Introduction
Non-small cell lung cancer (NSCLC) is the most
common type of lung cancer in the world, which accounts for
approximately 80–85% of all lung cancer cases while the remaining
20% is attributed to small-cell lung cancer (1–3). NSCLC
is generally resistant to chemotherapy and radiation, which often
leads to relapse and even mortality after the initial treatment
(4–6). Growing evidence has supported the role
of cancer stem cells (CSCs) in the expansion, progression and
chemoresistance of tumour cells, including those of lung cancer
(7). Current therapies of NSCLC are
less likely to affect CSCs, due to the characteristics of these
cells or their subpopulations that are intrinsically less sensitive
to common treatments (8–10). In addition, CSCs are largely
quiescent to treatments, which allows them to elude standard
chemotherapies. Therefore, failure to eradicate these
subpopulations are the underlying cause of cancer relapse and
fatality (11,12). The effort to identify new markers of
lung tumour CSCs that can predict tumour progression through early
stage diagnosis is an effective approach, since late detection may
significantly lead to higher metastasis and mortality (13). Furthermore, the development of
cancer as well as tumour cell metastasis is believed to be driven
by CSCs, which has been supported by many studies (14,15).
Therefore, in order to increase the efficacy of treatment and
detection of the disease, several CSC markers have been identified.
Although, many of these markers do exist in the context of lung
cancer, nevertheless we believe that a precise characterization of
the cells isolated by these markers, specifically for NSCLC is
still needed.
Several CSC markers have been identified from
various tumours including malignant cells derived from breast
(16), pancreas (17) prostate (18), colon (19) and head and neck (20) cancer, as well as leukaemia (21). Concerning lung cancer, putative CSCs
derived from various surface markers including CD24 (22), CD133 (23), CD44 (24), CD166 (25) and CD326 (EpCAM) (26) have also been extensively studied and
characterised. An example is the epithelial cell adhesion molecule
known as the EpCAM, mostly expressed in several human carcinomas
and in the majority of normal epithelial cells (27). Due to the strong association between
the high expression of EpCAM and the progression of tumour cells in
NSCLC, it is believed that this molecule would serve as a prominent
marker that can distinguish CSCs from non-CSCs in tumour specimens
which are mostly consisted of squamous cell carcinoma (28–30).
In addition, EpCAM was observed to be an important biomarker due to
its ability to detect circulating tumour cells in the blood of lung
cancer patients (31). Another
example is the leukocyte cell adhesion molecule (ALCAM) or CD166
that was also found to be a robust marker of lung CSCs, based on
the high expression detected in most lung cancer samples and the
capacity of the isolated cells to exhibit self-renewal capacity and
differentiation in vivo (32). The chemotherapy-resistant
characteristic is also one of the hallmarks that can specifically
discriminate a CSC from a non-CSC subpopulation. For instance, a
specific tumour subpopulation isolated from breast (33), colon (34) and gastric (35) cancer is believed to be a CSC
subpopulation based on the expression of the homing cell adhesion
molecule (HCAM) or CD44. The isolated cells positive for CD44
possess the ability for self-renewal and also the characteristic of
being resistant to common chemotherapy, indicating the utility of
CD44 as a marker for CSC (35).
Furthermore, CD44 was also believed to be crucial for initiating
and driving NSCLC stem cell mobility and metastasis (36). Hence, the aim of the present study
was to identify and characterise a novel CSC subpopulation from the
A549 cell line used as a model of NSCLC using a novel combination
of three markers, EpCAM, CD166 and CD44, rather than single markers
to strengthen the selection of the CSC population.
Materials and methods
Cell culture of NSCLC cell line
(A549)
The human NSCLC cell line A549, was obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
Cells were grown and maintained in a complete RPMI-1640 medium
(Invitrogen, Carlsbad, CA, USA) containing 10% foetal bovine serum
(FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin and were
grown at 37°C in a humidified atmosphere of 5% CO2. The
cells were maintained in a 75-cm2 tissue cultured flask
and were harvested using 0.25% trypsin-EDTA. All culture reagents
were obtained from Gibco (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) unless otherwise stated.
Sorting of triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−)
subpopulations
The A549 cells were harvested by incubating the
cells with 0.25% trypsin and followed by washing with
phosphate-buffered solution (PBS) containing 2% FBS. The suspension
cells were then labelled with antibodies (CD326/EpCAM-APC; 1:10
dilutions; cat. no. 347200; CD166-PE; 1:10 dilutions; cat. no.
560903; and CD44-FITC; 1:10 dilutions; cat. no. 347943) (BD
Biosciences, San Jose, CA, USA). Briefly, the cells were
transferred into 75-mm polystyrene round bottom test tubes (BD
Falcon; BD Biosciences) and were suspended in PBS (90 µl) added
with 2% FBS at a concentration of 1×106 cells/ml.
Subsequently, 10 µl of each antibody were added into the cell
suspension and were subsequently incubated for 30 min in the dark.
The cells were then washed and filtered through a 40-µm cell
strainer to obtain a single cell suspension before sorting. The
expression of the CSC markers, EpCAM, CD166 and CD44 was analysed
and sorted using a Fluorescence Activated Cell Sorter (FACSAria
III; BD Biosciences). Gating was used for sorting out
triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−) population
(Fig. 1).
Cell proliferation assay
MTS assay [3-(4,
5-dimethylthiazol-2-yl)-2H-tetrazolium, inner salt] was purchased
from Promega (Madison, WI, USA) and was used to quantify the
proliferation of both sorted and unsorted A549 cells at different
time-points (24, 48 and 72 h). The cells were seeded at a density
of 1×104 cells/well in a 96-well plate and were
incubated for the appropriate length of time (24, 48 and 72 h) in a
humidified 5% CO2 incubator at 37°C. Following that, the
MTS solution (15 µl) was added to each individual well for further
incubation for 4 h. Subsequently, solubilisation solution (100 µl)
was added to the cells and absorbance was assessed using
Odyssey® Sa Imaging system (LI-COR Biosciences, Lincoln,
NE, USA) at 570 nm, using wells without cells as blank. The
viability of cells at different time-points was calculated
according to the following formula: cell viability (%) = cells
(sample)/cells (control) × 100.
Clonogenic assay
Both sorted [triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−)] and
unsorted A549 cells were seeded in triplicates (1,000 cells/ml)
into a 6-well plate containing 2 ml of complete RPMI-1640 medium
per well. The sorted cells were kept in a culture at 37°C in a 5%
CO2 humidified incubator for 14 days. After 14 days, the
colonies were observed using an inverted microscope (Olympus,
Tokyo, Japan) at ×200 magnification and images from 20 fields of
view were captured to evaluate, characterise and manually count the
different clones, namely, holoclones, meroclones and paraclones
(Barrandon and Green) (37). For
the analysis on the number of colonies formed, the cells were
stained using Giemsa (Sigma-Aldrich, St. Louis, MO, USA) and were
manually counted. In brief, the supernatant was aspirated and the
wells were rinsed twice with PBS, followed by subsequent fixing
with 4% formaldehyde for 2 min at 24°C. Following fixation, the
colonies were stained with Giemsa (Sigma-Aldrich) for 15 min in the
dark at 24°C. Following that, the excessive dye was removed and the
stained colonies were set to air-dry before analysis and
counting.
Spheroid assay
Both sorted [triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−)] and
unsorted A549 cells were suspended in serum-free sphere medium
(1.0×103 cells/ml) containing DMEM-F12 (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10 ng/ml basic
fibroblast growth factor (bFGF; Life Technologies, Foster City, CA,
USA), 1% of B27 supplement (Life Technologies), 20 ng/ml of
epidermal growth factor (EGF; Life Technologies), 100 IU/ml
penicillin and 100 µg/ml streptomycin (Life Technologies). The
cells were re-suspended in a ratio of 1:1 (v/v) of growth
factor-reduced Matrigel (BD Biosciences) and serum-free sphere
medium. The cells were then seeded in a 96-well ultra-low
attachment (ULA) dish (Corning, Inc., Oneonta, NY, USA). The size
and number of the formed spheroids were assessed after 20 days of
culture by inverted phase contrast microscopy (Olympus).
Chemotherapy-resistance assay
The viability of both sorted and unsorted A549 cells
was assessed by MTS assay following treatment with 5-fluouracil
(5-FU) and cisplatin. Briefly, the cells were seeded at a
concentration of 1×104 cells/well in a 96-well plate and
were incubated overnight at 37°C in humidified air containing 5%
CO2. Subsequenlty, 5-FU and cisplatin were added at
different concentrations (5, 10, 30, 50, 70 and 90 µM) and
incubated for 48 h, and then, 15 µl of MTS solution (CellTiter
96® AQueous One Solution; Promega) was added to each
well and further incubated for another 4 h. Solubilisation solution
(100 µl) was added to the cells and absorbance at 570 nm was
assessed using Odyssey® Sa Imaging System (LI-COR
Biosciences). Cell viability was calculated using the formula as
aforementioned.
Aldehyde dehydrogenase detection
(Aldefluor assay)
The Aldefluor assay (StemCell Technologies,
Vancouver, Canada) was performed according to the manufacturer's
instructions. Briefly, a total of 2×105 cells were
collected, centrifuged, pelleted and re-suspended in 500 µl of
assay buffer before the staining. Aldefluor stain (2.5 µl) was
added into the samples followed by the addition of 5 µl ALDH1
inhibitor (diethylaminobenzaldehyde/DEAB) to the control tube. The
cells were vortexed and incubated for 30 min at 37°C in the dark.
Subsequently, the cells were centrifuged at 180 × g for 5 min and
the collected pellet was re-suspended in 500 µl assay buffer before
being subjected to flow cytometry (BD FACS Calibur; BD Biosciences)
and analysed using BD CellQuest™ Pro software (BD Biosciences).
