Introduction
Hepatocellular carcinoma (HCC) is the sixth major
type of cancer and the third most common cause of cancer-related
deaths worldwide, mainly associated with hepatitis B and C virus
infections (1). The global
incidence of HCC is increasing, with surgical resection and liver
transplantation being the current major treatment strategies in
patients diagnosed in the early stages (2). The overall survival rate of patients
diagnosed with HCC is <10% due to high recurrence and metastasis
rates and failure of early detection. While the cellular and
molecular mechanisms underlying HCC have been extensively studied
over the last decades, the pathogenesis process of this type of
tumor remains poorly understood.
PRMT5, a type II protein arginine methyltransferase,
is a 72 kDa protein that catalyzes methylation of arginine residues
in various substrate proteins, including chromatin-associated
proteins and transcriptional factors (3). The function of PRMT5 as a
methyltransferase has been extensively studied in relation to
cancer. PRMT5 acts as cancer-inducing gene by promoting cell
proliferation (4–8), inhibiting transcription of tumor
suppressor genes (4,9–13) and
inducing metastatic predisposition via epithelial-mesenchymal
transition (14). The symmetric
dimethylation of the arginine residues of histones H3 and H4 by
PRMT5 triggers the modification of the chromatin structure and
alterations in the expression patterns of diverse genes (3,15–17).
Additionally, PRMT5 induces transcriptional inhibition by directly
methylating the tumor suppressor proteins p53 and E2F1, thus
bestowing advantages for cancer cell survival (9,13).
Recent studies have demonstrated that PRMT5 suppresses the
expression of E-cadherin, the hallmark of EMT transition, through
interactions with Ajuba and the E-cadherin transcription factor
Snail (6,14).
PRMT5 has been extensively characterized in relation
to various types of cancer and its emerging role as a potential
oncoprotein is of significant clinical interest. However, the
expression of PRMT5 in relation to invasion phenotypes in HCC and
colon cancer has rarely been documented. Earlier invasion studies
using siRNAs were conducted under conditions of decreased
proliferation and increased cell death, which potentially
contributed to the decrease in the invasion activity of cancer
cells (17–20). Using HCC microarray datasets, we
have evaluated several molecular markers differentially expressed
in HCC (21–23). In the present study, we revealed
that PRMT5 was overexpressed in HCC and colorectal cancer tissues
and its depletion suppressed the invasion of cancer cell lines
without affecting the colony survival. Consistent with its
correlation with the invasive phenotype, the overexpression of
PRMT5 in HCC patient tissues was associated with aggressive
clinicopathological parameters, including poorer differentiation
and greater invasion, tumor size and α-fetoprotein level.
Materials and methods
Patients and tissue samples
HCC and colon cancer tissues were collected from
surgically resected patient specimens who underwent surgery between
April 1992 and December 2004, and between March 2014 and August
2014, respectively, at the Korea Cancer Center Hospital. A total of
120 HCC (including 33 pair-matched samples) and 10 pair-matched
colon cancer samples were used. This study was approved by the
Institutional Review Board, Korea Cancer Center Hospital. Written
informed patient consent was either waived for HCC or obtained for
colon cancer tissues.
Cell culture and gene silencing via
siRNA transfection
Huh7 (from the Japanese Cancer Research Resources
Bank), SW480 (from the American Type Culture Collection) and
SNU-709 cells (from the Korean Cell Line Bank) were cultured in
RPMI-1640 supplemented with 10% (v/v) fetal bovine serum (FBS) and
1% (w/v) antibiotics. The cells were transfected with control or
PRMT5 siRNA at a concentration of 10 nM using Lipofectamine RNAiMAX
reagent (13778-150; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's instructions. The
following human PRMT5 siRNA oligonucleotide sequences were used:
PRMT5 siRNA#1, 5′-UGUGACUAUAGUAAGAGGAUUGCAGUG-3′ and PRMT5 siRNA#2,
5′-AGGGACUGGAAUACGCUAAUUGUGGGA-3′.
RNA extraction and cDNA synthesis
Total RNA was extracted from cells using the RNeasy
Mini kit (cat. no. 304-150; GeneAll Biotechnology Co., Ltd., Seoul,
Korea). The concentration and quality of total RNA were assessed
using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies,
Wilmington, DE, USA) at 260 and 280 nm. For cDNA synthesis, total
RNA was reverse-transcribed using the iScript cDNA synthesis kit
(cat. no. 170-8891; Bio-Rad Laboratories, Inc., Hercules, CA, USA)
according to the manufacturer's protocol.
Semi-quantitative reverse
transcriptase-polymerase chain reaction (RT-PCR)
Semi-quantitative RT-PCR was performed using the
Maxime PCR PreMix kit (i-StarTaq; Intron Biotechnology, Seongnam,
Korea) and the following primer sequences: PRMT5,
5′-CTCCTACCTCCAATACCTGG-3′ (sense) and 5′-CATTCCCTCATGTCTGATGA-3′
(antisense); matrix metalloproteinase-2 (MMP-2),
5′-ATCTTTGCTGGAGACAAATTC-3′ (sense) and 5′-AACTTCACGCTCTTCAGACTT-3′
(antisense); β-actin, 5′-GGACTTCGAGCAAGAGATGG-3′ (sense) and
5′-AGCACTGTGTTGGCGTACAG-3′ (antisense); β2-microglobulin,
5′-GTGCTCGCGCTACTCTCTCT-3′ (sense) and 5′-CGGCAGGCATACTCATCTTT-3′
(antisense).
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed using
the iQTM SYBR® Green Supermix (cat. no. EBT-1801) and
CFX96 real-time RT-PCR detection system (both from Bio-Rad
Laboratories). The following primers were used: PRMT5#1,
5′-TTTCCCATCCTCTTCCCTATTAAG-3′ (sense) and
5′-CCCACTCATACCACACCTTC-3′ (antisense); PRMT5#2,
5′-CCGGCTACTTTGAGACTGG-3′ (sense) and 5′-TTTGGCCTTCACGTACCG-3′
(antisense); β2-microglobulin, 5′-AAGGACTGGTCTTTCTATCTCTTGTA-3′
(sense) and 5′-ACTATCTTGGGCTGTGACAAAGTC-3′ (antisense). Relative
gene expression was analyzed using the comparative threshold cycle
(2-ΔΔC(t)) method.
