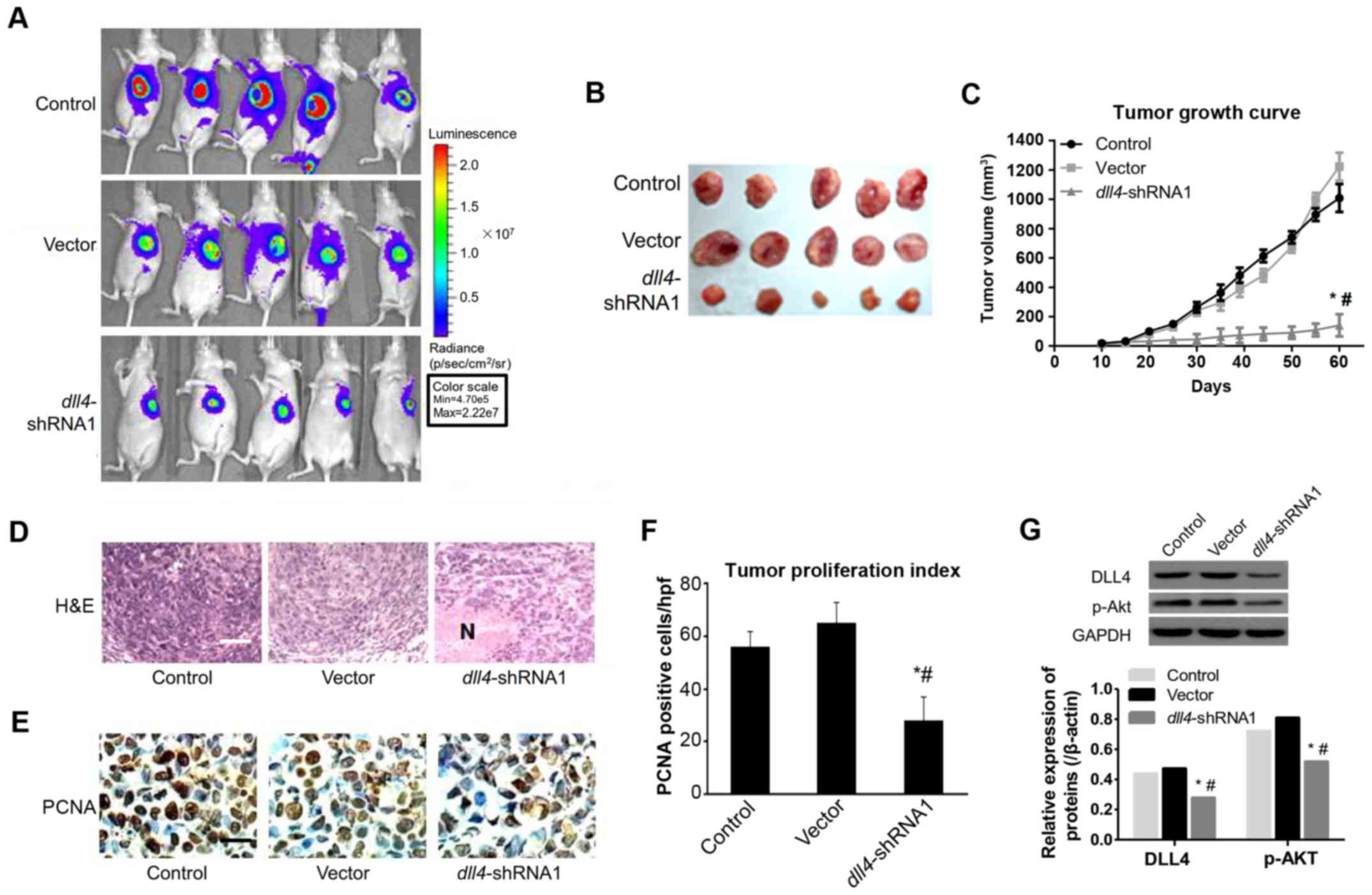 | Figure 4.Tumor growth properties and mice
survival rate after inoculation with Eca109
dll4−shRNA cells. (A) Bioluminescence
imaging of implanted, luciferase-expressing Eca109 vector,
shScramble or dll4-shRNA1 cells. (B) Macroscopic appearance
of stripped tumors, whose volumes were recorded every 5 days during
experiment, dll4-shRNA1 group shown more smaller volume than
control or vector group (P<0.01). (C) Tumor growth curve
after inoculation with Eca109
dll4−shRNA cells. (D) Hematoxylin and
eosin (H&E) staining of tumors derived from xenograft, N,
indicated extensive necrosis. Upper panel, the original images of
mice lungs section was acquired under the light microscope after
H&E staining, scale bar indicated 100 µm. Down panel, the
percentage of pulmonary nodules and the number of large foci of
lung tissue. (E) Immunohistochemistry (IHC) was performed to
measure the expression of proliferating cell nuclear antigen (PCNA)
in 3 groups, images were captured under the light microscope. Scale
bar, indicated 20 µm. Proliferative index of tumor 60 days after
inoculation. Proliferative index is estimated by the number of
PCNA-positive cells per high-power field. Positive staining for
PCNA is mainly located within the nuclei (magnification, ×400). (F)
The proliferative index of each tumor was calculated by averaging
the number of PCNA-positive cells in 5 random bright fields of
microscope (magnification, ×400), counted in a blinded fashion by a
single observer under the supervision of a pathologist;
*P<0.05 vs. vector group, #P<0.05
vs. control group. (G) Western blotting detected the expression of
DLL4 and p-Akt in xenografts. The tumor tissue lysates were probed
with polyclonal anti-DLL4, monoclonal anti-p-Akt (ser473), and