Mitochondrial membrane potential
(∆ψm) assay
Analysis of mitochondrial membrane potential was
conducted using the BD MitoScreen (BD Biosciences). To determine
the amount of cells undergoing mitochondrial membrane potential
(∆ψm) in response to cisplatin, the cells were incubated
in JC-1 stain
(5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine
iodide) for 15 min at 37°C in the dark, and then were washed two
times with 1X assay buffer. Analysis on the ∆ψm was
conducted based on the intensity of the fluorescence detected in
both the FL-1 and FL-2 channels by using a flow cytometer (BD
FACSCalibur; BD Biosciences).
Scratch-wound/migration assay
Briefly, both sorted and unsorted A549 cells were
seeded at a density of ~3-4×105 cells/well (6-well
plate) in complete RPMI-1640 medium and were grown overnight to 90%
confluency. Then, the cells were incubated with colcemid (10 µg/ml)
for 2 h for synchronization. After incubation, a scratch was
created using a 200-µl pipette tip and the wells were gently washed
using PBS to remove any residual debris. Fresh complete RPMI-1640
medium (2 ml) was added followed by incubation for another 48 h.
The images of the migrated cells from five fields of view were
captured using a relief-phase contrast microscope (Olympus IX71;
Olympus) at ×400 magnification. The cells migrated into the wound
area were evaluated using the following formula: Percentage of
cells migrated = [Initial scratch (0 h) - Final scratch (48 h)])/
Initial (0 h) × 100.
Quantitative real time-polymerase
chain reaction (qRT-PCR)
The mRNA expression of CSC markers in both sorted
and unsorted A549 cells was determined by quantitative RT-PCR
(qRT-PCR) using a LightCycler 480 (Roche, Mannheim, Germany).
Initially, the total RNA was extracted from both cells using an
RNAeasy extraction kit (Qiagen, Hamburg, Germany) according to the
manufacturer's protocol. Subsequently, cDNA was synthesised from 1
µg of total RNA using a Transcriptor First Strand cDNA Synthesis
kit (Roche Applied Science, Mannheim, Germany). The qRT-PCR
reaction was prepared using a SYBR 1 Master Mix (Roche) as well as
different sets of primers (REX1, SOX2, ABCG2, SSEA4, NANOG,
OCT4 and GAPDH) as displayed in Table I. The PCR reactions were run under
the following cycle conditions; a pre-denaturation step for 4 min
at 95°C followed by 40 cycles of denaturation at 95°C for 15 sec,
annealing at 60°C for 30 sec and extension at 72°C for 30 sec. The
basic relative gene expression (RQ) was calculated using the ∆∆Ct
formula and the efficiency (E) of a primer binding equal to 2.
 | Table I.List of primers used in qRT-PCR for
the expression of stem cell genes. |
Table I.
List of primers used in qRT-PCR for
the expression of stem cell genes.
| Gene | Accession | Forward primer | Reverse primer |
|---|
| REX1 | NM_174900 |
GCGTACGCAAATTAAAGTCCAGA |
ATCCTAAACCAGCTCGCAGAAT |
| SOX2 | NM_0031063 |
CCCCCGGCGGCAATAGCA |
TCGGCGCCGGGGAGATACAT |
| ABCG2 | NM_001257386 |
GTTTATCCGTGGTGTGTCTGG |
CTGAGCTATAGAGGCCTGGG |
| SSEA4 | NM_006927 |
TGGACGGGCACAACTTCATC |
GGGCAGGTTCTTGGCACTCT |
| NANOG | NM_001297698 |
TCCTCCATGGATCTGCTTATTCA |
CAGGTCTTCACCTGTTTGTAGCTGAG |
| OCT4 | NM_001173531 |
CGACCATCTGCCGCTTTGAG |
CCCCCTGTCCCCCATTCCTA |
Statistical analysis
Results are expressed as the means ± standard
deviation (SD) of three independent experiments. Statistical
analysis was performed using the IBM SPSS statistics version 21
(IBM Corp., Armonk, NY, USA). Comparisons between two groups were
performed using the two-tailed t-test; P-values <0.01 were
considered to indicate statistically significant differences.
Comparisons among groups were performed using one factor analysis
of variance (ANOVA) followed by the Tukey's post hoc test.
Results
Expression of triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−)
subpopulations in the A549 cell line
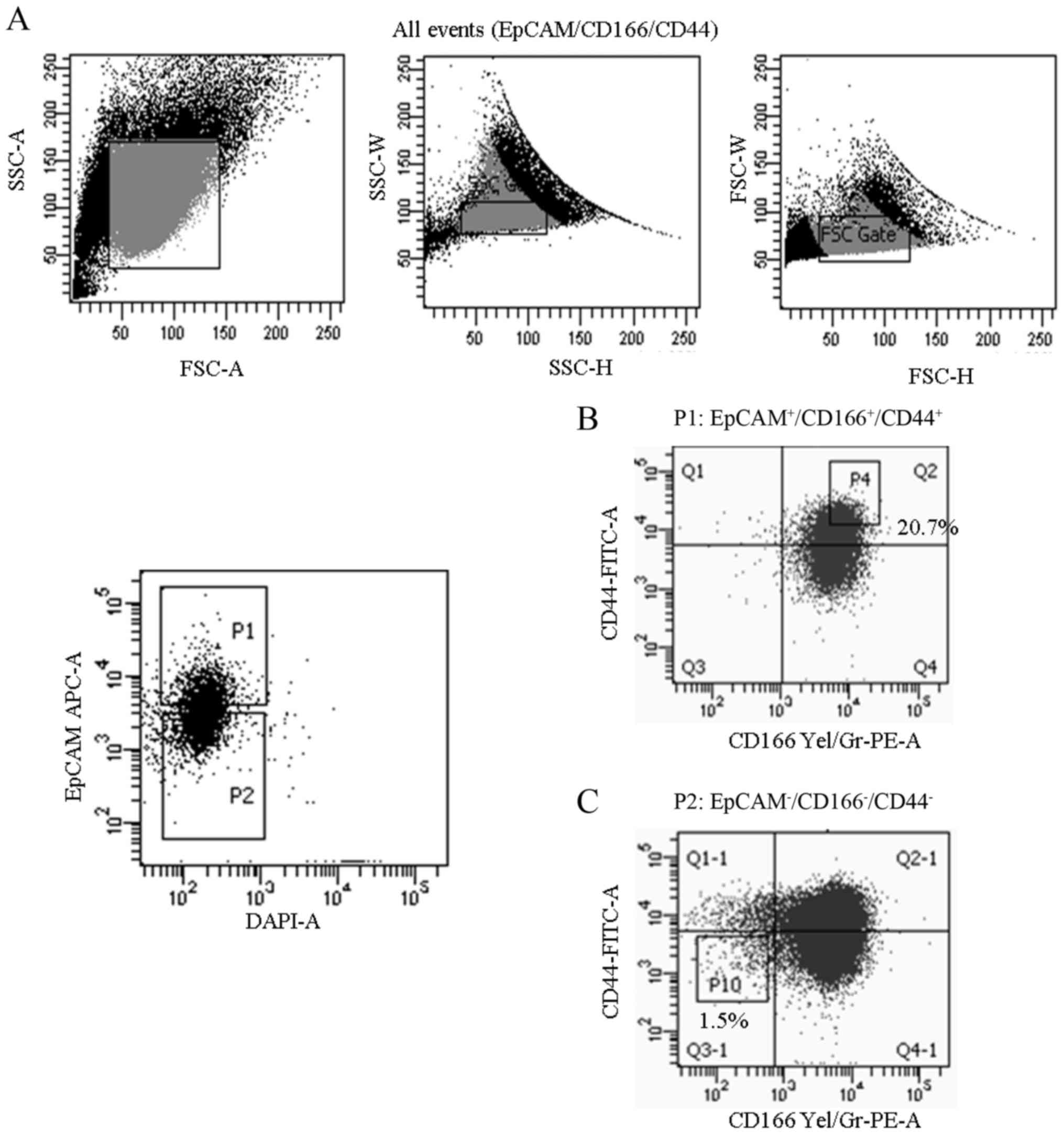
To identify the subpopulation believed to exhibit
the putative CSC characteristics, we analysed and sorted
subpopulations in the A549 cell line that expressed all three stem
cell surface markers (EpCAM, CD166 and CD44), namely the
triple-positive
(EpCAM+/CD166+/CD44+), and the
triple-negative
(EpCAM−/CD166−/CD44−) as controls
(Fig. 1). Our findings indicated
that the expression of EpCAM, CD166 and CD44 markers in the A549
cell line that was positive for all three markers
(EpCAM+/CD166+/CD44+) was 20.7%
(Fig. 1B), compared to only 1.5%
concerning the expression of the triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation (Fig. 1C). Both the
triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−)
subpopulations were sorted from the parental A549 cell line and
culture-expanded for further downstream analysis.