Protein extraction and western blot
analysis
Total cell lysates was lysed with TNN buffer [120 mM
NaCl, 40 mM Tris-HCl, pH 8.0, 0.5% (w/v) NP-40, 1 mM
phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 100 mM
sodium fluoride, 5 mM EDTA] containing protease inhibitor
(P3100-010; GenDEPOT, Katy, TX, USA) and quantified using Bio-Rad
protein assay based on the Bradford method (cat. no. 500-0006;
Bio-Rad Laboratories). Equivalent amounts of protein were
electrophoresed via 10% (w/v) SDS-PAGE and transferred onto
nitrocellulose membrane (cat. no. 10600002; GE Healthcare Life
Sciences, NJ, USA). The membrane was subsequently placed in TBST
buffer containing 5% (w/v) skim milk and blocked at room
temperature for 1 h, followed by treatment with primary antibodies,
including MMP-2 (diluted to 1:2,000; cat. no. sc-10736), β-actin
(diluted to 1:3,000; cat. no. sc-47778) and PRMT5 (diluted to
1:3,000; cat. no. sc-376937) (all from Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) for 2 h and further treatment with
horseradish peroxidase-conjugated secondary antibody (diluted to
1:3,000; cat. no. A120-101P and ct. no. A90-116P; Bethyl
Laboratories, Montgomery, TX, USA) for 1 h. Target proteins were
detected using a chemiluminescence kit (cat. no. sc-204806; Santa
Cruz Biotechnology, Inc.).
Matrigel invasion assay
The precoated filter chamber (6.5 mm in diameter, 8
µm pore size) of a polycarbonate Transwell membrane (Corning Inc.
Corning NY, USA) was coated with Matrigel. Cells (2×104)
suspended in serum-free medium were added into the upper
compartment of the chamber and FBS [10% (w/v)] medium placed in the
bottom of the chamber as a chemoattractant. After 24 and 32 h of
incubation, the cells were fixed, stained with Hemacolor solution
(cat. no. 111674; Merck Millipore, Darmstadt, Germany), visualized
under the microscope and quantified by counting four different
fields. All experiments were performed in triplicate.
Colony formation assay
After 24 h of transfection, the cells were seeded at
a density of 1,000 cells/plate on a 60-mm culture dish.
Subsequently, the cells were incubated for 10–13 days, fixed with
3.7% (w/v) formalin for 15 min, washed with distilled water and
stained with 0.5% (w/v) crystal violet for 30 min at room
temperature.
Statistical analysis
Statistical analysis was performed using the SPSS
software (version 23.0; IBM Corporation, Armonk, NY, USA). The
Mann-Whitney U test was applied to compare the expression of PRMT5
in non-tumor and tumor tissues. For comparisons of
clinicopathological parameters according to low and high expression
of PRMT5, the Chi-square or the Fisher's exact test were used as
deemed appropriate. P<0.05 was considered to indicate a
statistically significant difference.
Results
PRMT5 is overexpressed in human HCC
and colon cancer tissues
To ascertain whether PRMT5 is associated with human
cancer, we assessed PRMT5 mRNA levels in surgically removed frozen
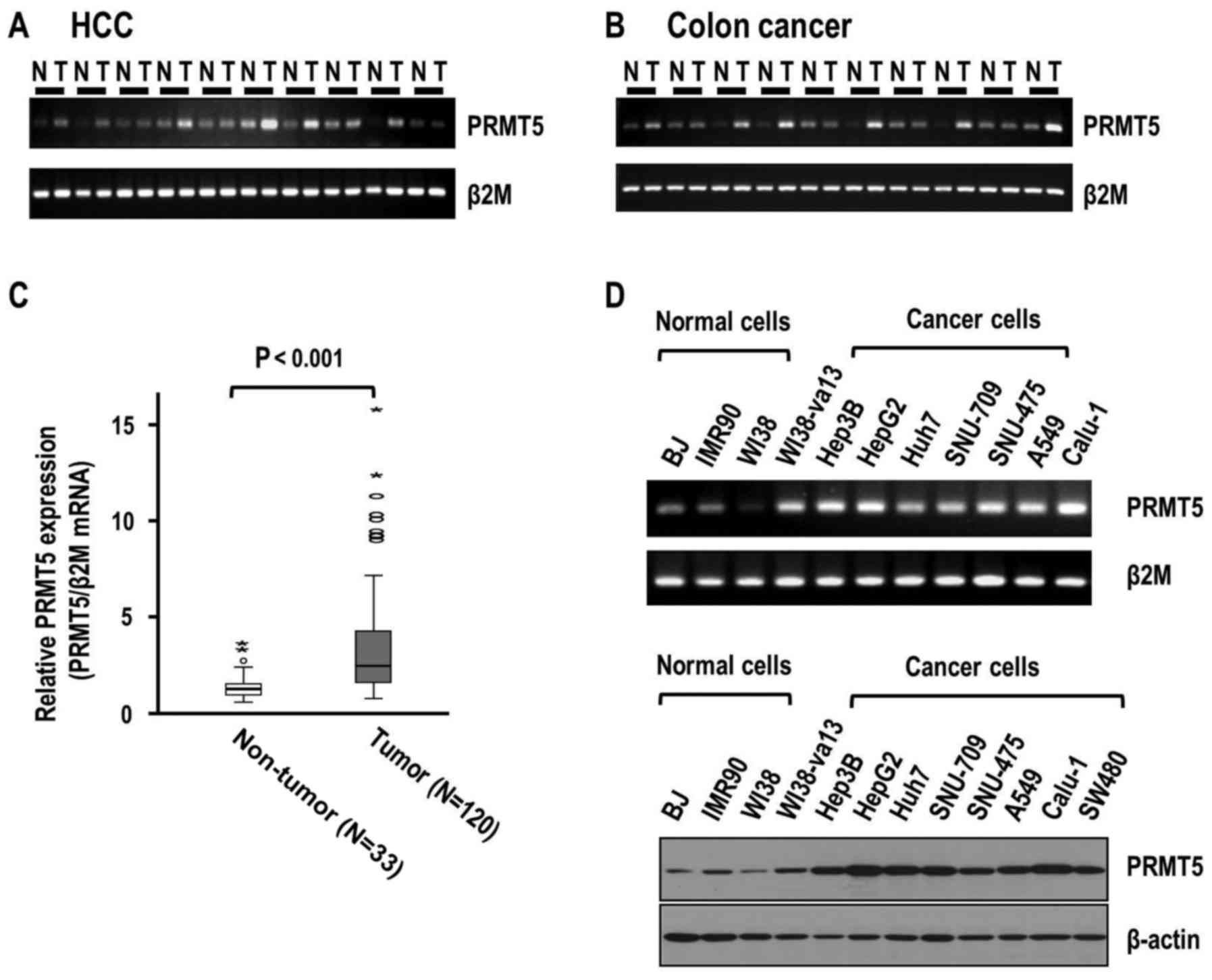
HCC and colon cancer tissue specimens. Semi-quantitative RT-PCR
analyses revealed higher expression of PRMT5 in HCC than that in
the corresponding adjacent liver tissues (Fig. 1A). Similar to the HCC tissues, colon
cancer tissues exhibited higher expression of PRMT5 compared to
non-tumor tissues (Fig. 1B). To
further validate the overexpression of PRMT5, we performed
quantitative real-time RT-PCR analysis using 120 HCC tissues
(n=120) (Table I) and normalized
expression to that in normal liver tissues, that were obtained from
metastatic cancers with no background fibrosis and cirrhosis.