anti-β-actin as internal control. A representative of 3additional
experiments is shown. Data depicted were derived from 3 independent
experiments using at least 5 animals for each cell line injected;
*P<0.05 vs. vector group, #P<0.05
vs. control group. |
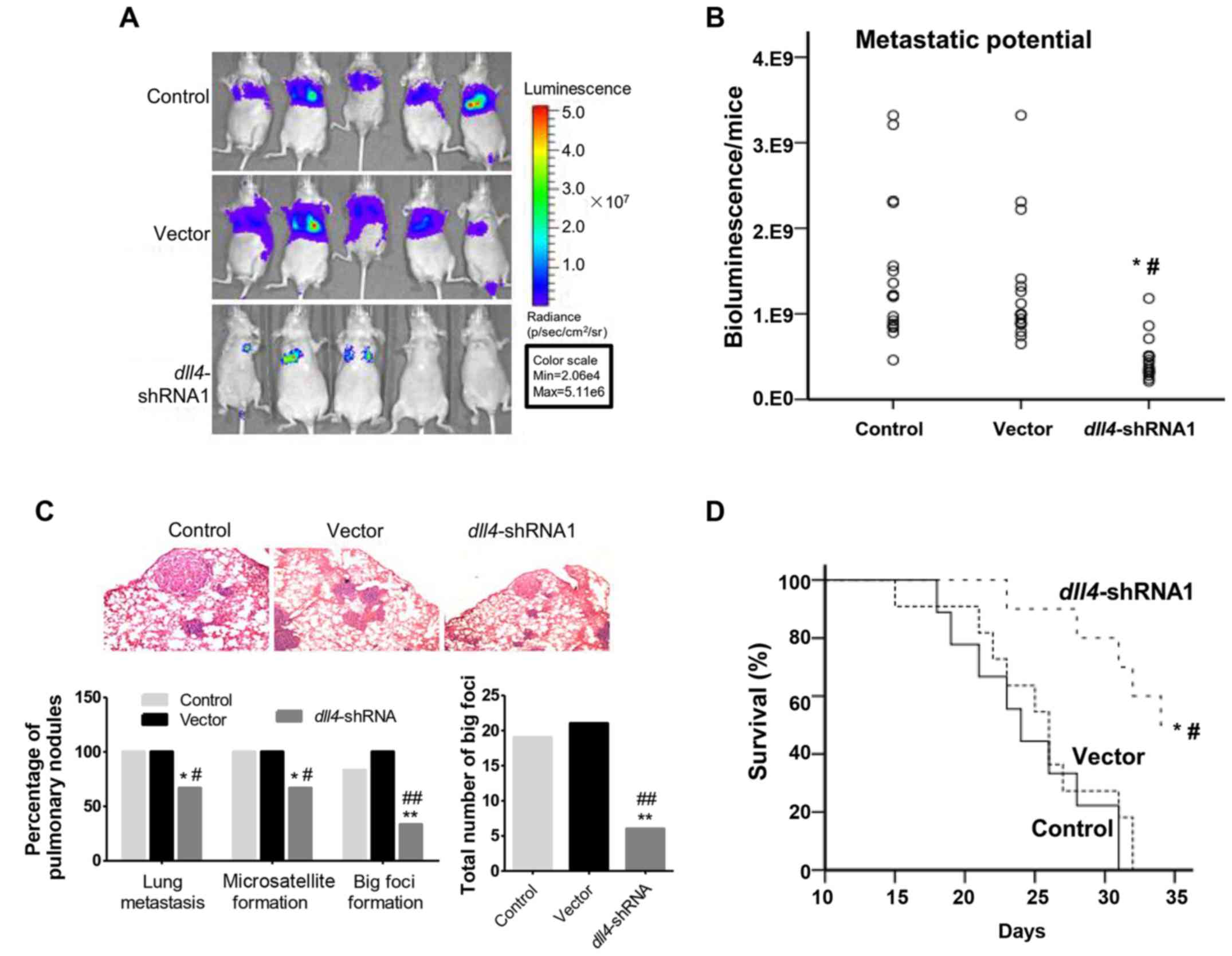
The lung colonization ability of the Eca109
dll4−shRNA cells was then investigated
after mice tail vein injection. Survival rates were examined and
observed by small animal imaging systems (Fig. 5A). The control and vector groups,
presented a significant amount of bioluminescence signal in the
lung area, whereas in the dll4-shRNA group, little signals
were detected in some mice. Subsequently, we quantified the
bioluminescence signal in mice lung regions (Fig. 5B). In the dll4-shRNA group,
the signal intensity was significantly lower, than that of the
control vector group (P<0.05). At the end of the
experiment, mice lungs were fixed with formalin and stained with
H&E staining. In the control and vector groups, pulmonary
nodules were observed at a higher percentage, while in the
dll4-shRNA group pulmonary nodules were found in 66.6% of
mice. In addition, we counted the number of large foci which were 6
in the Eca109 dll4−shRNA group, and 19
and 21 in the control and vector group, respectively (Fig. 5C). Finally, we observed that at the
end of experiment, ~50% of the nude mice survived in the Eca109
dll4−shRNA group (Fig. 5D), which indicated that mice
lifespan was prolonged in Eca109
dll4−shRNA group. Conversely, the nude
mice in the control and vector group all died at the end of the
experiment. The overall survival rate in the Eca109
dll4−shRNA group was higher than that
in the control and vector group and the difference was