Proliferation activity of sorted and
unsorted A549 cells
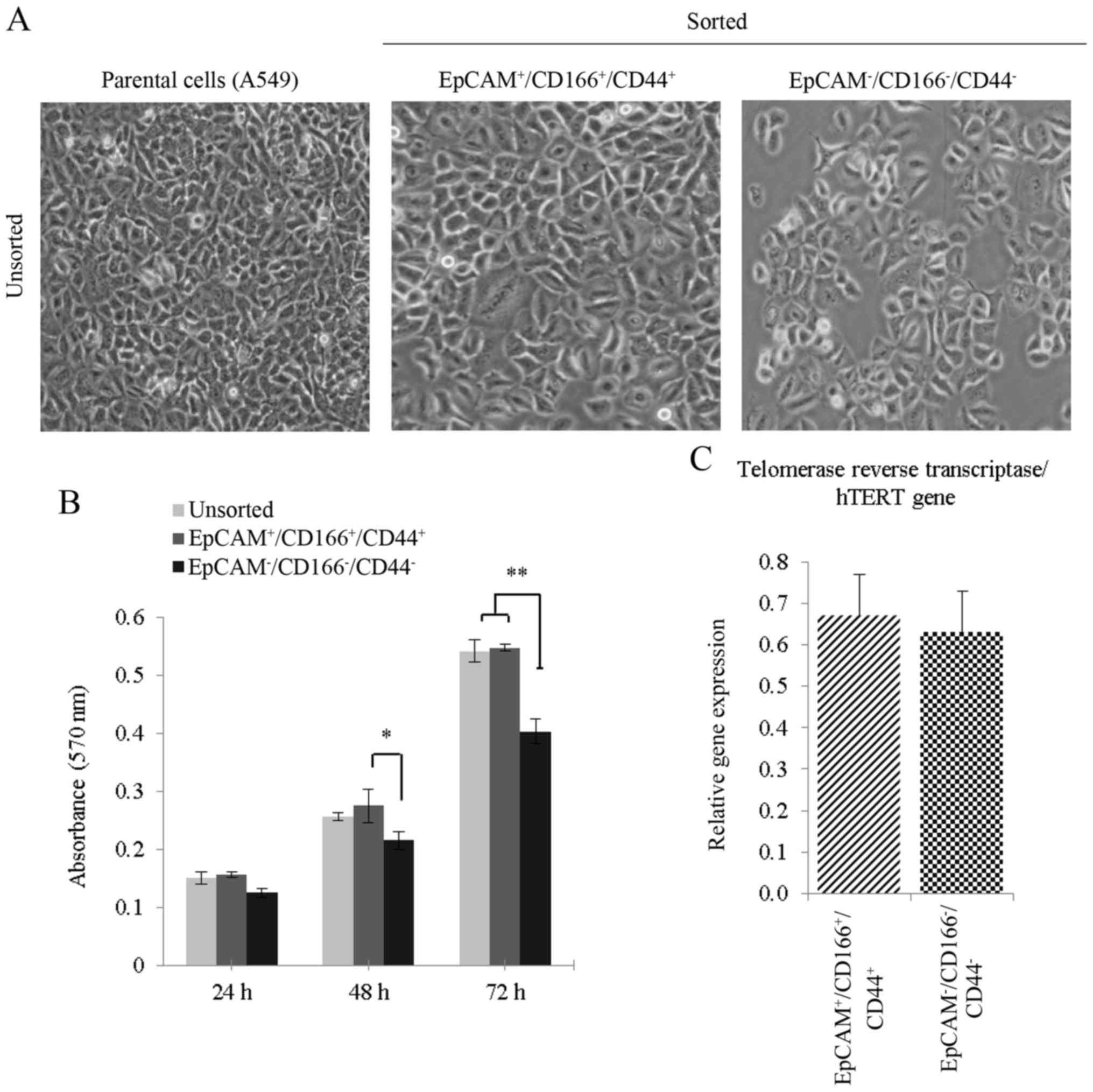
Higher density of cells was observed for both the
unsorted and triple-positive subpopulation as compared to the
triple-negative subpopulation (Fig.
2A). Based on the MTS activity, the triple-positive
subpopulation
(EpCAM+/CD166+/CD44+) demonstrated
higher cellular proliferative activities compared to the
triple-negative subpopulation
(EpCAM−/CD166−/CD44−) between 48
and 72 h (Fig. 2B). In addition,
the proliferative activity of the unsorted cells, which comprised
of a heterogeneous population was almost similar to that of the
triple-positive subpopulation (Fig.
2B), indicating that the triple-positive subpopulation is the
main contributor to the cell proliferation activity. Analysis on
the hTERT gene expression between both the triple-positive
and negative subpopulations revealed an equal hTERT activity
indicating high proliferative activity in both subpopulations
(Fig. 2C).
Triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation reveals compact clone formation as an indicator of
‘stemness’
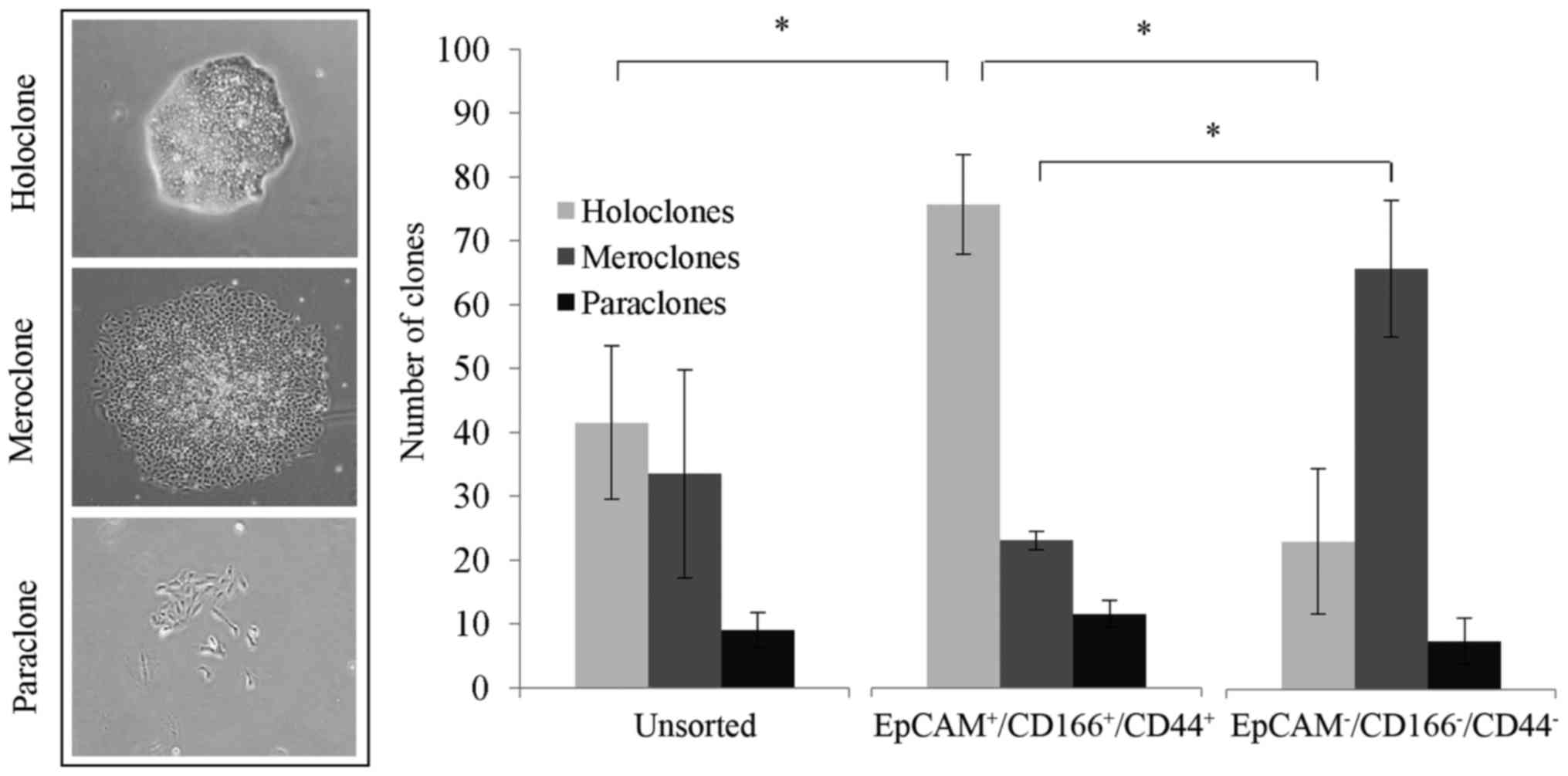
The results revealed that the formation of
holoclones was significantly higher (P<0.05), with 75.5±7.8
clones identified in the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation, compared to only 23.0±11.3 clones in the
triple-negative subpopulation and 41.5±12.0 clones in the unsorted
cells (Fig. 3). Notably, we noticed
that the triple-negative subpopulation presented with substantially
higher meroclones with 65.5±10.6 clones detected, compared to the
triple-positive subpopulation and unsorted cells with only 23.0±1.4
clones and 33.5 ±16.3 clones detected, respectively (Fig. 3).
Triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation demonstrates the ability to form spheres
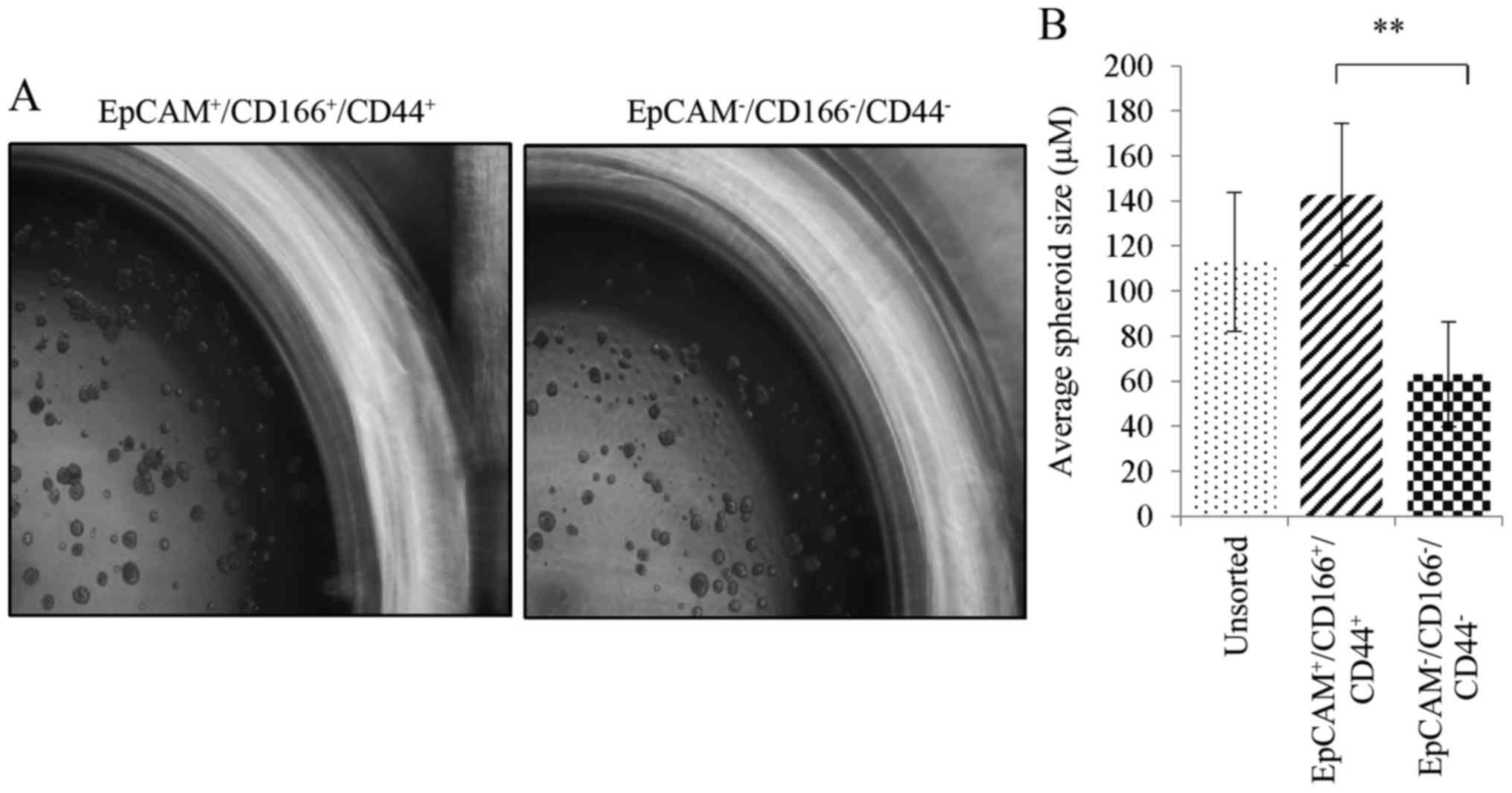
The study was further validated through the
investigation of the self-renewal capability of the sorted cells
using a spheroid formation assay. Bigger spheroids reaching an
average diameter of 142.7±31.6 µm in size were found in the
triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation (Fig. 4A). In
addition, there were smaller spheroids in the unsorted and the
triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation corresponding to an average spheroid diameter of
112.6±30.7 and 63.0±23.2 µm, as depicted in Fig. 4B. Although both the unsorted and the
triple-negative subpopulations were able to form spheroids, which
indicated the self-renewal capability of these cells, these
characteristics were more noticeable in the triple-positive
subpopulation which developed larger sizes of formed spheroids.
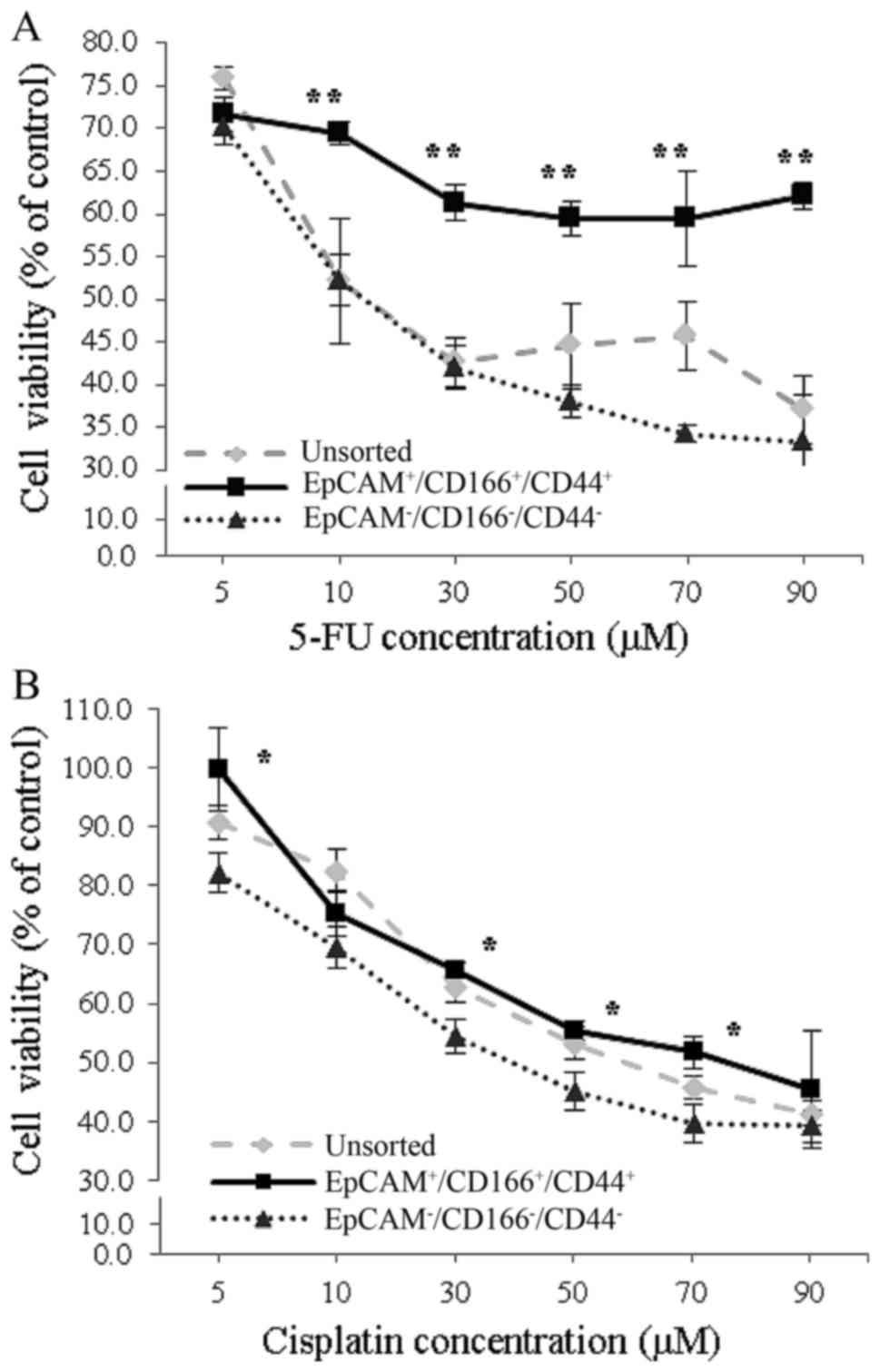
Chemotherapy-resistant properties of
triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation
To investigate the chemotherapy-resistant properties
of the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation, the cells were separately exposed to increasing
doses of 5-FU and cisplatin, while both the triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation and the unsorted cells were used as controls. When
the cells were exposed to an increased concentration of 5-FU, the
percentage of proliferation in the triple-positive subpopulation
was markedly higher (P<0.01), compared to both the
triple-negative subpopulation and the unsorted cells (Fig. 5A). Similarly, higher proliferation
rate in the triple-positive subpopulation was noticed compared to
the triple-negative subpopulation upon exposure to increasing
cisplatin concentrations (Fig. 5B).
Our findings also indicated a significant difference (P<0.01) in
the IC50 values of the triple-positive subpopulation
with 876.3±260.7 and 65.3±6.9 µM detected compared to the
triple-negative subpopulation with only 9.5±1.2 and 35.6±4.1 µM
after exposure of both subpopulations to 5-FU and cisplatin,
respectively (Table II).
 | Table II.IC50 values of the A549
cell line subpopulations. |
Table II.
IC50 values of the A549
cell line subpopulations.