Consistently, real-time RT-PCR data demonstrated greater expression
of PRMT5 in HCC than in adjacent liver tissues (P<0.001)
(Fig. 1C). The mean increase in
PRMT5 expression in adjacent liver (n=33) and tumor tissues
(n=120), was 3.58-fold (median, 2.47-fold). Our results clearly
indicated that PRMT5 was significantly overexpressed in human HCC
and colon cancer tissues.
 | Table I.Demographic and pathological data of
patients. |
Table I.
Demographic and pathological data of
patients.
| Variables | Classification | Distribution |
|---|
| Sex | Male/female | 101/19 |
| Age | Years, mean ± SD
(range) | 52.9±10.64
(25–77) |
| Etiology | Non B non
C/B/C | 21/93/6 |
| AST | IU/l, mean ± SD
(range) | 53.1±34.6
(15–182) |
| ALT | IU/l, mean ± SD
(range) | 46.0±25.1
(8–141) |
| AFP | ng/ml, mean ± SD
(range) | 9852.8±66103.9
(1–690,400) |
| Child-Pugh
score | A/B/C | 99/9/0 |
| Tumor size | cm, mean ± SD
(range) | 5.9±3.6
(1.2–18.5) |
| Number of
tumors |
Single/Multiple | 86/20 |
| Gradea | 1/2/3/4 | 14/67/36/1 |
| Fibrosis | No/Yes | 40/70 |
| Cirrhosis | No/Yes | 64/46 |
| Microscopic hepatic
vein invasion | No/Yes | 94/14 |
PRMT5 overexpression is associated
with invasion, differentiation, tumor size and α-fetoprotein
levels
To further determine the clinicopathological
correlations of PRMT5, patients were divided into low (n=68) and
high (n=52) PRMT5 expression groups according to mRNA levels. Based
on a 2.0-fold criterion in terms of the expression of PRMT5, the
high expression group was significantly associated with higher
α-fetoprotein (AFP; P=0.020), larger tumor size (P=0.011), poorer
differentiation (P=0.004) and more frequent microscopic hepatic
invasion (P=0.019), compared to the low expression group (Table II). The association of PRMT5 with
AFP was significant, irrespective of the cut-off criterion
(Table III). Our results
collectively indicated that the overexpression of PRMT5 contributed
to the aggressive phenotype of HCC. To further evaluate the
significance of PRMT5 in human cancer, we examined PRMT5 expression
patterns in other normal and cancer cell lines. Expression of PRMT5
in normal fibroblast cell strains (BJ, IMR90 and WI38) was clearly
lower than that in various cancer cell lines, including HCC (Hep3B,
HepG2, Huh7, SNU-709 and SNU-475), lung (A549 and Calu-1) and colon
cancer (SW480) (Fig. 1D). Notably,
WI38-va13 cells transformed with SV-40 exhibited higher PRMT5
expression, compared to parental WI38 cells.
 | Table II.Correlations between the expression
of PRMT5 and the clinicopathological parameters. |
Table II.
Correlations between the expression
of PRMT5 and the clinicopathological parameters.
|
| PRMT5
expression |
|
|---|
|
|
|
|
|---|
| Variables | <2 fold | ≥2 fold |
P-valuea |
|---|
| Sex |
|
Male | 60 | 41 | 0.163 |
|
Female | 8 | 11 |
|
| HBsAg |
|
Negative | 18 | 9 | 0.234 |
|
Positive | 50 | 43 |
|
| AST (U/l) |
|
<100 | 64 | 46 | 0.724 |
|
≥100 | 4 | 6 |
|
| ALT (U/l) |
|
<100 | 65 | 49 | 1.000 |
|
≥100 | 3 | 3 |
|
| AFP (ng/ml) |
|
<20 | 35 | 15 | 0.020b |
|
≥20 | 33 | 35 |
|
| Child-Pugh
score |
| A | 54 | 44 | 0.295 |
| B and
C | 7 | 2 |
|
| Tumor size
(cm) |
|
<5 | 40 | 18 | 0.011b |
| ≥5 | 28 | 33 |
|
| Tumor number |
|
Solitary | 52 | 34 | 0.096 |
|
Multiple | 8 | 12 |
|
| Grade |
| 1 | 13 | 1 | 0.004b |
|
2,3,4 | 54 | 50 |
|
| Fibrosis |
| No | 20 | 20 | 0.188 |
|
Yes | 44 | 26 |
|
| Cirrhosis |
| No | 40 | 24 | 0.279 |
|
Yes | 24 | 22 |
|
| Macroscopic
vascular invasion |
| No | 62 | 46 | 0.402 |
|
Yes | 2 | 4 |
|
| Microscopic hepatic
vein invasion |
| No | 58 | 36 | 0.019b |
|
Yes | 4 | 10 |
|
 | Table III.Correlation between the expression of
PRMT5 and AFP levels. |
Table III.
