Introduction
Oral cancer is a major public health issue and a
social challenge. Tongue squamous cell carcinoma (TSCC) is the most
aggressive type of oral cancer. Environmental factors, genetics and
immune status have been confirmed to be related to the
tumorigenesis of tongue cancer (1).
The tumor microenvironment, which is composed of complex components
including stem cells, fibroblasts, immune cells and their secreted
factors, is critical to the initiation, development and maintenance
of tumorigenesis (1). As the
‘residence niche’ of cancer cells, the composition and structure of
the tumor microenvironment not only affects the bio-behavior, but
also determines the survival and elimination of tumor cells
(1–3). Targeting the tumor microenvironment by
remodeling its components and construction could be a useful a
strategy to treat and prevent cancer (4).
Low pH and hypoxia are unique characteristics of the
tumor microenvironment (5,6). Glucose metabolism supplies energy for
the growth and maintainence of mammalian cells (7), and cancer cells are well-known to
employ glycolysis for energy metabolism (8,9). As
soon as the primary tumor outgrows its blood supply and creates a
hypoxic microenvironment, cancer cells typically switch to a
glycolytic metabolism and secrete lactic acid, creating a low pH
microenvironment (10–14). An acidic and hypoxic
microenvironment is not only toxic to the surrounding normal
somatic cells, but also induces the expression of HIF-1α, inhibits
antitumor immune responses and promotes proliferation, invasion,
metastases and recurrence (12,15–17).
Investigation and elucidation of the signaling mechanism of hypoxic
metabolism of cancer cells is critical to explore new cancer
treatment strategies (18).
Protein kinase D (PKD) is an evolutionarily
conserved serine/threonine protein kinase with structural,
enzymological and regulatory properties distinct from those of PKC
(19). PKD belongs to the
calmodulin-dependent protein kinase (CAMK) superfamily that also
includes PKD1/PKCμ, PKD2 and PKD3/PKCν (19,20).
PKDs can be activated by a number of external stimuli, including
G-protein-coupled receptor agonists, hormones, chemokines,
bioactive lipids and growth factors (19,21).
PKDs mediate diverse and complex biological functions throughout
the body, including cell proliferation and differentiation, signal
transduction, membrane trafficking, secretion, immunoregulation and
angiogenesis (19,21,22).
PKDs play important roles in tumor proliferation and
differentiation, migration, apoptosis, epithelial-to-mesenchymal
transition (EMT), angiogenesis, MDR, oxidative stress, autophagy
and transcriptional regulation (20,23–25).
PKD1, one of the most well-studied PKD family members, is expressed
and activated to varying degrees in cancer cells from many
different tissues of origin, and has distinct roles based on the
tissue of origin (19,24,26).
It is overexpressed in pancreatic cancer and skin cancers. However,
the expression of PKD1 is downregulated in prostate, breast and
gastric cancer (19,23,27,28).
Abnormal expression of PKD1 is often associated with tumorigenesis,
progression, apoptosis, invasion and poor prognosis (19,25,29).
In the present study, we investigated the role of
PKD1 in hypoxic glycolysis in SCC25 cancer cells. We found that
hypoxia not only induced the expression of HIF-1α, but also
promoted PKD1 expression and activation, through phosphorylation.
PKD1 not only regulated the growth and apoptosis of SCC25 cells
under hypoxic conditions, but also promoted glycolytic metabolism
of SCC25 cells by promoting the expression of HIF-1α, GLUT1, LDHA
and p38 MAPK within a hypoxic microenvironment.
Materials and methods
Antibodies and reagents
Antibodies against PKD1 rabbit mAb (cat. no. 90039),
phospho-PKD1 rabbit Ab (cat. no. 2051), p38 MAPK rabbit mAb (cat.
no. 8690), phospho-p38 MAPK (Thr180/Tyr182) rabbit mAb (cat. no.
4511), LDHA rabbit mAb (cat. no. 3582) and β-actin rabbit mAb (cat.
no. 8457) were obtained from Cell Signaling Technology, Inc.
(Beverly, MA, USA). Antibodies against HIF-1α rabbit mAb (cat. no.
ab51608), GLUT-1 rabbit pAb (cat. no. ab51608) and LC3 rabbit pAb
(cat. no. ab51520) were obtained from Abcam (Cambridge, MA, USA).
Annexin V-FITC apoptosis detection and lactate assay kits were
purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). We
purchased 2-NBDG
[2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose]
from Cayman Chemicals (Ann Arbor, MI, USA). Human PKD1 shRNA and
control shRNA plasmids were obtained from Thermo Fisher Scientific,
Inc. (Waltham, MA, USA). Human pEZ-Lv105-PKD1-overexpressing
plasmid vectors were purchased from GeneCopoeia, Inc. (Rockville,
MD, USA). The Cell Counting Kit (CCK-8) was obtained from Dojindo
Molecular Technologies, Inc. (Kumamoto, Japan).
Cell culture and hypoxia
treatment
Human oral squamous cell carcinoma (SCC25) cells
were cultured in DME/F-12 (1:1) medium supplemented with 10%
(vol/vol) fetal bovine serum (FBS), 1% (vol/vol)
penicillin/streptomycin mixture and 10μM hydrocortisone (both from
Gibco, Thermo Fisher Scientific, Inc.). H1975, SCC25 and HSC-4
cells used in this study were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA) and cultured at the
State Key Laboratory of Oral Diseases in a humidified atmosphere of
5% CO2 at 37°C. To induce hypoxia, SCC25 cells were
placed in a chamber filled with a gas mixture of 1% O2,
5% CO2 and 94% N2.
Establishment of stable PKD1-knockdown
and -overexpressing SCC25 cells
Human LVRH1GP-PKD1 shRNA (clone ID NM_002742.2) and
LVRU6GP-control shRNA plasmids were obtained from Thermo Fisher
Scientific, Inc. The PKD1-1-SH target sequences are as follows:
gcaacaatat cccactcatga. PKD1-2-SH target sequences are as follows:
gcaggtactacaaggaaattc. The shRNAs were transfected into the SCC25
cells using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Positive SCC25 clones were selected for at least two passages by
using puromycin treatment at a concentration of 0.5 µg/ml. After
two weeks of growth in selective medium, the cells were harvested
and the expression level of PKD1 was determined by western blot
analysis. Stable PKD1-overexpressing SCC25 cells were generated
similarly.
Cell viability assay
The CCK-8 was used to detect the cell viability of
the SCC25 cell group (as the normal control group), the
PKD1-knockdown group (PKD1-SH), PKD1-overexpressing group (PKD1-OE)
and SCC25 control groups (Con-SH and Con-OE). SCC25 cells were
seeded at a density of 1×103 cells/well in 96-well
plates and cultured at 37°C in 1% O2, 5% CO2
and 94% N2. Cell viability was assessed by measuring the
absorbance of the converted dye at 450 nm.
Annexin V-FITC apoptosis assay
SCC25 cells (wild-type SCC25, PKD1-SH, Con-SH,
PKD1-OE and Con-OE) were seeded in 6-well plates at a density of
2×105 cells/well and cultured at 37°C in 1%
O2, 5% CO2 and 94% N2. Annexin
V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for
flow cytometry was used to identify the percentage of apoptotic
cells following the manufacturer's protocol [Annexin V-FITC
apoptosis detection kit (Sigma-Aldrich; Merck KGaA)]. Annexin
V+/PI− apoptotic cells were identified via
flow cytometry (FCM) using Summit 5.2 software (Beckman Coulter,
Miami, FL, USA).
Glucose uptake assay
To analyze glucose uptake, the fluorescent glucose
analog 2-NBDG was used for direct quantification of glucose
incorporation in living cells by FCM (Beckman Coulter) (7,30).