|
| IC50
values (µM) for the treatments |
|---|
|
|
|
|---|
| A549 cell
subpopulation | 5-fluouracil
(5-FU) | Cisplatin |
|---|
| Unsorted |
27.8±6.3 | 59.2±7.9 |
|
EpCAM+/CD166+/CD44+ | 876.3±260.7 | 65.3±6.9 |
|
EpCAM−/CD166−/CD44− |
9.5±1.2 | 35.6±4.1 |
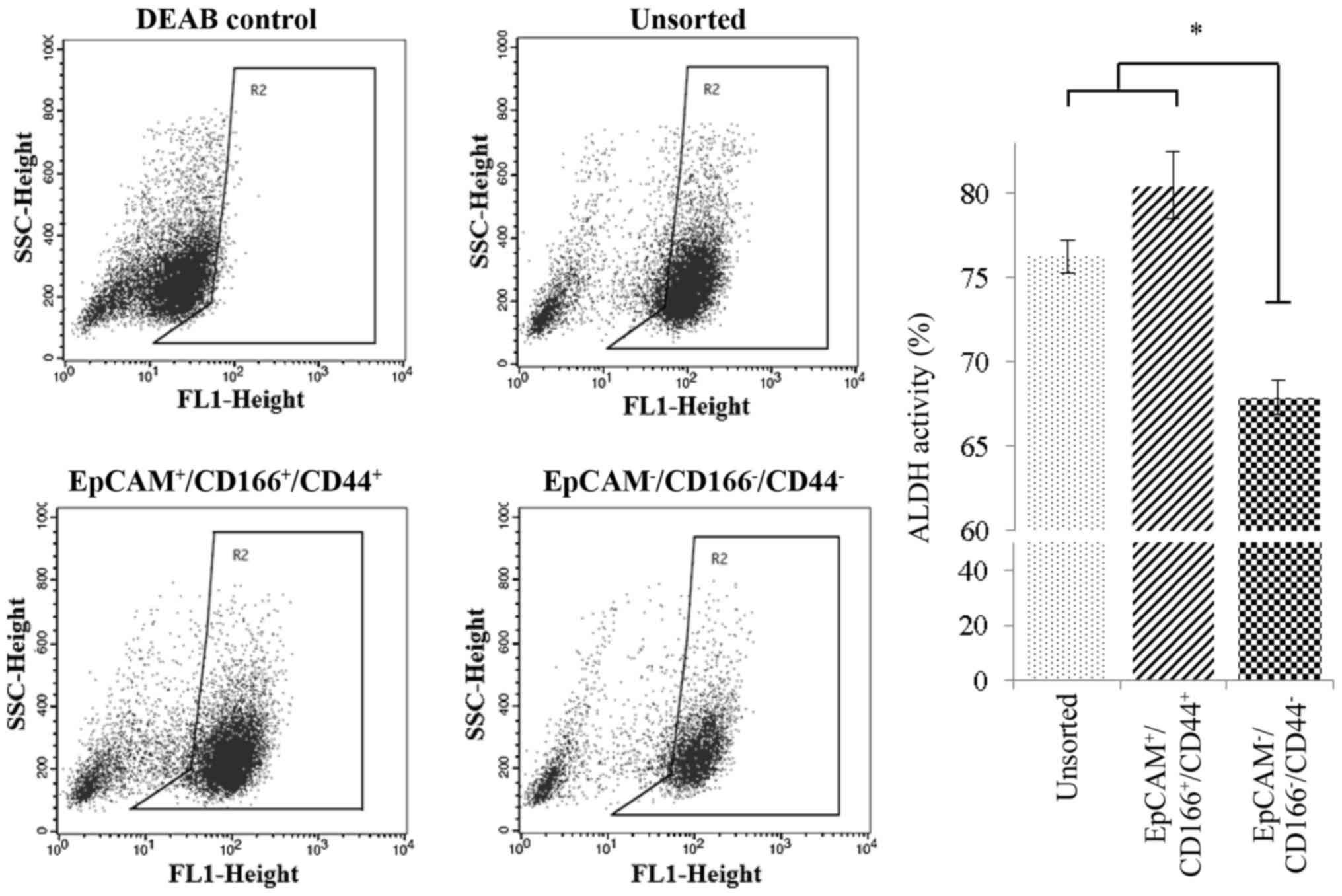
High ALDH1 activity in the
triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation
The chemo-resistance characteristic was further
investigated through analysis of ALDH activity on triple-positive
(EpCAM+/CD166+/CD44+) using the
Aldefluor™ kit (StemCell Technologies, Vancouver, BC, Canada). Our
analysis revealed that the level of ALDH1 activity was
significantly higher (P<0.05) in the triple-positive
subpopulation with 80.47±1.9% expression level detected, compared
to the triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation (67.88±0.7%) as displayed in Fig. 6. Furthermore, the expression level
of ALDH1 in the unsorted cells was 76.26±0.1%, which was almost
equal to the expression observed in the triple-positive
subpopulation indicating that the ALDH1 activity of the unsorted
cells was mostly contributed to the chemo-resistant characteristic
of the triple-positive subpopulation (Fig. 6).
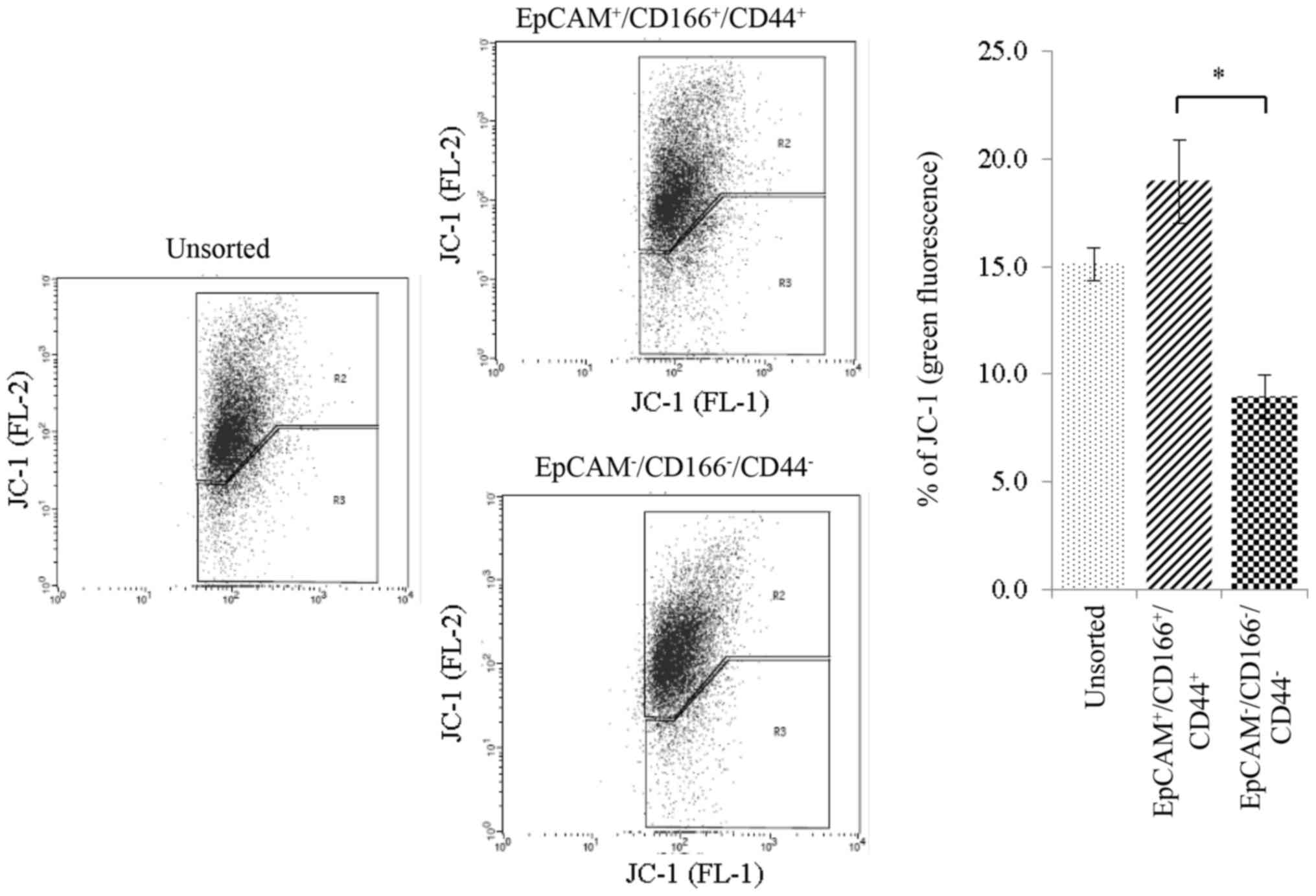
Mitochondrial membrane potential
(∆ψm) activity of the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation
Chemotherapeutic drugs were able to stimulate
changes of the inner mitochondrial membrane and subsequently cause
the loss of mitochondrial membrane potential (∆ψm).
Using the JC-1 stain, both unsorted and sorted cells were treated
with cisplatin and the ∆ψm was subsequently evaluated
analysing the JC-1 monomers (green fluorescence) on the FL-2
channel. Our findings indicated that cisplatin treatment resulted
in a greater percentage of FL-2 value of 18.9±1.9% as detected in
the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation. This was followed by the triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation and unsorted cells at 8.9±1.0 and 15.1±0.76%,
respectively (Fig. 7).
Depolarizarion of the ∆ψm indicated a strong activity of
the cells undergoing early apoptosis which was detected at a high
level in the triple-positive subpopulation.
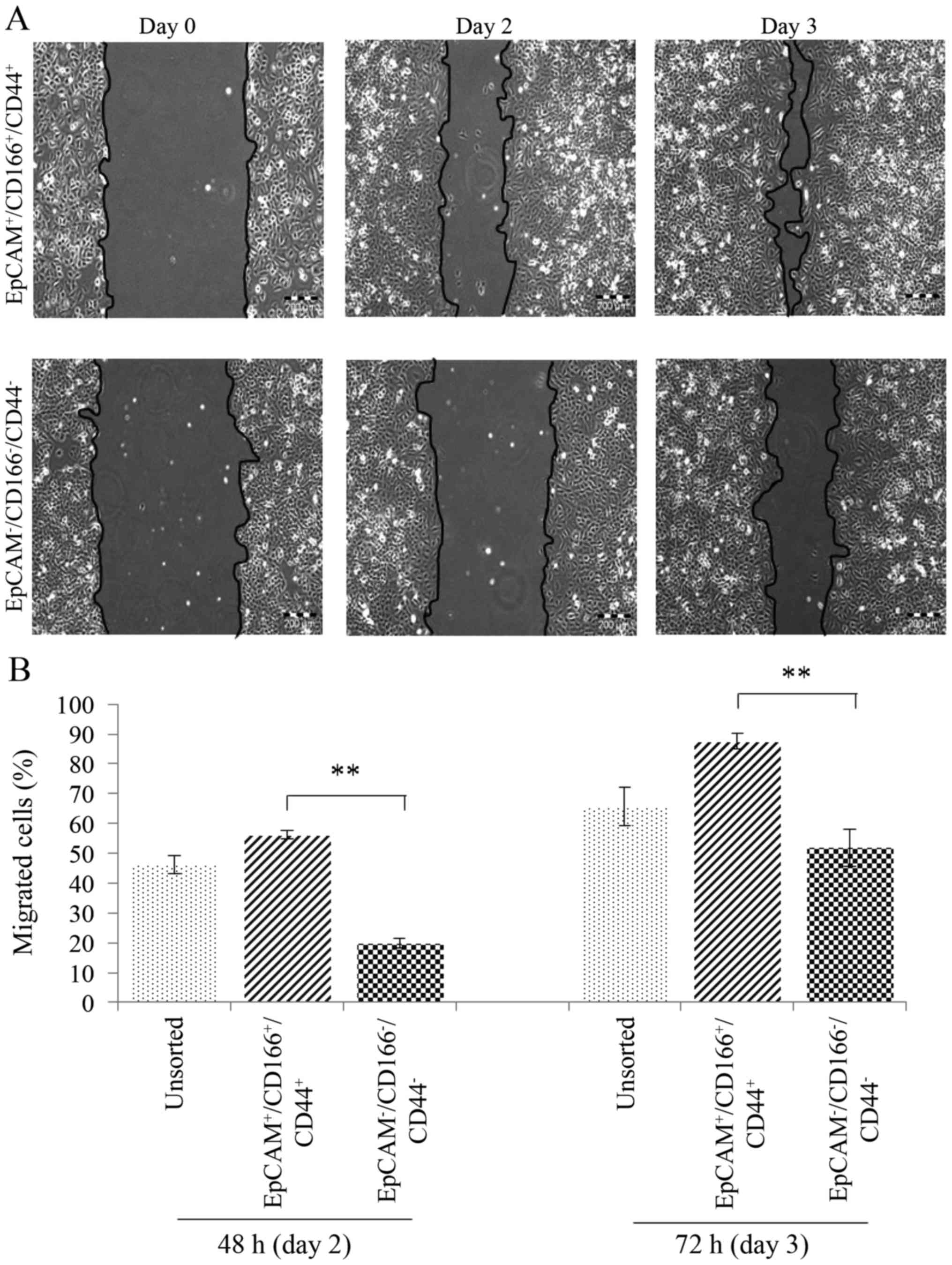
Triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation exhibits high epithelial to mesenchymal transition
(EMT) characteristics
The following assay was conducted by evaluating the
migration ability of the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation compared to the triple-negative
(EpCAM−/CD166−/CD44−)
subpopulation and unsorted cells. Using an in vitro
migration model, scratch assay was analysed on day 2 and 3
(Fig. 8). The triple-positive
subpopulation demonstrated significantly higher migration
(P<0.01) with 56.1±1.4 and 87.6±2.5% wound closure detected on
day 2 and day 3, respectively. The findings were compared to the
triple-negative subpopulation (with only 19.9±1.7% wound closure on
day 2 and 51.9±6.3% on day 3) and indicated that the
triple-positive subpopulation had a higher EMT activity.
Triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation of the A549 cell line exhibits the profile of stem
cell-associated genes
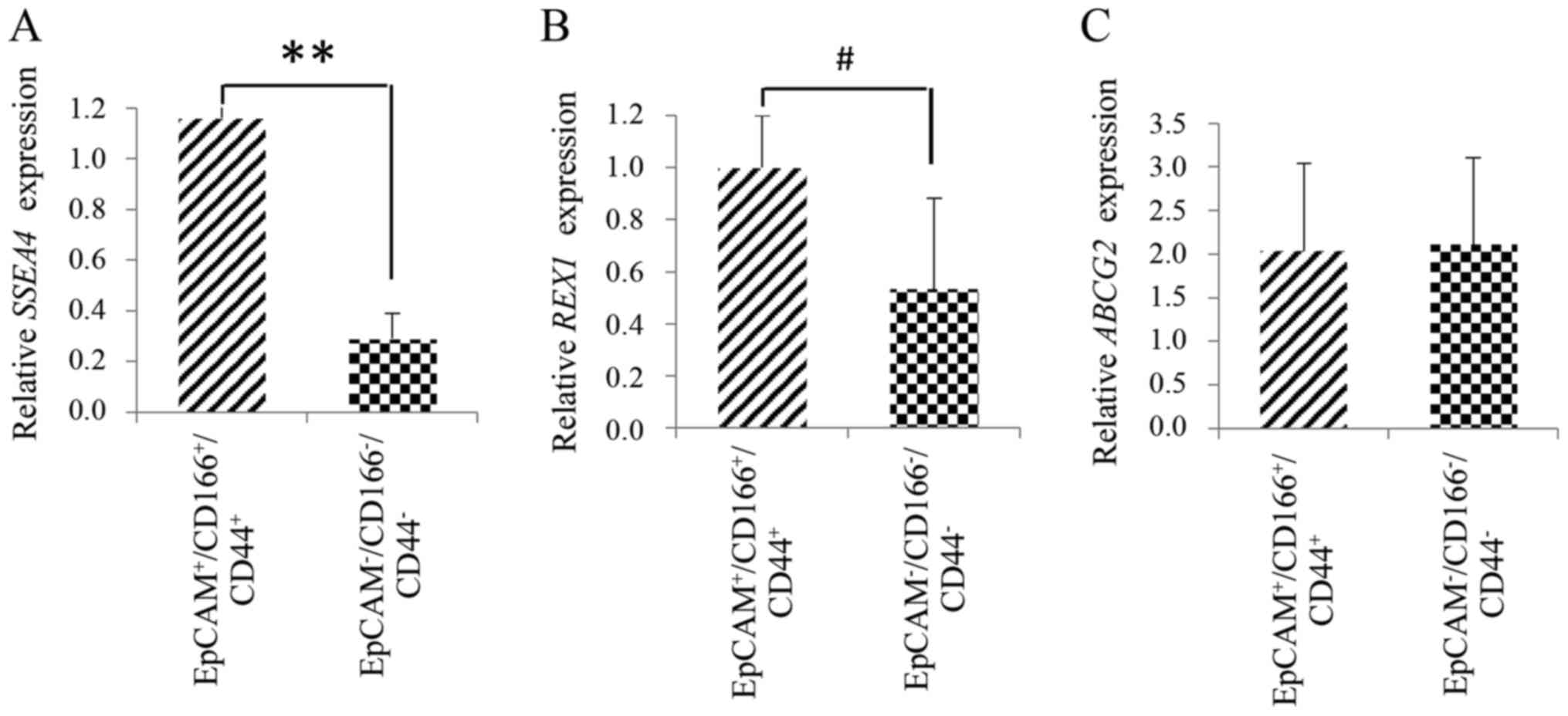
Finally, to further confirm the intrinsic stem cell
properties of the triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation, the expression of SSEA4, REX1, and
ABCG2 was analysed using the quantitative gene analysis. The
triple-positive subpopulation exhibited significantly higher
(P<0.001) expression of the specific pluripotent gene,
SSEA4 with 4.9±2.8-fold increase observed compared to the
triple-negative subpopulation (Fig.
9A). Slight increase (P<0.05) was observed for the specific
stem cell gene, REX1 in the triple-positive subpopulation
compared to the triple-negative subpopulation as depicted in
Fig. 9B. This event indicated that
the triple-positive subpopulation possessed the features of stem
cells as evidenced by the expression of markers related to CSC
development. However, no significant differences were detected for
the ABCG2 gene expression between the triple-positive and
triple-negative subpopulations (Fig.
9C).
Discussion
To the best of our knowledge, the present study is
the first to investigate and confirm the existence of a novel CSC
subpopulation based on the presence of three surface markers,
EpCAM, CD166 and CD44 combined, in which these markers can
specifically identify and discriminate a distinctive subpopulation
of putative CSCs in NSCLC. It has been hypothesised that by
eliminating this subpopulation, treatment efficacy would be
enhanced and chances of tumour relapse reduced. However, the
heterogeneity of CSCs is the major impediment that needs to be
overcome in order to achieve a better treatment goal. Therefore, a
marker that is both robust and specific to identify the major CSC
subpopulation is an effective approach for lung cancer treatment.
In our previous study, we have identified a potential CSC
subpopulation using the combination of two markers from three
surface markers (CD166, CD44 and EpCAM) that significantly
discriminated CSCs from the non-CSCs in NSCLC (38). Furthermore, we also concluded that
these subpopulations (CD166+/EpCAM+ and
CD166+/CD44+) exhibited the transcriptomic
profile of multipotent cells including the self-renewal capacity,
ability for adipogenic, as well as osteogenic differentiation and
upregulation of the CSC gene pathways, that further confirmed the
stemness characteristics of these subpopulations. In addition, the
in vivo study also confirmed the tumourigenic capacity
presented by the ability of these subpopulations to generate bigger
tumour bulk, compared to their non-CSCs counterpart
(CD166−/EpCAM− and
CD166−/CD44−) (38). Therefore, these findings have paved
the way to the present study of using a combination of all three
markers simultaneously in order to have a stringent selection of
CSCs in NSCLC.
Isolating CSCs from the A549 cell line using a
combination of all three markers (EpCAM, CD166 and CD44) resulted
in two subpopulations known as the triple-positive
(EpCAM+/CD166+/CD44+) and
triple-negative
(EpCAM−/CD166−/CD44−). In the
present study, we found that the triple-positive subpopulation
presented with 20.7% expression of the combined markers (Fig. 1), which was higher than the expected
percentage of CSCs reported in other studies (39,40).
Although the expression was high and may indicate that the
combination marker was unspecific, numerous studies have reported
the validity of these markers in detecting specific CSC
subpopulations, either as single markers, e.g., CD166 (41) and CD44 (42) or co-expressed with other known CSC
markers such as CD90 (43) and
CD133 (44). Our findings indicated
that the triple-positive markers, although highly expressed in the
A549 cell line, may serve as specific and yet robust markers that
can discriminate a large population of lung CSCs from the non-CSCs
due to their high specificity detected in several tumours.