Correlation between the expression of
PRMT5 and AFP levels.
|
| PRMT5
expression |
|
|---|
|
|
|
|
|---|
| Variables | <2-fold | ≥2-fold |
P-valuea |
|---|
| AFP (ng/ml) |
|
<20 | 35 | 15 | 0.020b |
|
≥20 | 33 | 35 |
|
| AFP (ng/ml) |
|
<50 | 41 | 20 | 0.029b |
|
≥50 | 27 | 30 |
|
| AFP (ng/ml) |
|
<100 | 49 | 22 | 0.002b |
|
≥100 | 19 | 28 |
|
| AFP (ng/ml) |
|
<200 | 51 | 26 | 0.010b |
|
≥200 | 17 | 24 |
|
| AFP (ng/ml) |
|
<400 | 55 | 29 | 0.007b |
|
≥400 | 13 | 21 |
|
PRMT5 depletion induces a decrease in
HCC invasion and the expression of matrix metalloproteinase
(MMP)-2
The clinicopathological correlation of PRMT5
overexpression with hepatic invasion in HCC tissues prompted us to
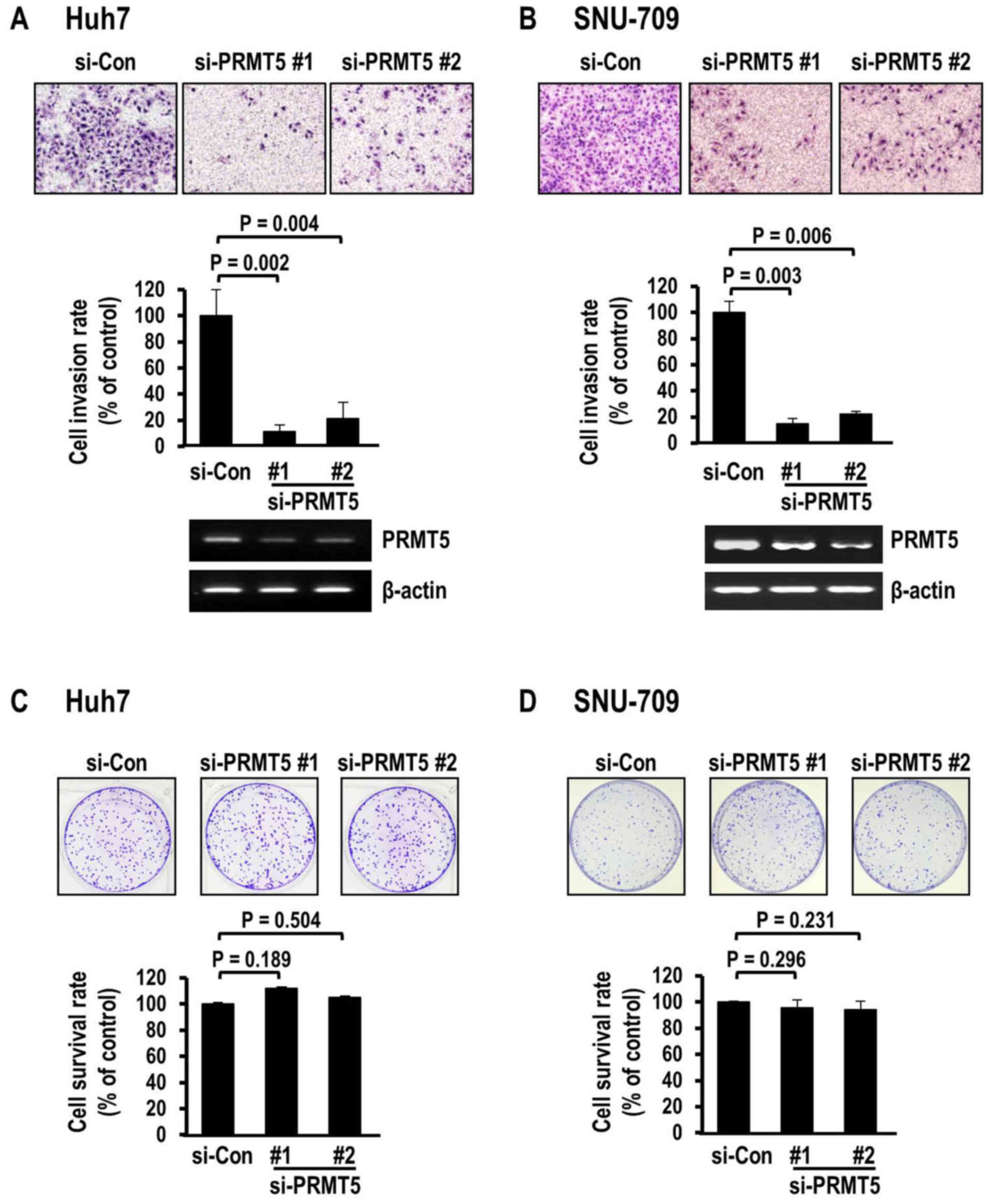
examine the effect of PRMT5 depletion on HCC invasion. Depletion of
PRMT5 achieved by transfection with specific siRNAs against coding
sequences led to a marked decrease in the invasion of the Huh7 HCC
cells, as determined using the Matrigel invasion assay (Fig. 2A). Suppression with two different
siRNAs exerted similar significant effects on invasion (siRNA#1,
P=0.002; siRNA#2, P=0.004), compared to the control siRNA, with
decreases of 88.6 and 78.8%, respectively. Decreased invasion
induced by PRMT5 depletion was consistently observed in another HCC
cell line, SNU-709 (Fig. 2B).
Similar to the Huh7 cells, the SNU-709 cells exhibited severely
decreased invasion rates of 85.2% (siRNA#1) and 77.8% (siRNA#2),
respectively, clearly demonstrating that PRMT5 depletion markedly
attenuated the invasive activity of HCC cells. The PRMT5-mediated
decrease in invasion may be attributable to alterations in
proliferation and apoptosis. In fact, the PRMT family has been
shown to be involved in protein arginine methylation during cell
death and proliferation (17–20).