SCC25 cells were seeded in 6-well plates and subjected to hypoxia
(1% O2, 5% CO2 and 94% N2). After
treatment for 24 h, the medium was removed and new medium
containing 10 µM 2-NBDG was added. The cells were then incubated at
37°C in 1% O2, 5% CO2 and 94% N2
for 30 min. Cells were washed twice with Dulbecco's
phosphate-buffered saline (DPBS; Invitrogen; Thermo Fisher
Scientific, Inc.) then trypsinized, centrifuged 1,200 rpm for 10
min and resuspended in DPBS. A flow cytometer was used to analyze
the uptake of 2-NBDG in the different cell groups.
Lactate assay
SCC25 cells were seeded in 6-well plates at a
density of 2×105 cells/well and cultured at 37°C in 1%
O2, 5% CO2 and 94% N2 for 24 h.
Lactate in the medium was measured using a lactate assay kit
according to the manufacturer's instructions (Sigma-Aldrich; Merck
KGaA).
Western blot analysis
Cells were collected and protein was collected using
a Protein Extraction kit (Sigma-Aldrich; Merck KGaA). The protein
concentration was measured using a BCA protein assay kit (Thermo
Fisher Scientific, Inc.). Total protein (20 µg) was separated using
6 or 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad
Laboratories, Hercules, CA, USA). The membranes were blocked with
5% skim milk and incubated with primary antibodies against PKD1,
phospho-PKD1, HIF-1α, LC3, GLUT-1, LDHA, phospho-p38 MAPK, p38 MAPK
or β-actin at a dilution of 1:1,000 at 4°C overnight. Membranes
were washed with TBST and then incubated with horseradish
peroxidase (HRP)-conjugated secondary antibodies at a dilution of
1:2,000 for 2 h at room temperature. Luminescent signals were
detected by ECL Western Blotting Substrate (Millipore, Billerica,
MA, USA). Band intensity was quantified using Quantity One Software
(Bio-Rad Laboratories). Phospho-p38 MAPK/p38 MAPK was defined as
the phosphorylated index (PI) of p38 MAPK.
Statistical analysis
All data are presented as mean ± standard deviation
(SD). Significant differences between groups were analyzed by
one-way analysis of variance (ANOVA), followed by a Bonferroni's
post hoc test. A P-value <0.05 was considered to indicate a
statistically significant difference. Graphs were plotted and
analyses were performed using GraphPad Prism 7 software (GraphPad
Software, Inc., La Jolla, CA, USA).
Results
Hypoxia induces PKD1 expression in
SCC25 cancer cells
The SCC25 cell line is the most common type of human
oral squamous cell carcinoma. In our pre-experiment, we
investigated the expression of PKD1 in H1975, SCC25 and HSC-4
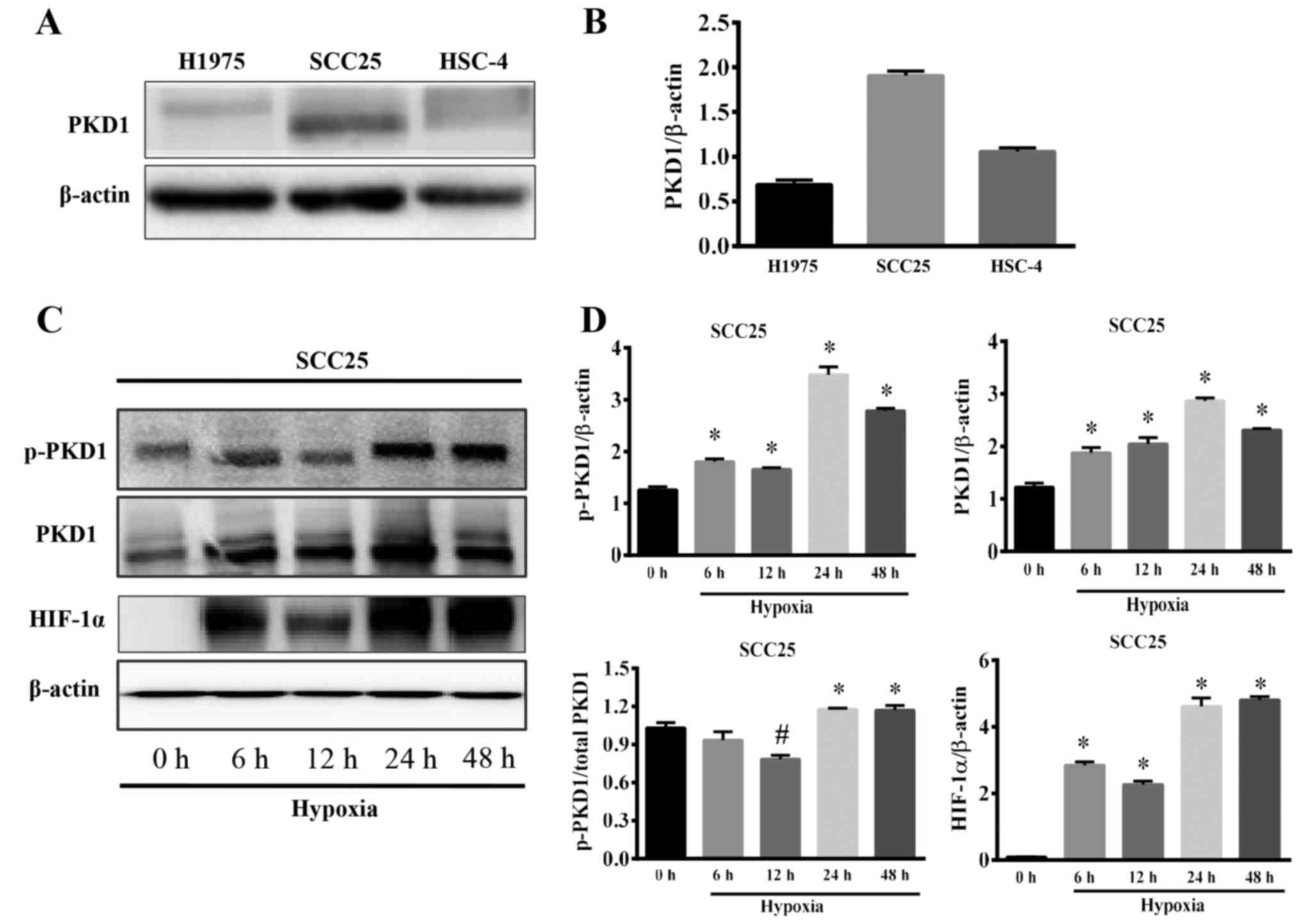
cancer cell lines. The results showed that PKD1 was expressed in
SCC25 cells at a high level (Fig. 1A
and B); therefore, we selected SCC25 as the target cell line
for further study.
Hypoxia is a common characteristic of the tumor
microenvironment. Hypoxia induces cancer cells to express HIF-1α to
initiate the expression of tumor growth factor and provide a growth
signal for cancer cells. On the other hand, hypoxia promotes cancer
cell switch to glycolysis for energy generation. To determine the
role of PKD1 in regulating glycolytic metabolic progression of
tumor cells, we first analyzed the expression and activation of
PKD1 in tumor cells under a hypoxic microenvironment. SCC25 cells
were cultured in a humidified atmosphere of 5% CO2, 1%
O2 and 94% N2 at 37°C to simulate a hypoxic
tumor microenvironment. The expression of PKD1, p-PKD1 and HIF-1α
were detected by western blot analysis. As shown in Fig. 1C, SCC25 cells exposed to 6 h of
hypoxia exhibited enhanced expression of p-PKD1, PKD1 and HIF-1α.