statistically significant (P<0.05).
Discussion
At the early stages of esophageal cancer, certain
changes in cell-cell and cell-matrix interactions lead to abnormal
cell behavior and result in invasive and malignant transformation.
In the late stages of tumor development, metastasis is the
predominant complication and makes esophageal cancer difficult to
control, which has been identified as the main cause of high
mortality (17).
DLL4 plays a significant role in normal vascular
development, which has been hypothesized as a potential target for
anti-angiogenic therapy (18). In a
previous study (19) it was
observed in a mouse model of tumor-xenograft, that YW152F (DLL4
mAb) significantly increased vascular density, followed by a
decrease in tumor growth rate, which clearly established the
potential role of DLL4 in inhibiting tumor growth. However, the
molecular mechanism through which DLL4 regulates vessel patterning
remained unclear until recently. Despite extensive studies on
vascular endothelial growth factor (VEGF) and DLL4 signaling,
little is known on the role of DLL4 on cancer metastasis (20). In order to further elucidate the
above-mentioned observations, an shRNA lentiviral vector was
constructed to knockdown the expression of DLL4 in the Eca109
cells. Subsequently, the migration and invasion ability of
dll4-shRNA1 cells downregulated significantly. The benefit
of the lentiviral vector transfection is that the induced cells
have heritable changes characteristics for subsequent study. Gene
silencing caused by siRNA interference methods is unstable, which
leads to genetic trait recovery. Of course, there are also some
defects in the lentiviral technique. It could be time-consuming in
the process of puromycin screening in requiring the target cells.