Accordingly, we performed colony survival analysis with a view to
observe phenotypic changes, including cellular proliferation and
death. Notably, in contrast to the data obtained from the invasion
analysis, PRMT5 depletion did not affect colony survival in either
cell line. Specifically, PRMT5 siRNA (#1 and #2)-transfected Huh7
(Fig. 2C) and SNU-709 cells
(Fig. 2D) did not exhibit
differences in colony survival, compared to corresponding cells
transfected with control siRNA. These findings excluded the
possibility that the suppression of invasion mediated by PRMT5 is
due to alterations in cell proliferation or death. Actually, the
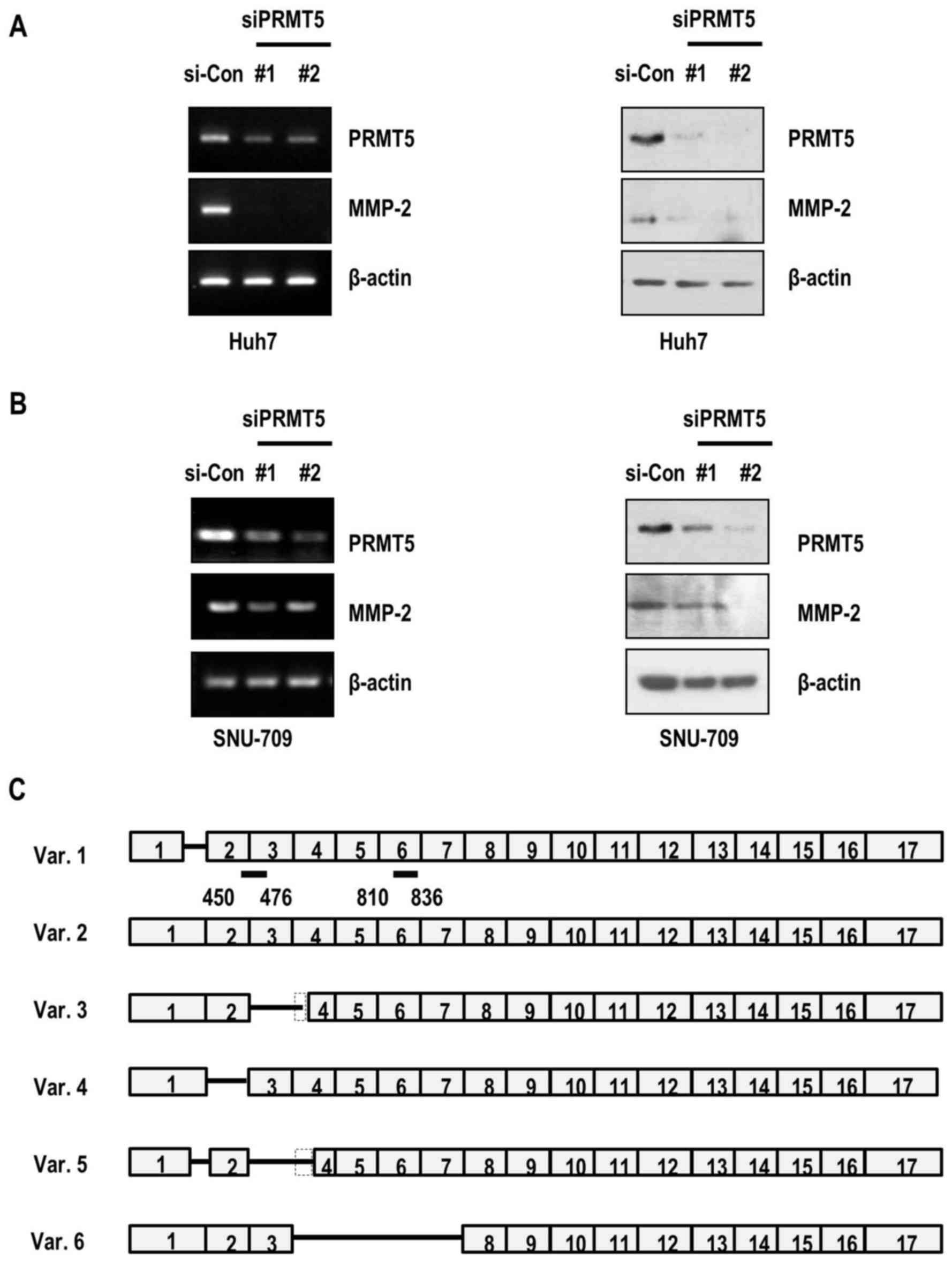
siRNAs used in the present study recognized the coding sequences of
several isoforms (siRNA#1, isoforms 1–5; siRNA#2, isoforms 1, 2, 4
and 6) (Fig. 3C). To further
determine whether the PRMT5-controlled invasion was associated with
matrix metalloproteinase-2 (MMP-2), a protease that cleavages
extracellular matrix and promotes cancer cell invasion (24–26),
we evaluated the expression of MMP-2 under conditions of PRMT5
depletion in HCC cancer cell lines. Notably, MMP-2 mRNA as well as
MMP-2 protein levels were suppressed in PRMT5-depleted Huh7 cancer
cells (Fig. 3A). Consistently, the
expression of MMP-2 in SNU-709 cells was decreased upon PRMT5
knockdown (Fig. 3B). Our results
indicated that PRMT5 depletion-mediated decrease in cell invasion
occured through the reduction of the expression of MMP-2 in the HCC
cells.