According to Barrandon and Green (1987) (45) who described different clonogenicity
to multiplication characteristics, only colonies known as
holoclones (colonies with well-defined border) with cellular
characteristics of pluripotent stem cells (i.e., high nuclear to
cytoplasmic ratio, prominent nucleolus and small cell size) were
capable of extensive proliferation, self-renewal and cellular
growth. However, the meroclones and paraclones (colonies without a
well-defined border) had a limited proliferation capacity and were
incapable of self-renewal. From our findings, we observed that in
contrast to the triple-negative subpopulation, the triple-positive
subpopulation could give rise to higher number of holoclones
(Fig. 3). Furthermore, the
triple-negative subpopulation consisted of mostly meroclones,
indicating that the subpopulation was predominantly governed by
differentiated types of CSCs. We also observed that the
triple-positive subpopulation had the ability to form bigger
spheroids (Fig. 4) compared to its
counterpart. Significance in the size of spheroids between the
putative CSCs and non-CSCs by in vitro analysis that may
also reflex the bulk of tumours in xenografts has been reported as
an indicator of CSC stemness in numerous studies. The
triple-negative subpopulation may also form spheroids as these
cells are also tumour cells, however the observation that they were
significantly smaller than the triple-positive subpopulation cells
indicated its level of tumourigenicity (46). Collectively, the results indicated
that the triple-positive subpopulation has the fundamental
characteristic of stemness (Figs. 3
and 4) (47).
In addition to clonogenicity, chemoresistance is
also an equally important characteristic that can clearly define a
typical feature of CSC (48). Since
the expression of ALDH1 was high in the triple-positive
subpopulation (Fig. 6), it was
plausible that the subpopulation was resistant to both cisplatin
and 5-FU treatments when compared to the triple-negative
subpopulation (Fig. 5).
Furthermore, enriched CSCs such as the triple-positive
subpopulation would have a lower sensitivity to common chemotherapy
as reflected by the higher cell viability post treatment, compared
to the unsorted cells where the sensitivity to chemotherapy was
higher due to the existence of non-CSCs in their population
(49,50). This finding was in line with
previous studies that have revealed a similar observation in
CD133+ subpopulation derived from colon cancer cells and
pretreated with both oxaliplatin and 5-FU (51). The chemotherapy-resistant activity,
as confirmed by gene expression analysis, has recently been
reported as a potential marker that can specifically discriminate
CSCs from non-CSCs (52–55) in different tumour types (52,53,56)
However, our observation in the expression of ABCG2 mRNA (a
multidrug resistance family gene) indicated no differences between
the triple-positive and triple-negative subpopulations (Fig. 9). Nonetheless, it is believed that
the ABCG2 may not directly influence the chemoresistant
characteristic of this subpopulation. Therefore, screening for
several other multidrug resistance genes such as the ABCG1,
P-glycoprotein and MDR family were necessary in order to
fully characterise the chemo-resistant activity of this
subpopulation.
There was higher loss of mitochondrial membrane
potential (∆ψm) in the triple-positive subpopulation
following cisplatin treatment, than the triple-negative
subpopulation indicating that this particular subpopulation was
more sensitive to the treatment. The treatment also induced loss of
the ∆ψm activity which eventually led to an activation
of the intrinsic apoptotic pathways in both subpopulations
(57,58). A higher ∆ψm activity in
the triple-positive subpopulation following treatment was possibly
due to a greater proliferation activity as seen in the
triple-positive subpopulation (Fig.
2), all of which may lead to the production of a higher number
of proliferative cells for apoptosis. It is also known that due to
the cisplatin mechanism of action that binds to DNA, the activity
of cisplatin is greater in highly proliferative cells since the
inhibition activity of cisplatin only peaks during cellular
division and mitosis (59).
In addition to the stemness and high resistance to
drug treatments, high metastatic and EMT activity are also part of
the hallmarks of CSCs (60). EMT
would normally result in the migration of CSCs with invasive
characteristics, induced by the transforming growth factor-B
(TGF-B) and activation of hedgehog signalling pathway (61) under a hypoxic condition (62). Our findings clearly indicated that
the triple-positive subpopulation exhibited significantly higher
migratory phenotype on day 2 and 3 when compared to the
triple-negative subpopulation. The EpCAM and CD44 target molecules,
that have been reported to regulate cell migration and metastasis
through the Wnt/β-catenin signalling pathway (63–66),
may be the key reason for cellular motility and invasion that
contribute to the characteristics of CSCs, and ultimately tumour
metastasis (67).
CSCs preferentially express genes relevant for
stemness such as SOX2, NANOG, REX1, SSEA4 and OCT4
which have been shown to be important for human embryonic stem cell
development (68). The mRNA
expression of REX1 and SSEA4, were relatively higher
(Fig. 9) in the triple-positive
subpopulation, indicating the contribution of these pluripotent
genes in the functions and maintenance of CSCs. We believed that
these genes were not only important markers that can determine a
pure population of CSCs, but also suitable candidate genes for
targeted therapy to disrupt the regulation of CSCs for an enhanced
treatment efficacy of lung cancer.
In conclusion, we revealed for the first time, that
the triple-positive subpopulation derived from the A549 cell line
possessed CSC characteristics, including being highly
proliferative, having greater clonogenicity, ability for
self-renewal through spheroid formation, being chemoresistant, and
exhibiting high ALDH activity and migratory potential. To better
understand the chemoresistance of this subpopulation, an in
vivo drug-resistant assay can be performed to further verify
the characteristics of this subpopulation. Furthermore, the gene
expression analysis confirmed that the triple-positive
subpopulation expressed higher pluripotent genes (REX1 and SSEA4)
compared to the triple-negative subpopulation. Collectively, the
present study indicated that the identified triple-positive
(EpCAM+/CD166+/CD44+)
subpopulation may be a potential candidate of CSCs for tumour
samples derived from NSCLC and the population may also represent a
significant group of chemoresistant cells that can be used for drug
screening and chemosensitivity testing.
Acknowledgments
The authors wish to thank the Director General of
Health of Malaysia for his permission to publish the present
study.
Funding
The present study was supported by a grant from the
Ministry of Health (MOH), Malaysia (no.
JPP-IMR-16-038/NMRR-16-869-30708).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KSF conceived, designed and performed the
experiments. MNL performed part of the experiment and edited the
manuscript. NZ, NAF and AZAR analysed the data. PB and ZZ provided
the reagents and tools. NAS and KSF wrote the paper. BHY and PLM
edited and reviewed the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board, Institute for Medical Research (IMR),
Malaysia and the Medical Research and Ethics Committee (MREC),
Ministry of Health (MOH), Malaysia.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Yagui-Beltrán A and Jablons DM: A
translational approach to lung cancer research. Ann Thorac
Cardiovasc Surg. 15:2132009.PubMed/NCBI
|
|
2
|
Okudela K, Nagahara N, Katayama A and
Kitamura H: Cancer Stem Cells in Lung Cancer: Distinct Differences
between Small Cell and Non-Small Cell Lung Carcinomas. Cancer Stem
Cells Theories and Practice. InTech. 2011 https://www.intechopen.com/.../cancer-stem-cells-theorieMarch
22–2011. View
Article : Google Scholar
|
|
3
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caramori G, Casolari P, Cavallesco GN,
Giuffrè S, Adcock I and Papi A: Mechanisms involved in lung cancer
development in COPD. Int J Biochem Cell Biol. 43:1030–1044. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buttery RC, Rintoul RC and Sethi T: Small
cell lung cancer: The importance of the extracellular matrix. Int J
Biochem Cell Biol. 36:1154–1160. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pei J, Lou Y, Zhong R and Han B: MMP9
activation triggered by epidermal growth factor induced FoxO1
nuclear exclusion in non-small cell lung cancer. Tumour Biol.
35:6673–6678. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J, Li ZH, White J and Zhang LB: Lung
cancer stem cells and implications for future therapeutics. Cell
Biochem Biophys. 69:389–398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vermeulen L, Sprick MR, Kemper K, Stassi G
and Medema JP: Cancer stem cells - old concepts, new insights. Cell
Death Differ. 15:947–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freitas DP, Teixeira CA, Santos-Silva F,
Vasconcelos MH and Almeida GM: Therapy-induced enrichment of
putative lung cancer stem-like cells. Int J Cancer. 134:1270–1278.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dingli D and Michor F: Successful therapy
must eradicate cancer stem cells. Stem Cells. 24:2603–2610. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen X, Liao R, Li D and Sun J: Induced
cancer stem cells generated by radiochemotherapy and their
therapeutic implications. Oncotarget. 8:17301–17312.
2017.PubMed/NCBI
|
|
12
|
Tang Q-L, Liang Y, Xie XB, Yin JQ, Zou CY,
Zhao ZQ, Shen JN and Wang J: Enrichment of osteosarcoma stem cells
by chemotherapy. Chin J Cancer. 30:426–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liloglou T, Bediaga NG, Brown BR, Field JK
and Davies MP: Epigenetic biomarkers in lung cancer. Cancer Lett.
342:200–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Allan AL, Vantyghem SA, Tuck AB and
Chambers AF: Tumor dormancy and cancer stem cells: Implications for
the biology and treatment of breast cancer metastasis. Breast Dis.
26:87–98. 2006-2007. View Article : Google Scholar
|
|
15
|
Lobo NA, Shimono Y, Qian D and Clarke MF:
The biology of cancer stem cells. Annu Rev Cell Dev Biol.
23:675–699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Velasco-Velázquez MA, Popov VM, Lisanti MP
and Pestell RG: The role of breast cancer stem cells in metastasis
and therapeutic implications. Am J Pathol. 179:2–11. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maitland NJ and Collins AT: Prostate
cancer stem cells: A new target for therapy. J Clin Oncol.
26:2862–2870. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kozovska Z, Gabrisova V and Kucerova L:
Colon cancer: Cancer stem cells markers, drug resistance and
treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prince ME, Sivanandan R, Kaczorowski A,
Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles
LE: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang JC and Dick JE: Cancer stem cells:
Lessons from leukemia. Trends Cell Biol. 15:494–501. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kristiansen G, Schlüns K, Yongwei Y,
Denkert C, Dietel M and Petersen I: CD24 is an independent
prognostic marker of survival in nonsmall cell lung cancer
patients. Br J Cancer. 88:231–236. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tirino V, Camerlingo R, Franco R, Malanga
D, La Rocca A, Viglietto G, Rocco G and Pirozzi G: The role of
CD133 in the identification and characterisation of
tumour-initiating cells in non-small-cell lung cancer. Eur J
Cardiothorac Surg. 36:446–453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leung EL-H, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang WC, Shyh-Chang N, Yang H, Rai A,
Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al: Glycine
decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin S, Sun JG, Wu JB, Long HX, Zhu CH,
Xiang T, Ma H, Zhao ZQ, Yao Q, Zhang AM, et al: Aberrant microRNAs
expression in CD133+/CD326+ human lung
adenocarcinoma initiating cells from A549. Mol Cells. 33:277–283.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laimer K, Fong D, Gastl G, Obrist P, Kloss
F, Tuli T, Gassner R, Rasse M, Norer B and Spizzo G: EpCAM
expression in squamous cell carcinoma of the oral cavity: Frequency
and relationship to clinicopathologic features. Oral Oncol.