It would be better if the upstream promoter of DLL4 was modified,
which may be related to an inducible function of tetracycline, to
make the process of gene silencing more credible.
The existing studies on DLL4 are mostly focusing on
the role of DLL4 in regulating angiogenesis. Few studies aimed to
investigate DLL4 in tumor metastasis and tumor growth. Previous
research on cervical cancer indicated that DLL4 expression at both
the mRNA and protein level in cervical cancer tissues was
significantly higher than that in normal cervical tissues.
Univariate and multivariate logistic regression analyses
demonstrated that overexpression of DLL4 was strongly associated
with lymph node metastasis. Furthermore, survival analysis revealed
that the expression of DLL4 was an independent factor of an
unfavorable overall survival. The study indicated that DLL4 may be
a potential clinical diagnostic marker for patients with early
stage cervical cancer (21). In
contrast, evidence showed that Notch signaling drives a cancer stem
cell phenotype by regulating genes that establish stemness. Using
patient derived xenograft models the authors demonstrated that
inhibition of the Notch signaling cascades is efficacious in
decreasing tumor growth. Furthemore, the Notch activity in a
patient EUS-derived biopsy was also found to predict the outcome of
chemotherapy (22).
In the present study, we evaluated the role of DLL4
in the esophagus cancer cell line Eca109.
The scratch and Transwell assays demonstrated that
after the dll4 silencing, the invasive and migration ability
of the Eca109 cells significantly decreased, with attenuation of
the Akt signaling pathway and increase of the expression
E-cadherin. Furthermore, in vivo studies, our results
indicated that the ability of dll4-shRNA cells in migration
and invasion significantly decreased compared with the control and
vector control. the expression of both DLL4 and p-Akt was
significantly lower in the dll4 knocked-down cells. However,
the present study still has some limitations; for instance, the
relationship between DLL4/Notch/Akt signaling cascades has not been
identified. Future studies should focus on the exact molecular
signaling pathway that plays a role in regulating the apoptosis and
metastasis of esophageal carcinoma. Finally, in the present study
we used one cell line, Eca109, to explore the role of DLL4 in the
progression of esophagus cancer, which may be a limitation of this
study. These data are preliminary and further research is required
to verify them in other esophageal cancer cell lines. Furthermore,
the underlying mechanism could be further identified and elucidated
in future studies.
In conclusion, the present study indicated that DLL4
could be viewed as a novel target on esophageal carcinoma treatment
and targeted therapy. Further understanding focusing on the
molecular pathway and the protein which DLL4 targeted would be
performed to better understand the role of DLL4 in the progress of
esophagus cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Academic Leaders
Training Program of Pudong Health Bureau of Shanghai (no.
PWRd2012-15) and the Natural Science Foundation of Shanghai
Municipal Science and Technology Commission (no. 13ZR1437300).
Availability of data and materials
We declared that materials described in the
manuscript, including all relevant raw data, will be freely
available to any scientist wishing to use them for non-commercial
purposes, without breaching participant confidentiality.
Authors' contributions
XG, YD and X Y contributed the central idea,
analysed most of the data, and wrote the initial draft of the
paper. The remaining authors contributed to refining the ideas,
carrying out additional analyses and finalizing this paper. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board of the Department of Laboratory Animal
Science of Fudan University Pudong Medical Center (Shanghai,
China).
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Barzi A and Thara E: Angiogenesis in
esophageal and gastric cancer: A paradigm shift in treatment.
Expert Opin Biol Ther. 14:1319–1332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37 Suppl
8:S4–S66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liotta LA and Kohn E: Anoikis: Cancer and
the homeless cell. Nature. 430:973–974. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagaprashantha LD, Vatsyayan R, Lelsani
PC, Awasthi S and Singhal SS: The sensors and regulators of
cell-matrix surveillance in anoikis resistance of tumors. Int J
Cancer. 128:743–752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bailey JM, Singh PK and Hollingsworth MA:
Cancer metastasis facilitated by developmental pathways: Sonic
hedgehog, Notch, and bone morphogenic proteins. J Cell Biochem.