PRMT5 depletion weakens the invasion
of colon cancer cells accompanied by decreased expression of
MMP-2
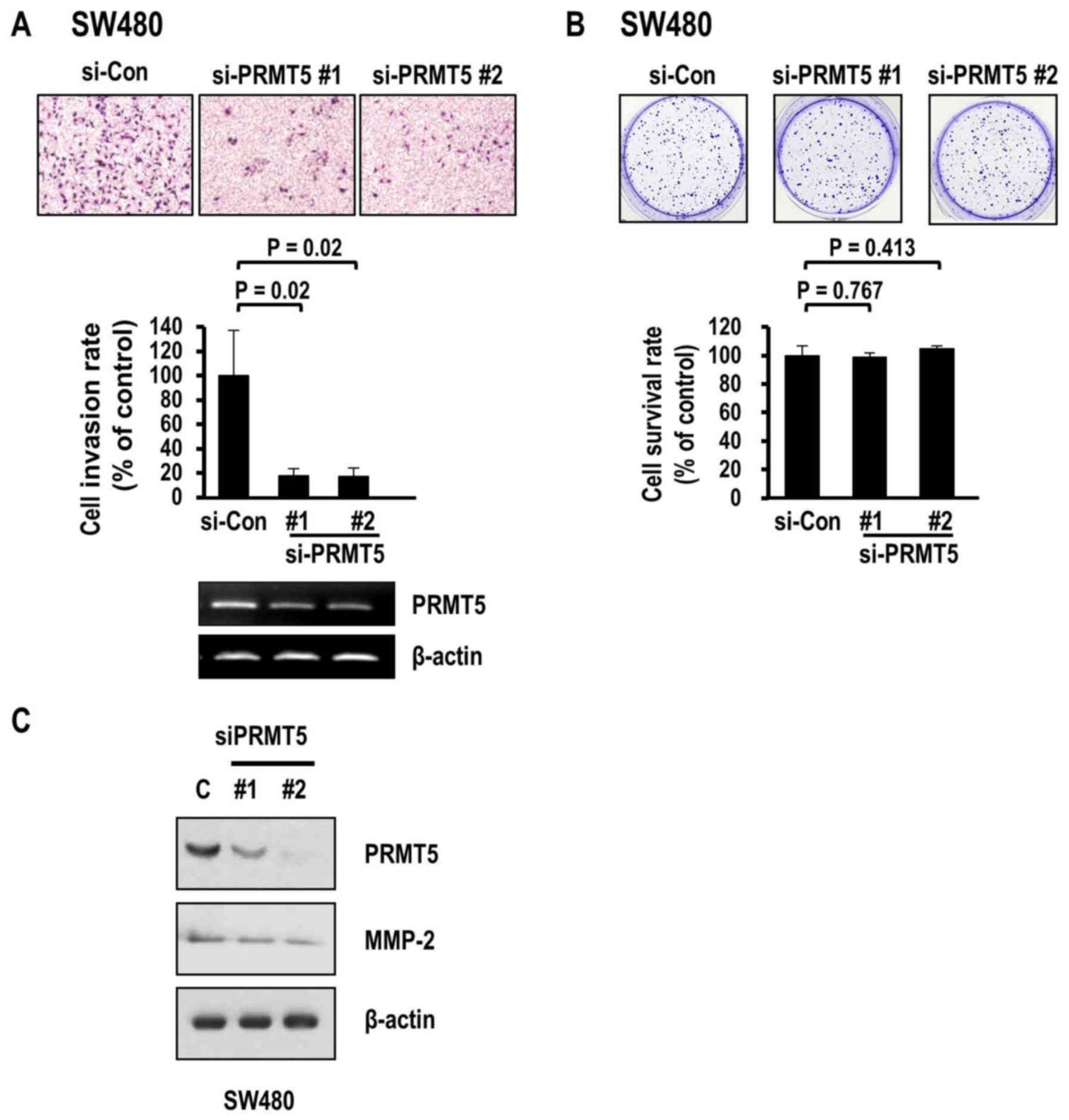
Our finding that PRMT5 depletion induced a decrease
in the invasive activity of HCC further raised the question of
whether PRMT5 exerts similar effects on colon cancer cell invasion.
To resolve this issue, we examined the invasive activity of human
colon cancer cell lines depleted of PRMT5. Similar to the results
obtained with HCC cells, SW480 colon cancer cells exhibited
decreased invasion following the knockdown of PRMT5 (Fig. 4A). Transfection with two different
PRMT5 siRNAs (#1 and #2) induced significant decreases in the
invasion rate (80.2 and 79.8%, respectively). However, the
clonogenic survival of colon cancer cells was not affected
(Fig. 4B) while MMP-2 levels were
consistently decreased upon PRMT5 depletion (Fig. 4C). Collectively, the results clearly
indicated that PRMT5 depletion weakened the invasive activity of
both colon cancer and HCC cell types through the inhibitory effects
on MMP-2 expression and activity. Furthermore, PRMT5 contributed to
the aggressive characteristics of human cancer cells by promoting
their invasive activity.
Discussion
Extensive analysis of PRMT5 in relation to human
cancer has revealed its significant overexpression in various types
of cancer, including breast (27)
and gastric cancer (28), lymphoma
(5,7,29)
leukemia (7,30) and prostate cancer (31–33).
Earlier clinicopathological analyses demonstrated that higher
expression of PRMT5 was correlated with advanced tumor grade,
presence of lymph node metastasis and poor prognosis (19,34,35).
Consistent with previous studies by Zhang et al (17,18)
and Shimizu et al (19),
PRMT5 was significantly overexpressed in our HCC patient tissue
samples, confirming the validity of these findings. Notably, our
clinicopathological findings indicated the association among higher
expression of PRMT5 with more frequent presence of microscopic
hepatic invasion and higher AFP levels. To our knowledge, this is
the first study to report such a correlation and our results
further highlight the importance of PRMT5 as a potential HCC
biomarker.
In addition to in vivo studies, several in
vitro experiments have revealed that depletion of PRMT5 induced
a decrease in cancer cell survival (36) and proliferation (19,28,34,36–38).
While these phenotypes have been observed in various cancer cell
lines, the exact molecular mechanisms remain to be established. In
the present study, we used two different siRNAs to deplete the
expression of PRMT5 in two HCC cell lines and one colon cancer cell
line. Consistently, the knockdown of PRMT5 led to a significant
decrease in the invasiveness of cancer cells, based on data from
the invasion assay and the expression patterns of MMP-2. These
results were in accordance with the previous finding that the
depletion of PRMT5 modulated the expression of E-cadherin through
interactions with Ajuba, to regulate the transcription factor Snail
(6). However, earlier invasion
analyses were performed under conditions whereby the PRMT5 siRNAs
used induced a decrease in cell proliferation and/or survival
(17,18,38).