In particular, when SCC25 cells were subjected to 24 h of hypoxia,
the expression of p-PKD1, PKD1 and HIF-1α reached the highest level
(Fig. 1D). Moreover, the ratio of
p-PKD1/PKD1 was significantly increased (Fig. 1D, left lower panel). Based on the
above-mentioned research results, treatment with 24 h hypoxia was
selected for the subsequent experiments. This result indicates that
hypoxia not only induces the expression of PKD1, but also induces
the phosphorylation and activation of PKD1.
PKD1 promotes the growth and inhibits
the apoptosis of SCC25 cells in a hypoxic microenvironment
Toleration of a hypoxic microenvironment is a
characteristic of tumor cells. To investigate the role of PKD1 in
the growth and apoptosis of tumor cells grown under hypoxia, SCC25
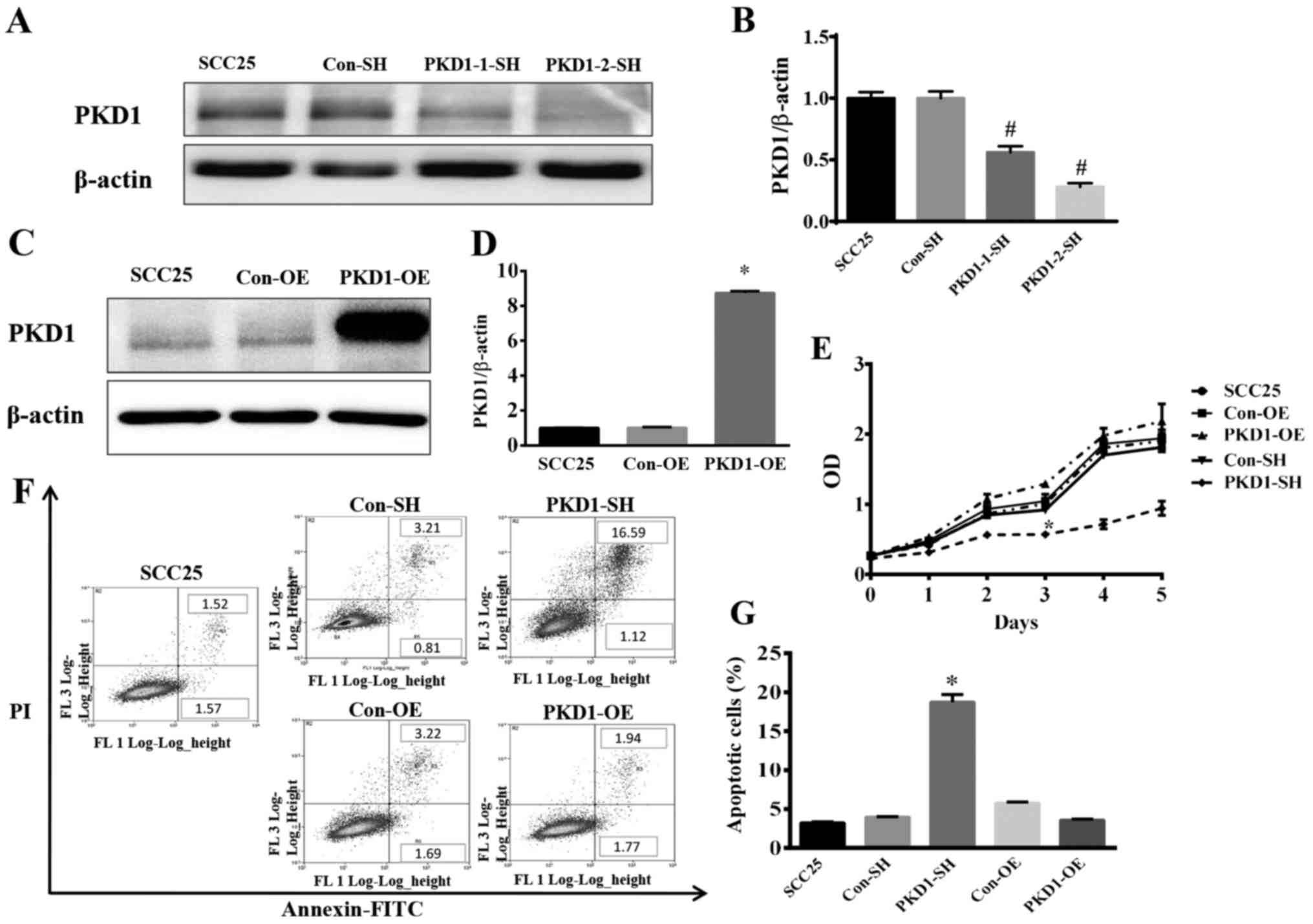
cells were transfected with PKD1 shRNA (PKD1-1-SH and PKD1-2-SH) to
knockdown PKD1. Compared with the negative control (Con-SH), the
expression of PKD1 was significantly reduced, as shown by western
blot analysis (Fig. 2A and B). As
shown in Fig. 2A and B, PKD1-2-SH
was found to be the most effective shRNA plasmid with which to
knockdown PKD1. Thus, we used PKD1-2-SH to knockdown PKD1 in the
following experiments. Additionally, SCC25 cells were transfected
with a pCDNA3.1/NT-PKD1 plasmid with a pre-inserted PKD1 gene for
PKD1 overexpression (PKD1-OE) (Fig. 2C
and D). PKD1-SH, PKD1-OE and wild-type SCC25 cells were
cultured in a humidified atmosphere of 1% O2, 5%
CO2 and 94% N2 at 37°C. Live cells were
analyzed using the CCK-8 cell counting kit; apoptotic cells were
analyzed via FCM. As shown in Fig.
2E, the cell growth curve demonstrated a significantly highest
cell death at 3 days in the PKD1-SH group when compared with the
other 4 tested groups. Importantly, SCC25 cell apoptosis was
significantly increased after PKD1 knockdown with PKD1-SH (Fig. 2F and G). However, the results
revealed that there was no significant difference in growth between
the PKD1-Con and PKD1-OE group, and growth of the
PKD1-overexpressing cells was not markedly affected according to
the cell growth curve (Fig. 2E).
These results indicate that PKD1 plays an important role in
maintaining the survival and growth of SCC25 cells in a hypoxic
microenvironment.