102:829–839. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang JP, Li N, Bai WZ, Qiu XC, Ma BA,
Zhou Y, Fan QY and Shan LQ: Notch ligand Delta-like 1 promotes the
metastasis of melanoma by enhancing tumor adhesion. Braz J Med Biol
Res. 47:299–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sethi N, Dai X, Winter CG and Kang Y:
Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast
cancer by engaging notch signaling in bone cells. Cancer Cell.
19:192–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tao J, Erez A and Lee B: One NOTCH
further: Jagged 1 in bone metastasis. Cancer Cell. 19:159–161.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xing F, Okuda H, Watabe M, Kobayashi A,
Pai SK, Liu W, Pandey PR, Fukuda K, Hirota S, Sugai T, et al:
Hypoxia-induced Jagged2 promotes breast cancer metastasis and
self-renewal of cancer stem-like cells. Oncogene. 30:4075–4086.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W,
Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA,
et al: The Notch ligand Jagged2 promotes lung adenocarcinoma
metastasis through a miR-200-dependent pathway in mice. J Clin
Invest. 121:1373–1385. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu Y, Su H, Li X, Guo G, Cheng L, Qin R,
Qing G and Liu H: The NOTCH ligand JAGGED2 promotes pancreatic
cancer metastasis independent of NOTCH signaling activation. Mol
Cancer Ther. 14:289–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Liu M, Feng Y, Huang YF, Xu YF, Che
JP, Wang GC and Zheng JH: High expression of Notch ligand Jagged2
is associated with the metastasis and recurrence in urothelial
carcinoma of bladder. Int J Clin Exp Pathol. 6:2430–2440.
2013.PubMed/NCBI
|
|
14
|
Huang QB, Ma X, Li HZ, Ai Q, Liu SW, Zhang
Y, Gao Y, Fan Y, Ni D, Wang BJ and Zhang X: Endothelial Delta-like
4 (DLL4) promotes renal cell carcinoma hematogenous metastasis.
Oncotarget. 5:3066–3075. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kontomanolis E, Panteliadou M,
Giatromanolaki A, Pouliliou S, Efremidou E, Limberis V, Galazios G,
Sivridis E and Koukourakis MI: Delta-like ligand 4 (DLL4) in the
plasma and neoplastic tissues from breast cancer patients:
Correlation with metastasis. Med Oncol. 31:9452014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao M, Yang S, Ning X and Huang Y:
Aberrant expression of δ-like ligand 4 contributes significantly to
axillary lymph node metastasis and predicts postoperative outcome
in breast cancer. Hum Pathol. 45:2302–2310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
NIH requests information on care of
laboratory animals. Am J Vet Res. 67:204–205. 2006.PubMed/NCBI
|
|
18
|
Reinacher-Schick A, Pohl M and Schmiegel
W: Drug insight: Antiangiogenic therapies for gastrointestinal
cancers - focus on monoclonal antibodies. Nat Clin Pract
Gastroenterol Hepatol. 5:250–267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ridgway J, Zhang G, Wu Y, Stawicki S,
Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I,
et al: Inhibition of DLL4 signalling inhibits tumour growth by
deregulating angiogenesis. Nature. 444:1083–1087. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sainson RC and Harris AL: Anti-DLL4
therapy: Can we block tumour growth by increasing angiogenesis?
Trends Mol Med. 13:389–395. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang S, Liu Y, Xia B, Deng J, Liu T, Li Q,
Yang Y, Wang Y, Ning X, Zhang Y and Xiao M: DLL4 as a predictor of
pelvic lymph node metastasis and a novel prognostic biomarker in
patients with early-stage cervical cancer. Tumour Biol.
37:5063–5074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Da Silva TG, Jin K, Han X,
Ranganathan P, Zhu X, Sanchez-Mejias A, Bai F, Li B, Fei DL, et al:
Notch signaling drives stemness and tumorigenicity of esophageal
adenocarcinoma. Cancer Res. 74:6364–6374. 2014. View Article : Google Scholar : PubMed/NCBI
|