Decreased proliferation and survival can affect the invasion of
cancer cells, thus reducing cell activity. Notably, in our
experiments, PRMT5 depletion-mediated decrease in invasion activity
was achieved with no adverse effects on colony survival. This
discrepancy may be attributed to differences in the efficacy of
siRNA or the rate of decrease in the expression of PRMT5. Indeed,
the PRMT5 siRNAs used depleted isoforms 1–5 (siRNA#1) and isoforms
1, 2, 4 and 6 (siRNA#2) (Fig.
3C).
Collectively with data from comprehensive earlier
studies using human cancer patient tissues, our present results
highlighted the utility of PRMT5 as a biomarker for various human
cancer types, including HCC. The finding that depletion of PRMT5
induced a marked decrease in cancer cell invasive activity further
supported its potential use as an anticancer therapeutic target.
Further research is warranted to clarify the mechanisms by which
PRMT5 regulates cancer cell invasion activity, including the
substrates involved.
Acknowledgements
The biospecimens and data used in the present study
were provided by the Radiation Tissue Resources Bank of Korea
Cancer Center Hospital (TB-2016-04-C/P20).
Funding
The present study was supported by grants from the
National Research Foundation of Korea (nos. 2012M3A9B6055346,
2017M3A9A8033561 and 2017R1A2B4008805) and the Korea Institute of
Radiological and Medical Sciences (nos. 50531-2018 and 50542-2018)
funded by the Ministry of Science, ICT and Future Planning,
Republic of Korea.
Availability of data and materials
The data and materials used in the present study are
available from the corresponding authors upon reasonable
request.
Authors' contributions
SBM, MBG and KHL designed and guided the study. JYJ,
ERP, YNS and MYK performed the experiments. JYJ, JSL and KHL wrote
the paper. JYJ and JSL performed statistical analysis. HJS and HYJ
reviewed and edited manuscript. EHC, SMM, USS, SHP, CJH, DWC and
SBK provided tissues and generated clinical data. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
This study was approved by the Institutional Review
Board of Korea Cancer Center Hospital. Patient consent was either
waived for liver or obtained for colon cancer tissues.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PRMT5
|
protein arginine methyltransferase
5
|
|
HCC
|
human hepatocellular carcinoma
|
|
MMP
|
matrix metalloproteinase
|
|
AFP
|
α-fetoprotein
|
|
AST
|
aspartate aminotransferase
|
|
ALT
|
alanine transaminase
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment, and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wolf SS: The protein arginine
methyltransferase family: An update about function, new
perspectives and the physiological role in humans. Cell Mol Life
Sci. 66:2109–2121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pal S, Vishwanath SN, Erdjument-Bromage H,
Tempst P and Sif S: Human SWI/SNF-associated PRMT5 methylates
histone H3 arginine 8 and negatively regulates expression of ST7
and NM23 tumor suppressor genes. Mol Cell Biol. 24:9630–9645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pal S, Baiocchi RA, Byrd JC, Grever MR,
Jacob ST and Sif S: Low levels of miR-92b/96 induce PRMT5
translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO
J. 26:3558–3569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou Z, Peng H, Ayyanathan K, Yan KP,
Langer EM, Longmore GD and Rauscher FJ III: The LIM protein AJUBA
recruits protein arginine methyltransferase 5 to mediate
SNAIL-dependent transcriptional repression. Mol Cell Biol.
28:3198–3207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang L, Pal S and Sif S: Protein arginine
methyltransferase 5 suppresses the transcription of the RB family
of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol.
28:6262–6277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stopa N, Krebs JE and Shechter D: The
PRMT5 arginine methyltransferase: Many roles in development, cancer
and beyond. Cell Mol Life Sci. 72:2041–2059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jansson M, Durant ST, Cho EC, Sheahan S,
Edelmann M, Kessler B and La Thangue NB: Arginine methylation
regulates the p53 response. Nat Cell Biol. 10:1431–1439. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo Z, Zheng L, Xu H, Dai H, Zhou M,
Pascua MR, Chen QM and Shen B: Methylation of FEN1 suppresses
nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol.
6:766–773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He W, Ma X, Yang X, Zhao Y, Qiu J and Hang
H: A role for the arginine methylation of Rad9 in checkpoint
control and cellular sensitivity to DNA damage. Nucleic Acids Res.
39:4719–4727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bandyopadhyay S, Harris DP, Adams GN,
Lause GE, McHugh A, Tillmaand EG, Money A, Willard B, Fox PL and
Dicorleto PE: HOXA9 methylation by PRMT5 is essential for
endothelial cell expression of leukocyte adhesion molecules. Mol
Cell Biol. 32:1202–1213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho EC, Zheng S, Munro S, Liu G, Carr SM,
Moehlenbrink J, Lu YC, Stimson L, Khan O, Konietzny R, et al:
Arginine methylation controls growth regulation by E2F-1. EMBO J.
31:1785–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar B, Yadav A, Brown NV, Zhao S,
Cipolla MJ, Wakely PE, Schmitt AC, Baiocchi RA, Teknos TN, Old M,
et al: Nuclear PRMT5, cyclin D1 and IL-6 are associated with poor
outcome in oropharyngeal squamous cell carcinoma patients and is
inversely associated with p16-status. Oncotarget. 8:14847–14859.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanaka H, Hoshikawa Y, Oh-hara T, Koike S,
Naito M, Noda T, Arai H, Tsuruo T and Fujita N: PRMT5, a novel
TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis
via nuclear factor-kappaB activation. Mol Cancer Res. 7:557–569.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nicholas C, Yang J, Peters SB, Bill MA,
Baiocchi RA, Yan F, Sïf S, Tae S, Gaudio E, Wu X, et al: PRMT5 is
upregulated in malignant and metastatic melanoma and regulates
expression of MITF and p27(Kip1.). PLoS One. 8:e747102013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y,
Zhao Q, Shao X, Bu Q, Li H, et al: Targeting protein arginine
methyltransferase 5 inhibits colorectal cancer growth by decreasing
arginine methylation of eIF4E and FGFR3. Oncotarget. 6:22799–22811.