PKD1 regulates HIF-1α expression
PKD1 is a protease kinase that is involved in the
bio-behavior of cancer cells. HIF-1α is an important regulator of
the glucose metabolism in cancer cells. To reveal the role of PKD1
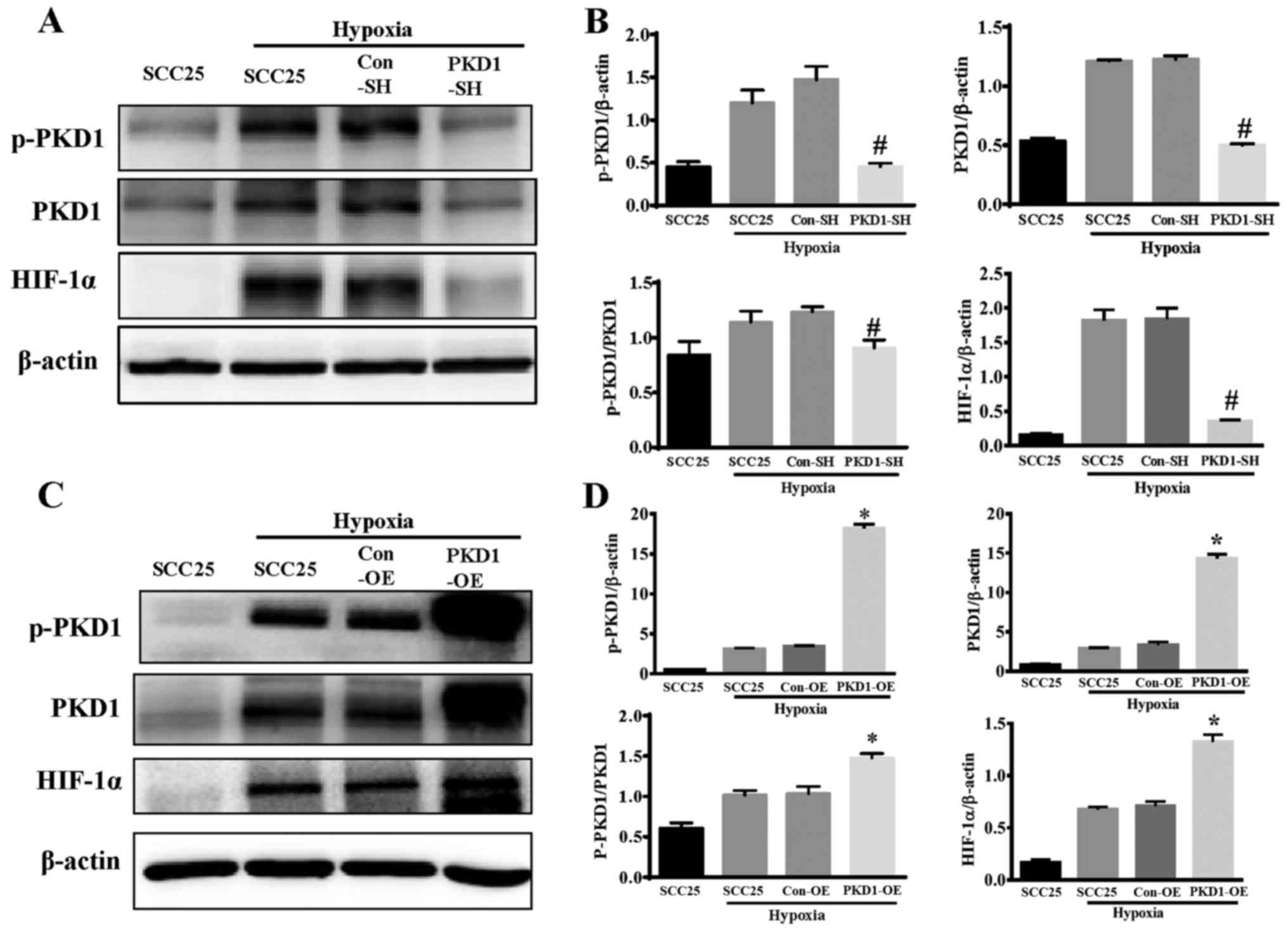
in the progression of hypoxia-induced HIF-1α expression, wild-type
SCC25, PKD1-SH and PKD1-OESCC25 cells were cultured in a humidified
atmosphere of 5% CO2, 1% O2 and 94%
N2 at 37°C for 24 h. SCC25 cells cultured in normal
condition as a control, PKD1-knockdown or -overexpressing SCC25
cells did not exhibit a significant difference in the expression of
HIF-1α under a normal condition (data not shown). PKD1 and HIF-1α
expression was evaluated by western blot analysis. As shown in
Fig. 3, the phosphorylation and
activation of PKD1 and HIF-1α expression was significantly
increased in the SCC25 cells under hypoxia. Moreover, the
expression of HIF-1α was significantly inhibited after PKD1
knockdown by PKD1-SH (Fig. 3A and
B). On the other hand, overexpression of PKD1 and
phosphorylated activation significantly promoted hypoxia-induced
HIF-1α expression (Fig. 3C and D).
These results indicate that PKD1 is an important regulator of
hypoxia-induced HIF-1α expression.
PKD1 promotes glycolysis progression
in SCC25 cells
The Warburg effect is a unique characteristic of
cancer cell glycometabolism. Cancer cells uptake glucose and
release lactate through glycolysis under hypoxic conditions.
Detection of the ability of cancer cells to uptake glucose and
release lactate in culture media can reveal the glycolytic activity
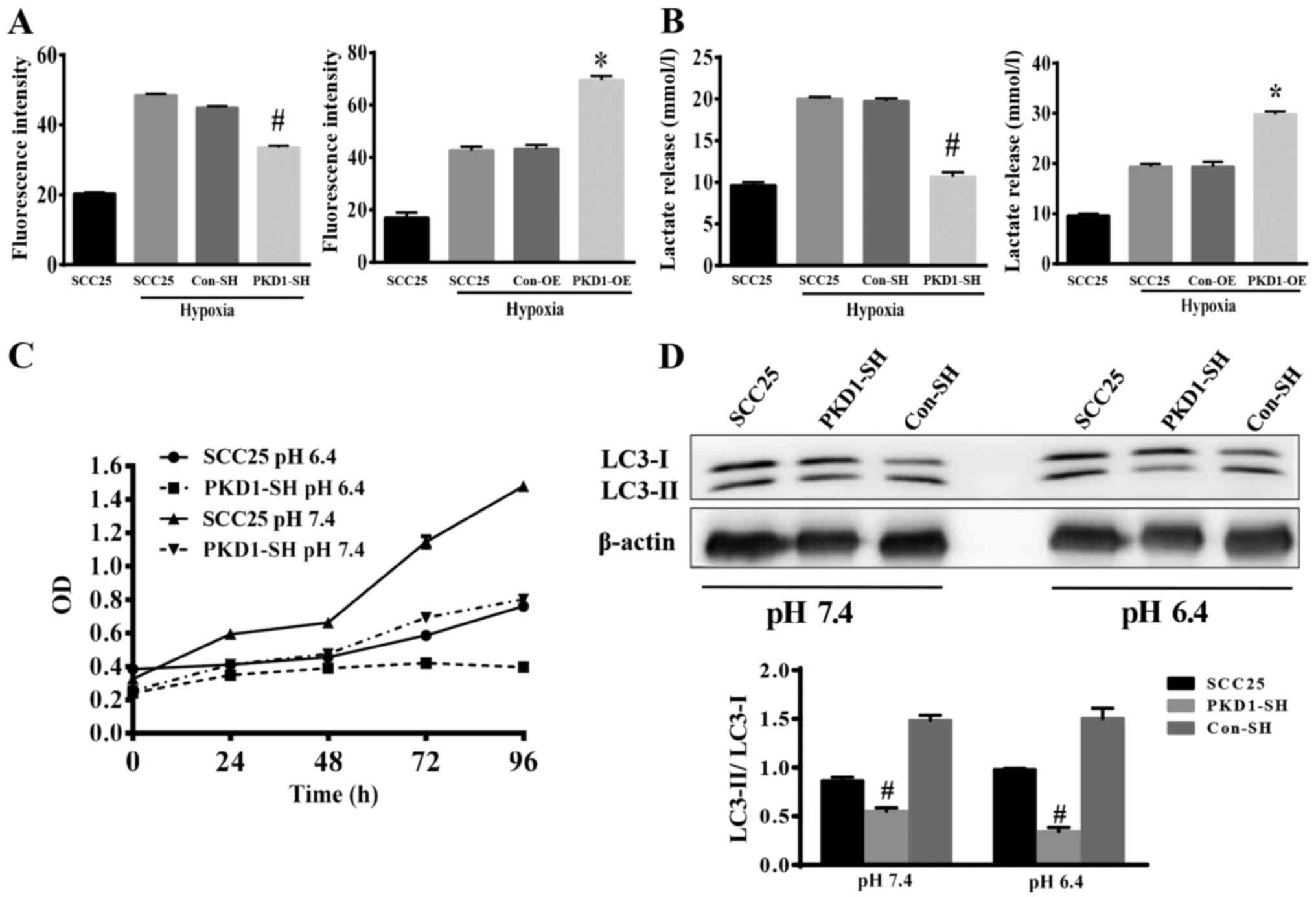
of cancer cells. Wild-type SCC25, PKD1-SH and PKD1-OESCC25 cells
were cultured in medium with 10 nM 2-NBDG for 30 min. Glucose
uptake and lactate release by cancer cells were analyzed via FCM
and lactate assay kits, respectively. As shown in Fig. 4A and B, hypoxia stimulated SCC25
cells to uptake glucose and release lactate. When PKD1 was knocked
down with PKD1-SH, the ability of SCC25 cells to uptake glucose was
significantly reduced (Fig. 4A,
left panel), as lactate release was also reduced (Fig. 4B, left panel). Conversely, when PKD1
was overexpressed, the ability of SCC25 cells to uptake glucose and
release lactate was significantly increased (Fig. 4A and B, right panels). These results
indicate that PKD1 is an important signaling molecule promoting
cancer cell glycolysis.