44:72–77. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Litvinov SV, van Driel W, van Rhijn CM,
Bakker HA, van Krieken H, Fleuren GJ and Warnaar SO: Expression of
Ep-CAM in cervical squamous epithelia correlates with an increased
proliferation and the disappearance of markers for terminal
differentiation. Am J Pathol. 148:865–875. 1996.PubMed/NCBI
|
|
29
|
Munz M, Baeuerle PA and Gires O: The
emerging role of EpCAM in cancer and stem cell signaling. Cancer
Res. 69:5627–5629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pak MG, Shin DH, Lee CH and Lee MK:
Significance of EpCAM and TROP2 expression in non-small cell lung
cancer. World J Surg Oncol. 10:53–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Skirecki T, Hoser G, Kawiak J, Dziedzic D
and Domagała-Kulawik J: Flow cytometric analysis of
CD133− and EpCAM-positive cells in the peripheral blood
of patients with lung cancer. Arch Immunol Ther Exp (Warsz).
62:67–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tachezy M, Zander H, Wolters-Eisfeld G,
Müller J, Wicklein D, Gebauer F, Izbicki JR and Bockhorn M:
Activated leukocyte cell adhesion molecule (CD166): An ‘inert’
cancer stem cell marker for non-small cell lung cancer? Stem Cells.
32:1429–1436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Beça FF, Caetano P, Gerhard R,
Alvarenga CA, Gomes M, Paredes J and Schmitt F: Cancer stem cells
markers CD44, CD24 and ALDH1 in breast cancer special histological
types. J Clin Pathol. 66:187–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH and
Kim HR: Colon cancer stem cell markers CD44 and CD133 in patients
with colorectal cancer and synchronous hepatic metastases. Int J
Oncol. 46:1582–1588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takaishi S, Okumura T, Tu S, Wang SS,
Shibata W, Vigneshwaran R, Gordon SA, Shimada Y and Wang TC:
Identification of gastric cancer stem cells using the cell surface
marker CD44. Stem Cells. 27:1006–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su J, Wu S, Wu H, Li L and Guo T: CD44 is
functionally crucial for driving lung cancer stem cells metastasis
through Wnt/β-catenin-FoxM1-Twist signaling. Mol Carcinog.
55:1962–1973. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barrandon Y and Green H: Cell size as a
determinant of the clone-forming ability of human keratinocytes.
Proc Natl Acad Sci USA. 82:5390–5394. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zakaria N, Yusoff NM, Zakaria Z, Lim MN,
Baharuddin PJ, Fakiruddin KS and Yahaya B: Human non-small cell
lung cancer expresses putative cancer stem cell markers and
exhibits the transcriptomic profile of multipotent cells. BMC
Cancer. 15:842015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi Y, Fu X, Hua Y, Han Y, Lu Y and Wang
J: The side population in human lung cancer cell line NCI-H460 is
enriched in stem-like cancer cells. PLoS One. 7:e333582012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang P, Gao Q, Suo Z, Munthe E, Solberg S,
Ma L, Wang M, Westerdaal NA, Kvalheim G and Gaudernack G:
Identification and characterization of cells with cancer stem cell
properties in human primary lung cancer cell lines. PLoS One.
8:e570202013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Levin TG, Powell AE, Davies PS, Silk AD,
Dismuke AD, Anderson EC, Swain JR and Wong MH: Characterization of
the intestinal cancer stem cell marker CD166 in the human and mouse
gastrointestinal tract. Gastroenterology. 139:2072–2082.e5. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li W, Ma H, Zhang J, Zhu L, Wang C and
Yang Y: Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem
cell markers in tumorigenesis and metastasis. Sci Rep. 7:138562017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bahnassy AA, Fawzy M, El-Wakil M, Zekri
AR, Abdel-Sayed A and Sheta M: Aberrant expression of cancer stem
cell markers (CD44, CD90, and CD133) contributes to disease
progression and reduced survival in hepatoblastoma patients: 4-year
survival data. Transl Res. 165:396–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang H, Lin XG, Hua P, Wang M, Ao X,
Xiong LH, Wu C and Guo JJ: The study of the tumor stem cell
properties of CD133+ CD44+ cells in the human
lung adenocarcinoma cell line A549. Cell Mol Biol (Noisy-le-grand).
56 Suppl:OL1350–8. 2010.PubMed/NCBI
|
|
45
|
Barrandon Y and Green H: Three clonal
types of keratinocyte with different capacities for multiplication.
Proc Natl Acad Sci USA. 84:2302–2306. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yan W, Chen Y, Yao Y, Zhang H and Wang T:
Increased invasion and tumorigenicity capacity of
CD44+/CD24− breast cancer MCF7 cells in vitro
and in nude mice. Cancer Cell Int. 13:622013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang C-P, Tsai MF, Chang TH, Tang WC,
Chen SY, Lai HH, Lin TY, Yang JC, Yang PC, Shih JY, et al:
ALDH-positive lung cancer stem cells confer resistance to epidermal
growth factor receptor tyrosine kinase inhibitors. Cancer Lett.
328:144–151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Izumiya M, Kabashima A, Higuchi H,
Igarashi T, Sakai G, Iizuka H, Nakamura S, Adachi M, Hamamoto Y,
Funakoshi S, et al: Chemoresistance is associated with cancer stem
cell-like properties and epithelial-to-mesenchymal transition in
pancreatic cancer cells. Anticancer Res. 32:3847–3853.
2012.PubMed/NCBI
|
|
51
|
Todaro M, Alea MP, Di Stefano AB,
Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G,
Medema JP, et al: Colon cancer stem cells dictate tumor growth and
resist cell death by production of interleukin-4. Cell Stem Cell.
1:389–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen Y-C, Chen YW, Hsu HS, Tseng LM, Huang
PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al: Aldehyde
dehydrogenase 1 is a putative marker for cancer stem cells in head
and neck squamous cancer. Biochem Biophys Res Commun. 385:307–313.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang EH, Hynes MJ, Zhang T, Ginestier C,
Dontu G, Appelman H, Fields JZ, Wicha MS and Boman BM: Aldehyde
dehydrogenase 1 is a marker for normal and malignant human colonic
stem cells (SC) and tracks SC overpopulation during colon
tumorigenesis. Cancer Res. 69:3382–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci
F, Jacquemier J, et al: Aldehyde dehydrogenase 1-positive cancer
stem cells mediate metastasis and poor clinical outcome in
inflammatory breast cancer. Clin Cancer Res. 16:45–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tanei T, Morimoto K, Shimazu K, Kim SJ,
Tanji Y, Taguchi T, Tamaki Y and Noguchi S: Association of breast
cancer stem cells identified by aldehyde dehydrogenase 1 expression
with resistance to sequential Paclitaxel and epirubicin-based
chemotherapy for breast cancers. Clin Cancer Res. 15:4234–4241.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bouchier-Hayes L, Muñoz-Pinedo C, Connell
S and Green DR: Measuring apoptosis at the single cell level.
Methods. 44:222–228. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ma D, Tremblay P, Mahngar K, Akbari-Asl P,
Collins J, Hudlicky T and Pandey S: Induction of apoptosis and
autophagy in human pancreatic cancer cells by a novel synthetic C-1
analogue of 7-deoxypancratistatin. Am J Biomed Sci. 3:278–291.
2011. View Article : Google Scholar
|
|
59
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang X, Zhu Y, Ma Y, Wang J, Zhang F, Xia
Q and Fu D: The role of cancer stem cells in cancer metastasis: New
perspective and progress. Cancer Epidemiol. 37:60–63. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang F, Ma L, Zhang Z, Liu X, Gao H,
Zhuang Y, Yang P, Kornmann M, Tian X and Yang Y: Hedgehog signaling
regulates epithelial-mesenchymal transition in pancreatic cancer
stem-like cells. J Cancer. 7:408–417. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ni J, Cozzi P, Hao J, Beretov J, Chang L,
Duan W, Shigdar S, Delprado W, Graham P, Bucci J, et al: Epithelial
cell adhesion molecule (EpCAM) is associated with prostate cancer
metastasis and chemo/radioresistance via the PI3K/Akt/mTOR
signaling pathway. Int J Biochem Cell Biol. 45:2736–2748. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Patriarca C, Macchi RM, Marschner AK and
Mellstedt H: Epithelial cell adhesion molecule expression (CD326)
in cancer: A short review. Cancer Treat Rev. 38:68–75. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wielenga VJ, Smits R, Korinek V, Smit L,
Kielman M, Fodde R, Clevers H and Pals ST: Expression of CD44 in
Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J
Pathol. 154:515–523. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hou J-M, Krebs M, Ward T, Sloane R, Priest
L, Hughes A, Clack G, Ranson M, Blackhall F and Dive C: Circulating
tumor cells as a window on metastasis biology in lung cancer. Am J
Pathol. 178:989–996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Mizrak SC, Chikhovskaya JV, Sadri-Ardekani
H, van Daalen S, Korver CM, Hovingh SE, Roepers-Gajadien HL, Raya
A, Fluiter K, de Reijke TM, et al: Embryonic stem cell-like cells
derived from adult human testis. Hum Reprod. 25:158–167. 2010.
View Article : Google Scholar : PubMed/NCBI
|