2015.PubMed/NCBI
|
|
18
|
Zhang B, Dong S, Li Z, Lu L, Zhang S, Chen
X, Cen X and Wu Y: Targeting protein arginine methyltransferase 5
inhibits human hepatocellular carcinoma growth via the
downregulation of beta-catenin. J Transl Med. 13:3492015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shimizu D, Kanda M, Sugimoto H, Shibata M,
Tanaka H, Takami H, Iwata N, Hayashi M, Tanaka C, Kobayashi D, et
al: The protein arginine methyltransferase 5 promotes malignant
phenotype of hepatocellular carcinoma cells and is associated with
adverse patient outcomes after curative hepatectomy. Int J Oncol.
50:381–386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prabhu L, Wei H, Chen L, Demir Ö, Sandusky
G, Sun E, Wang J, Mo J, Zeng L, Fishel M, et al: Adapting AlphaLISA
high throughput screen to discover a novel small-molecule inhibitor
targeting protein arginine methyltransferase 5 in pancreatic and
colorectal cancers. Oncotarget. 8:39963–39977. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim BY, Suh KS, Lee JG, Woo SR, Park IC,
Park SH, Han CJ, Kim SB, Jeong SH, Yeom YI, et al: Integrated
analysis of prognostic gene expression profiles from hepatitis B
virus-positive hepatocellular carcinoma and adjacent liver tissue.
Ann Surg Oncol. 19 Suppl 3:S328–S338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim BY, Choi DW, Woo SR, Park ER, Lee JG,
Kim SH, Koo I, Park SH, Han CJ, Kim SB, et al:
Recurrence-associated pathways in hepatitis B virus-positive
hepatocellular carcinoma. BMC Genomics. 16:2792015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park ER, Kim SB, Lee JS, Kim YH, Lee DH,
Cho EH, Park SH, Han CJ, Kim BY, Choi DW, et al: The mitochondrial
hinge protein, UQCRH, is a novel prognostic factor for
hepatocellular carcinoma. Cancer Med. 6:749–760. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seftor RE, Seftor EA, Gehlsen KR,
Stetler-Stevenson WG, Brown PD, Ruoslahti E and Hendrix MJ: Role of
the alpha v beta 3 integrin in human melanoma cell invasion. Proc
Natl Acad Sci USA. 89:1557–1561. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brooks PC, Strömblad S, Sanders LC, von
Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP and
Cheresh DA: Localization of matrix metalloproteinase MMP-2 to the
surface of invasive cells by interaction with integrin alpha v beta
3. Cell. 85:683–693. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muschel RJ and Yong J: The Gelatinases,
MMP-2 and MMP-9-Implications for invasion and metastasisProteases
and Their Inhibitors in Cancer Metastasis. Foidart JM and Muschel
RJ: Springer Netherlands; Dordrecht: pp. 39–52. 2002, View Article : Google Scholar
|
|
27
|
Wu Y, Wang Z, Zhang J and Ling R: Elevated
expression of protein arginine methyltransferase 5 predicts the
poor prognosis of breast cancer. Tumour Biol.
39:10104283176959172017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda M, Shimizu D, Fujii T, Tanaka H,
Shibata M, Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, et
al: Protein arginine methyltransferase 5 is associated with
malignant phenotype and peritoneal metastasis in gastric cancer.
Int J Oncol. 49:1195–1202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chung J, Karkhanis V, Tae S, Yan F, Smith
P, Ayers LW, Agostinelli C, Pileri S, Denis GV, Baiocchi RA, et al:
Protein arginine methyltransferase 5 (PRMT5) inhibition induces
lymphoma cell death through reactivation of the retinoblastoma
tumor suppressor pathway and polycomb repressor complex 2 (PRC2)
silencing. J Biol Chem. 288:35534–35547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y,
Du X, Li J, Li P, Ren R, et al: Targeting methyltransferase PRMT5
eliminates leukemia stem cells in chronic myelogenous leukemia. J
Clin Invest. 126:3961–3980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu Z, Li Y, Lee P, Liu T, Wan C and Wang
Z: Protein arginine methyltransferase 5 functions in opposite ways
in the cytoplasm and nucleus of prostate cancer cells. PLoS One.
7:e440332012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang HT, Zhang D, Zha ZG and Hu CD:
Transcriptional activation of PRMT5 by NF-Y is required for cell
growth and negatively regulated by the PKC/c-Fos signaling in
prostate cancer cells. Biochim Biophys Acta. 1839:1330–1340. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deng X, Shao G, Zhang HT, Li C, Zhang D,
Cheng L, Elzey BD, Pili R, Ratliff TL, Huang J, et al: Protein
arginine methyltransferase 5 functions as an epigenetic activator
of the androgen receptor to promote prostate cancer cell growth.
Oncogene. 36:1223–1231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bao X, Zhao S, Liu T, Liu Y, Liu Y and
Yang X: Overexpression of PRMT5 promotes tumor cell growth and is
associated with poor disease prognosis in epithelial ovarian
cancer. J Histochem Cytochem. 61:206–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shilo K, Wu X, Sharma S, Welliver M, Duan
W, Villalona-Calero M, Fukuoka J, Sif S, Baiocchi R, Hitchcock CL,
et al: Cellular localization of protein arginine
methyltransferase-5 correlates with grade of lung tumors. Diagn
Pathol. 8:2012013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen H, Lorton B, Gupta V and Shechter D:
A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through
histone H3 and H4 arginine methylation coupled transcriptional
activation and repression. Oncogene. 36:373–386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen D, Zeng S, Huang M, Xu H, Liang L and
Yang X: Role of protein arginine methyltransferase 5 in
inflammation and migration of fibroblast-like synoviocytes in
rheumatoid arthritis. J Cell Mol Med. 21:781–790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saha K, Fisher ML, Adhikary G, Grun D and
Eckert RL: Sulforaphane suppresses PRMT5/MEP50 function in
epidermal squamous cell carcinoma leading to reduced tumor
formation. Carcinogenesis. 38:827–836. 2017. View Article : Google Scholar : PubMed/NCBI
|