We also detected the growth and the expression of
autophagy proteins by SCC25 cells cultured under an acidic
environment after knockdown of PKD1. As shown in Fig. 4C, the growth of SCC25 cells was
significantly inhibited under an acidic condition. Fig. 4D shows that the expression of
autophagy proteins was significantly decreased (LC3-II/LC-3I).
These data indicated that PKD1 moderated autophagy to promote the
adaptation of tumor cells to an acidic microenvironment.
PKD1 promotes cancer cel lglycolysis
by promoting GLUT1 and LDHA expression
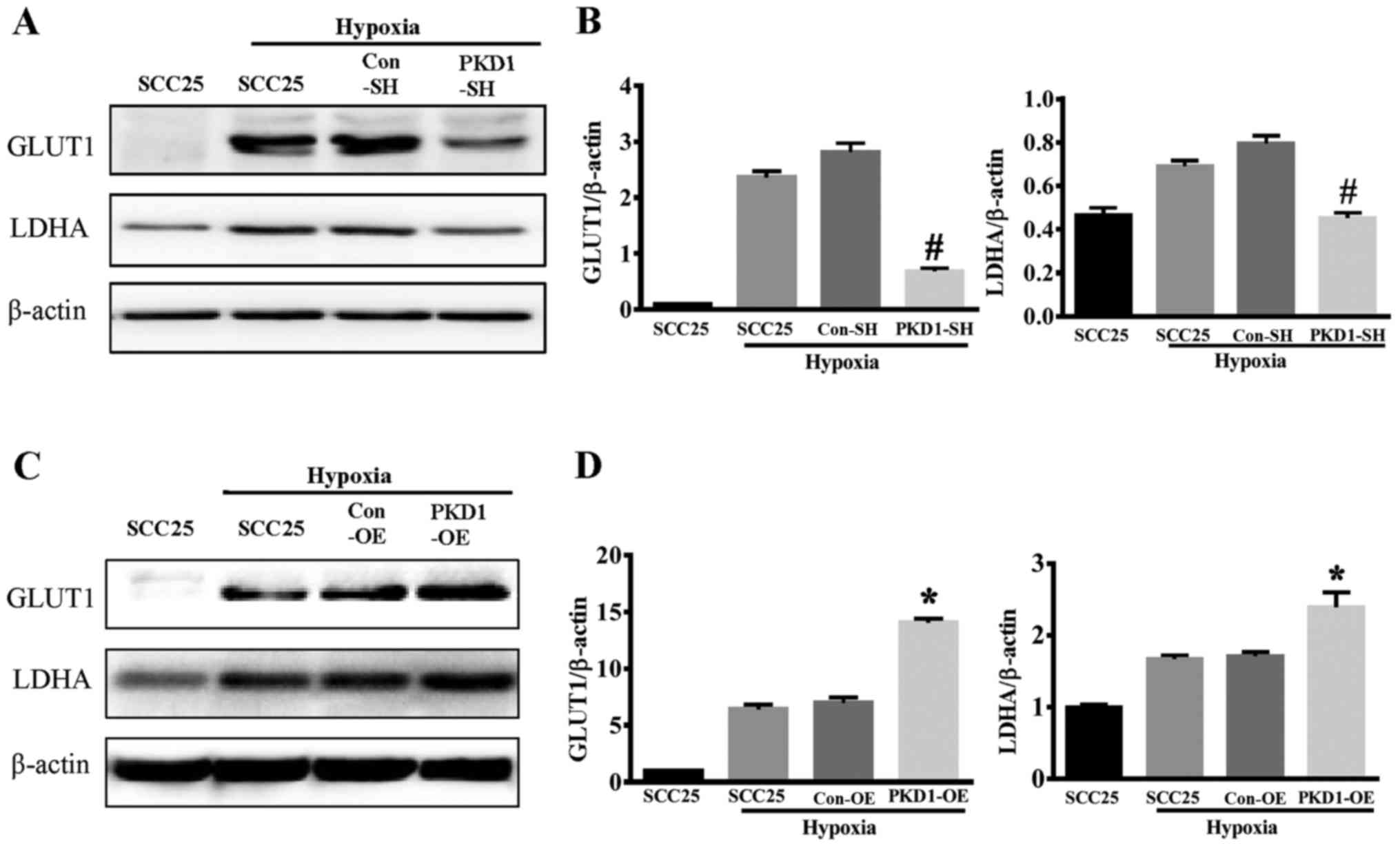
GLUT1 and LDHA are two key enzymes involved in the
process of glycolysis. Hypoxia induced both GLUT1 and LDHA
expression in SCC25 cancer cells (Fig.
5). When PKD1 was knocked down with PKD1-SH, the expression of
GLUT1 and LDHA was significantly reduced as compared to wild-type
SCC25 or control-SHSCC25 cells under hypoxia (Fig. 5A and B). Conversely,
PKD1-overexpressing SCC25 cells had significantly increased
expression of GLUT1 and LDHA under hypoxic conditions (Fig. 5C and D). These results indicate that
PKD1 promotes glycolysis in oral squamous cancer cells by inducing
the expression of glycolytic enzymes under hypoxic
microenvironments.
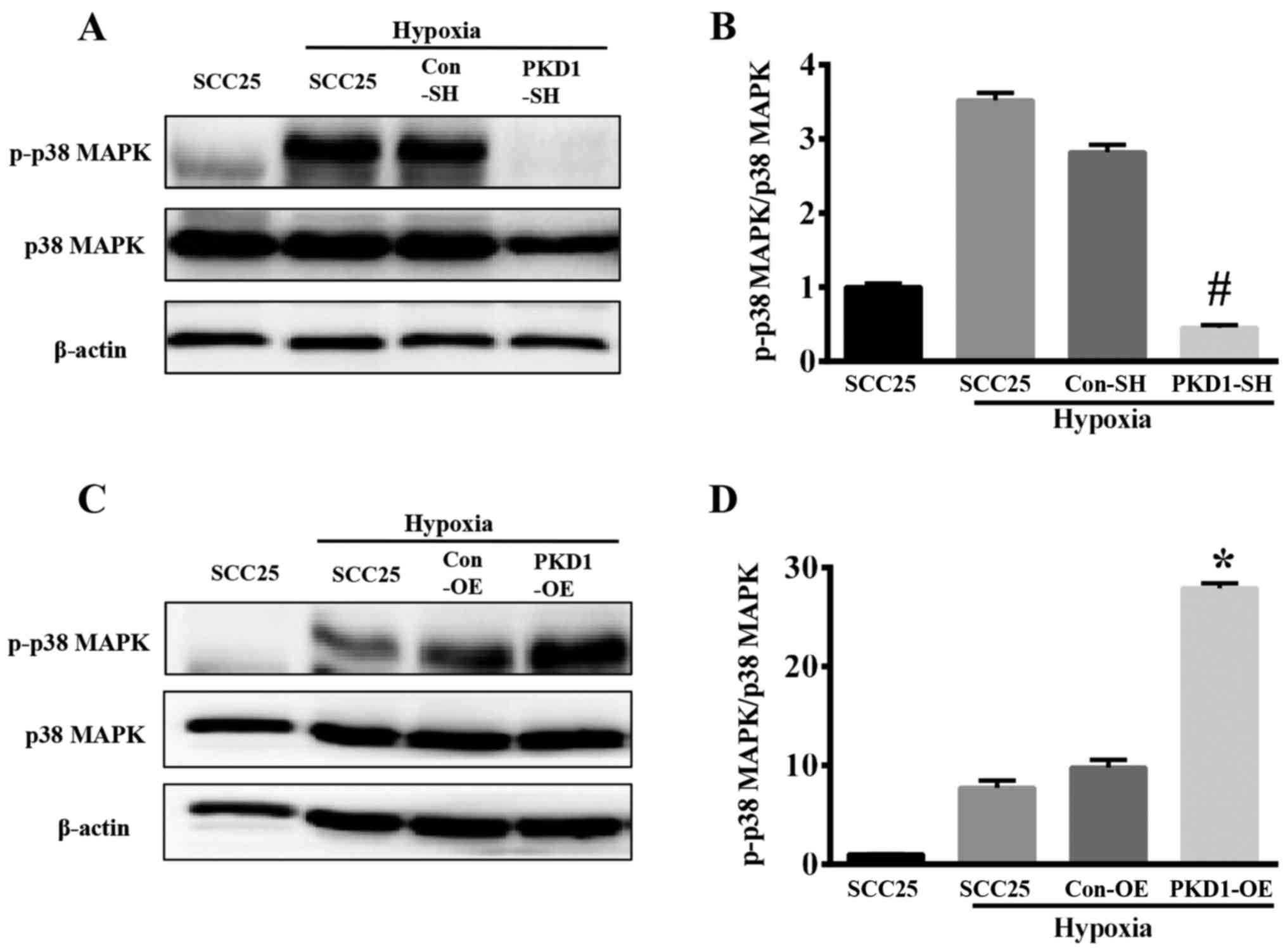
Hypoxia induces the expression and
activation of p-38 MAPK via PKD1
The p38 MAPKs are members of the MAPK family that
are activated by a variety of environmental stresses and
inflammatory cytokines. As shown in Fig. 6, hypoxia is an important
environmental stress that activates p38 MAPK. SCC25 cells
(wild-type SCC25, PKD1-SH and PKD1-OE) were cultured in a hypoxic
environment for 24 h, and the expression of p-p38 MAPK and p38 MAPK
was analyzed via western blot analysis. The phosphorylation level
of p38 MAPK was increased under hypoxia (Fig. 6A and C), whereas the expression
level of total p38 MAPK was not statistically significantly
different. The phosphorylation level of p38 MAPK and phosphorylated
index (PI) of p-p38 MAPK/p38 MAPK were significantly decreased when
SCC25 cells were transfected with PKD1-SH in a hypoxic environment
for 24 h (Fig. 6A and B). Moreover,
the level of p-p38 MAPK and the PI of p-p38 MAPK/p38 MAPK were
significantly increased when the PKD1-OE SCC25 cells were treated
with hypoxia (Fig. 6C and D),
compared with the expression level of total p38 MAPK.
Discussion
The tumor microenvironment is the ‘resident niche’
of cancer cells (1). A change in
the tumor microenvironment can determine the survival or
elimination of tumor cells (18).
Targeting the tumor microenvironment has thus become an attractive
strategy for cancer biotherapy (4).
Hypoxia and low pH are unique characteristics of the
tumor microenvironment (6). When
tumor growth outpaces angiogenesis within the tumor, the
O2 level can be entirely suppressed within cancerous
tissues (31,32). Thus, tumor cells switch to a
glycolytic metabolism to generate energy and secrete lactic acid,
creating an acidic, hypoxic microenvironment (12,17).
Hypoxia induces the expression of HIF-1α, which controls the
expression of hundreds of genes including tumor growth factors,
metabolic enzymes and pro-metastatic factors that can promote the
growth, metastasis and glucose metabolism of cancer cells,
respectively. This unique microenvironment is also toxic to
untransformed cells, inhibiting the antitumor immune response and
increasing cancer cell drug resistance (33–36).
As a result, cancer cells acquire an optimal ‘resident
microenvironment’ and escape predation of the immune system
(37,38).
In the past decade, cancer metabolism has received a
substantial amount of interest. Along with the progress of cancer
genomics, proteomics and metabolomics, the connections between
oncogenic signaling pathways and metabolic reprogramming in cancer
are being increasingly recognized (39,40).
Oncogenic signaling drives many of the metabolic
responses of normal cells to growth-promoting signals (41). For example, AKT activation can
increase glucose uptake, enhance activation and mitochondrial
localization of hexokinase, and increase glycolytic flux. Both
rapamycin complex 1 (mTORC1) and hypoxia-inducible factor (HIF)
contribute to the increased expression and activation of glycolytic
enzymes (35,42). Additionally, MYC can promote the
splicing of the pyruvate kinase gene PKM and enhance the expression
of PKM2 (43).
PKD1, one of the PKD family members, is
constitutively expressed in a variety of tumor cells (19). PKD1 signaling is involved in
multiple bio-behaviors of cancer cells, including proliferation,
apoptosis, migration, angiogenesis, oxidative stress response,
autophagy and invasion (19). The
expression and activation of PKD1 can be induced by multiple
stimuli, such as growth factors, oxidative stress and chemokines
(19,44). In the present study, we found that
hypoxia not only induced the expression of HIF-1α, but also induced
the expression and activation (via phosphorylation) of PKD1 in
SCC25 cells after culture for 6 h under hypoxic condition. Although
the phosphorylated PKD1 decreased at 12 h, it soon increased and
arrived at a peak at 24 h (Fig. 1).
Notably, although the phosphorylated PKD1 decreased at 6 h, the
expression of PKD1 and HIF-1α still maintained a high level. This
indicates that PKD1 is involved in the expression of HIF-1α which
is induced by hypoxia.
Both PKD1 and HIF-1α are important regulators of
cancer cell proliferation, differentiation, apoptosis and
angiogenesis (19,24). Furthermore, HIF-1α is also an
important regulator of cancer metabolism under hypoxic conditions
(35). This study presents the role
of PKD1 in the growth, apoptosis and metabolism under hypoxia in
the most common type of human oral squamous cell carcinoma cells,
SCC25. We cultured SCC25 cells in 5% CO2, 1%
O2 and 94% N2 to simulate a hypoxic tumor
microenvironment. When PKD1 expression was inhibited with shRNA,
cell growth was significantly inhibited. In contrast, SCC25 cells
expressed a high level of PKD1 under a hypoxic condition, thus, we
transfected the exogenous PKD1 gene into the cells. Overexpression
of PKD1 did not significantly affect the growth and apoptosis of
SCC25 cells (Fig. 2). This
indicates that PKD1 is a key regulator of cancer cell adaptation to
the hypoxic microenvironment.
Hypoxia induces the expression of HIF-1α to initiate
the expression of tumor growth factors and glycolytic enzymes
(45,46), which provide growth signals and
energy for tumor cells to adapt to and survive in a hypoxic
microenvironment (45). After our
previous finding that PKD1 promotes cancer cell growth and survival
under hypoxia, we became interested in the role of PKD1 in cancer
cell glycolysis under hypoxic conditions. We used RNA interference
and enforced expression to suppressor overexpress PKD1 in oral
squamous cell carcinoma cells (SCC25), cultured in 5%
CO2, 1% O2 and 94% N2 to simulate
the hypoxic microenvironment. PKD1 knockdown with shRNA
significantly decreased not only the expression of HIF-1α (Fig. 3), but also the expression of GLUT1
and LDHA (Fig. 5). Similarly, when
PKD1 was overexpressed in SCC25 cells, HIF-1α, GLUT1 and LDHA
expression was significantly increased (Figs. 3 and 5). It is clear that HIF-1α is a
transcription regulator under hypoxia conditions. HIF-1α regulates
the metabolism of cancer cells by upregulating the expressions of
glucose metabolism-related genes such as ENO1, pyruvate kinase 2
(PKM2), phosphoglycerate kinase 1 (PGK1), GLUT1 and LDHA in
response to hypoxia (30). GLUT1 is
the major carrier mediating glucose transport across mammalian cell
membranes (9,30). The function of LHDA is to convert
pyruvate, one of the products of glucose metabolism, to lactate,
which is then released into the extracellular milieu, lowering the
pH of the tumor microenvironment (6,47).
Following the decreased expression of GLUT1 and LDHA after PKD1
inhibition, the ability of SCC25 cells to uptake glucose and
release lactate was also decreased (Fig. 4). Conversely, when PKD1 was
overexpressed in SCC25 cells, the ability of cancer cells to uptake
glucose and release lactate was increased (Fig. 4). These results support the
hypothesis that PKD1 is an important regulator of cancer cell
glycolytic metabolism and the creation of an acidic
microenvironment.
We next analyzed the expression and phosphorylation
of p38 MAPK after PKD1 inhibition or overexpression under hypoxic
conditions. p38 MAPK, which belongs to the MAPK superfamily, is a
well-studied stress-activated kinase that transmits numerous
extracellular signals and is involved in multiple cellular events
(48,49). p38 MAPK acts as a double-edged
sword; in early stages, it acts as a tumor suppressor; however, it
is required for cell survival in later stages of tumor progression
(50). Hypoxia not only induces the
expression of HIF-1α, but also increases the phosphorylation and
activation of p38 MAPK in cancer cells (51). The activation of p38 MAPK signaling
is necessary for HIF-1α accumulation and nuclear translocation
(52). In the present study, we
observed that the knockdown and overexpression of PKD1 elevated the
expression of p-p38 MAPK in order to further ascertain whether
p-p38 MAPK is involved in the regulation of PKD1 in hypoxia. We
found that PKD1 is associated with the activation of p38 MAPK
signaling (Fig. 6). These results
indicate that PKD1 plays an important role in hypoxia-induced
expression and activation of HIF-1α through p38 MAPK.
In conclusion, low pH and hypoxia are unique
characteristics of the tumor microenvironment, which is a potential
target for cancer treatment. Adaptation to this unique
microenvironment is a basic requirement for cancer cells to
survive. Our results have shown that PKD1 not only mediates the
growth and apoptosis of cancer cells in the hypoxic environment,
but also regulates the glycolytic metabolism of cancer cells by
promoting glucose uptake and expression of HIF-1α and glycolytic
enzymes in a hypoxic environment. Inhibition of the expression and
activation of PKD1 can significantly inhibit glycolytic metabolism
of cancer calls as well as create an acidic tumor microenvironment.
Hence, PKD1 may be considered as a potential target associated with
microenvironment-directed tumor biotherapy.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
National Natural Science Foundation of China (nos. 81372892,
81621062 and 81520108009), the 111 Project of MOE China (B14038),
the Open Foundation from the State Key Laboratory of Oral Disease,
Sichuan University (nos. SKLOD201601 and SKLOD2016OF01) and for the
financial support, Sichuan Province Science and Technology
Innovation Team Program (no. JCPT 2011-9).
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PZ designed the outline of the paper. JC, BMC and
YPF conducted the experiments. XYL, QL and YD collected protein
samples for western blot analysis. JC and BMC analyzed data. JC and
PZ wrote the manuscript. JC and YPF prepared the figures. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Anastasiou D: Tumour microenvironment
factors shaping the cancer metabolism landscape. Br J Cancer.
116:277–286. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alfarouk KO: Tumor metabolism, cancer cell
transporters, and microenvironmental resistance. J Enzyme Inhib Med
Chem. 31:859–866. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Avantaggiati ML: Cancer metabolism as a
therapeutic target: Finding the right target(s) in the context of
tumor heterogeneity, evolution, and metabolic plasticity. Oncology.
27:474476–477. 2013.PubMed/NCBI
|
|
4
|
Bahrami A, Khazaei M, Hassanian SM,
ShahidSales S, Joudi-Mashhad M, Maftouh M, Jazayeri MH, Parizade
MR, Ferns GA and Avan A: Targeting the tumor microenvironment as a
potential therapeutic approach in colorectal cancer: Rational and
progress. J Cell Physiol. 233:2928–2936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ackerman D and Simon MC: Hypoxia, lipids,
and cancer: Surviving the harsh tumor microenvironment. Trends Cell
Biol. 24:472–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brahimi-Horn MC and Pouysségur J: Hypoxia
in cancer cell metabolism and pH regulation. Essays Biochem.
43:165–178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamada K, Saito M, Matsuoka H and Inagaki
N: A real-time method of imaging glucose uptake in single, living
mammalian cells. Nat Protoc. 2:753–762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Annibaldi A and Widmann C: Glucose
metabolism in cancer cells. Curr Opin Clin Nutr Metab Care.
13:466–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adekola K, Rosen ST and Shanmugam M:
Glucose transporters in cancer metabolism. Curr Opin Oncol.
24:650–654. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alfarouk KO, Verduzco D, Rauch C,
Muddathir AK, Adil HH, Elhassan GO, Ibrahim ME, David Polo Orozco
J, Cardone RA, Reshkin SJ, et al: Glycolysis, tumor metabolism,
cancer growth and dissemination. A new pH-based etiopathogenic
perspective and therapeutic approach to an old cancer question.
Oncoscience. 1:777–802. 2014.
|
|
11
|
Alfarouk KO, Verduzco D, Rauch C,
Muddathir AK, Bashir AH, Elhassan GO, Ibrahim ME, Orozco JD,
Cardone RA, Reshkin SJ, et al: Erratum: Glycolysis, tumor
metabolism, cancer growth and dissemination. A new pH-based
etiopathogenic perspective and therapeutic approach to an old
cancer question. Oncoscience. 2:3172014.
|
|
12
|
Frérart F, Sonveaux P, Rath G, Smoos A,
Meqor A, Charlier N, Jordan BF, Saliez J, Noël A, Dessy C, et al:
The acidic tumor microenvironment promotes the reconversion of
nitrite into nitric oxide: Towards a new and safe radiosensitizing
strategy. Clin Cancer Res. 14:2768–2774. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Doherty JR and Cleveland JL: Targeting
lactate metabolism for cancer therapeutics. J Clin Invest.
123:3685–3692. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Justus CR, Dong L and Yang LV: Acidic
tumor microenvironment and pH-sensing G protein-coupled receptors.
Front Physiol. 4:3542013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kato Y, Ozawa S, Miyamoto C, Maehata Y,
Suzuki A, Maeda T and Baba Y: Acidic extracellular microenvironment
and cancer. Cancer Cell Int. 13:892013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koukourakis MI, Kakouratos C, Kalamida D,
Bampali Z, Mavropoulou S, Sivridis E and Giatromanolaki A:
Hypoxia-inducible proteins HIF1α and lactate dehydrogenase LDH5,
key markers of anaerobic metabolism, relate with stem cell markers
and poor post-radiotherapy outcome in bladder cancer. Int J Radiat
Biol. 92:353–363. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marchiq I and Pouysségur J: Hypoxia,
cancer metabolism and the therapeutic benefit of targeting
lactate/H+ symporters. J Mol Med (Berl). 94:155–171.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bailey KM, Wojtkowiak JW, Hashim AI and
Gillies RJ: Targeting the metabolic microenvironment of tumors. Adv
Pharmacol. 65:63–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
LaValle CR, George KM, Sharlow ER, Lazo
JS, Wipf P and Wang QJ: Protein kinase D as a potential new target
for cancer therapy. Biochim Biophys Acta. 1806:183–192.
2010.PubMed/NCBI
|
|
20
|
Mikhalap SV, Kovalevska LM and Sydorenko
SP: The role of PKD family protein kinases in the regulation of
protein post-translational modification. Ukr Biokhim Zh. 80:16–24.
2008.(In Ukrainian).
|
|
21
|
Malhotra V and Campelo F: PKD regulates
membrane fission to generate TGN to cell surface transport
carriers. Cold Spring Harb Perspect Biol. 3:a0052802011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eisenberg-Lerner A and Kimchi A: PKD at
the crossroads of necrosis and autophagy. Autophagy. 8:433–434.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liou GY and Storz P: Protein kinase D
enzymes: Novel kinase targets in pancreatic cancer. Expert Rev
Gastroenterol Hepatol. 9:1143–1146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mikhalap SV, Shabelnyk MIu and Sydorenko
SP: Protein kinases of PKD family as a potential object of
translational research in oncology. Ukr Biokhim Zh (1999).
82:18–32. 2010.(In Ukrainian). PubMed/NCBI
|
|
25
|
No authors listed: Study reveals new drug
target for PKD. Nephrol News Issues. 30:162016.
|
|
26
|
Zhang T, Sell P, Braun U and Leitges M:
PKD1 protein is involved in reactive oxygen species-mediated
mitochondrial depolarization in cooperation with protein kinase Cδ
(PKCδ). J Biol Chem. 290:10472–10485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qin XJ, Gao ZG, Huan JL, Pan XF and Zhu L:
Protein kinase D1 inhibits breast cancer cell invasion via
regulating matrix metalloproteinase expression. Eur J Gynaecol
Oncol. 36:690–693. 2015.PubMed/NCBI
|
|
28
|
Ochi N, Tanasanvimon S, Matsuo Y, Tong Z,
Sung B, Aggarwal BB, Sinnett-Smith J, Rozengurt E and Guha S:
Protein kinase D1 promotes anchorage-independent growth, invasion,
and angiogenesis by human pancreatic cancer cells. J Cell Physiol.
226:1074–1081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rozengurt E: Protein kinase D signaling:
Multiple biological functions in health and disease. Physiology.
26:23–33. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jóźwiak P, Krześlak A, Bryś M and Lipińska
A: Glucose-dependent glucose transporter 1 expression and its
impact on viability of thyroid cancer cells. Oncol Rep. 33:913–920.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Teppo S, Sundquist E, Vered M, Holappa H,
Parkkisenniemi J, Rinaldi T, Lehenkari P, Grenman R, Dayan D,
Risteli J, et al: The hypoxic tumor microenvironment regulates
invasion of aggressive oral carcinoma cells. Exp Cell Res.
319:376–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pereira KM, Chaves FN, Viana TS, Carvalho
FS, Costa FW, Alves AP and Sousa FB: Oxygen metabolism in oral
cancer: HIF and GLUTs (Review). Oncol Lett. 6:311–316. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marín-Hernández A, Gallardo-Pérez JC,
Ralph SJ, Rodríguez-Enríquez S and Moreno-Sánchez R: HIF-1alpha
modulates energy metabolism in cancer cells by inducing
over-expression of specific glycolytic isoforms. Mini Rev Med Chem.
9:1084–1101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hay MP, Hicks KO and Wang J:
Hypoxia-directed drug strategies to target the tumor
microenvironment. Adv Exp Med Biol. 772:111–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Esteban MA and Maxwell PH: HIF, a missing
link between metabolism and cancer. Nat Med. 11:1047–1048. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leo C, Giaccia AJ and Denko NC: The
hypoxic tumor microenvironment and gene expression. Semin Radiat
Oncol. 14:207–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Labiano S, Palazon A and Melero I: Immune
response regulation in the tumor microenvironment by hypoxia. Semin
Oncol. 42:378–386. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu C, Gao S, Qu Z and Zhang L: Tumor
microenvironment: Hypoxia and buffer capacity for immunotherapy.
Med Hypotheses. 69:590–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
No authors listed: Metabolism in cancer.
JAMA. 310:24622013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Camarda R, Williams J and Goga A: In vivo
reprogramming of cancer metabolism by MYC. Front Cell Dev Biol.
5:352017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cantor JR and Sabatini DM: Cancer cell
metabolism: One hallmark, many faces. Cancer Discov. 2:881–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cornu M, Albert V and Hall MN: mTOR in
aging, metabolism, and cancer. Curr Opin Genet Dev. 23:53–62. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dang CV, Le A and Gao P: MYC-induced
cancer cell energy metabolism and therapeutic opportunities. Clin
Cancer Res. 15:6479–6483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liou GY, Storz P and Leitges M: A bright
future for protein kinase D1 as a drug target to prevent or treat
pancreatic cancer. Mol Cell Oncol. 3:e10354772015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Finger EC and Giaccia AJ: Hypoxia,
inflammation, and the tumor microenvironment in metastatic disease.
Cancer Metastasis Rev. 29:285–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Airley RE and Mobasheri A: Hypoxic
regulation of glucose transport, anaerobic metabolism and
angiogenesis in cancer: Novel pathways and targets for anticancer
therapeutics. Chemotherapy. 53:233–256. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cui XG, Han ZT, He SH, Wu XD, Chen TR,
Shao CH, Chen DL, Su N, Chen YM, Wang T, et al: HIF1/2α mediates
hypoxia-induced LDHA expression in human pancreatic cancer cells.
Oncotarget. 8:24840–24852. 2017.PubMed/NCBI
|
|
48
|
Matrone A, Grossi V, Chiacchiera F, Fina
E, Cappellari M, Caringella AM, Di Naro E, Loverro G and Simone C:
p38alpha is required for ovarian cancer cell metabolism and
survival. Int J Gynecol Cancer. 20:203–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
García-Cano J, Roche O, Cimas FJ,
Pascual-Serra R, Ortega-Muelas M, Fernández-Aroca DM and
Sánchez-Prieto R: p38 MAPK and Chemotherapy: We Always Need to Hear
Both Sides of the Story. Front Cell Dev Biol. 4:692016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brichkina A and Bulavin DV: Cancer
suppression by systemic inactivation of p38 MAPK. Oncotarget.
8:14275–14276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Koodie L, Ramakrishnan S and Roy S:
Morphine suppresses tumor angiogenesis through a HIF-1alpha/p38
MAPK pathway. Am J Pathol. 177:984–997. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yan L, Cao X, Zeng S, Li Z, Lian Z, Wang
J, Lv F, Wang Y and Li Y: Associations of proteins relevant to MAPK
signaling pathway (p38 MAPK-1, HIF-1 and HO-1) with coronary lesion
characteristics and prognosis of peri-menopausal women. Lipids
Health Dis. 15:1872016. View Article : Google Scholar : PubMed/NCBI
|