Introduction
The treatment of malignant brain tumors is one of
the most complex areas of modern medicine. Upon completing all
standard modern treatments, the median survival time of patients
with glioblastoma is 1 year (1).
Less than one-quarter of patients with metastatic brain tumors have
an 8–12-month survival rate (2).
Metastatic and invasive processes are the principal characteristics
of malignant tumors (3). The low
treatment success rate may be attributed to the use of outdated
methods that have limited effects on these processes (4).
Invasive and metastatic processes are now known to
be associated with cancer stem cells (CSCs) (5). A high dose of radiation does not
guarantee an effective elimination of these cells in the tumor
node. The heterogeneity and dynamic nature of the glioblastoma stem
cell population, in combination with the selective permeability of
the blood-brain barrier for medicinal substances, explain why
currently available agents remain unable to successfully eliminate
these cells (6). These cells are
unaffected by the external environment and respond to stimuli by
producing new and more resistant clones of neoplastic cells;
therefore, a combination of existing therapeutic methods, including
chemotherapy and radiotherapy, is required to control CSCs.
Biomedical cell products consisting of a complex of
a cell line and an adjuvant represent a novel type of antitumor
agent, and are being developed to address the problem of CSC
resistance to standard treatment. The most promising agent is a
leukoconcentrate of receptor-type tyrosine-protein phosphatase C
(CD45)+ and hematopoietic progenitor cell antigen CD34
(CD34)+ mononucleocytes, with a high content of
hematopoietic stem cells (HSCs). This class of cells is known for
having a high propensity to migrate towards CSCs (7). As the evidence demonstrates, these
cells are able to reach CSCs and affect certain signaling pathways
associated with proliferation and invasion (7). In previous research, it was reported
that intercellular cooperation with HSCs involves the transmission
of the cytoplasmic content into glioma and carcinoma cells, which
has a pronounced inhibitory effect on tumor cell activity (8). This phenomenon may be used for the
regulation of CSCs by inhibiting the mechanisms that determine
their invasive abilities.
However, the aggressiveness of glioblastoma does not
only depend on the degree of tumor cell differentiation; it also
depends on their specific molecular phenotype, formed during
coordinated intercellular interactions under the influence of
clonal selection factors. The key pathogenic mechanism for the
transformation of the molecular phenotype of tumor cells is
epithelial-mesenchymal transition (EMT), in which epithelial cells
lose their apical-basal polarity, the cytoskeleton is reorganized,
components of the extracellular matrix are secreted, and the cells
become more mobile and capable of invasion into the surrounding
tissues and migration to distant organs (9–11). In
relation to neuroepithelial tumors, this mechanism allows for the
transformation of a primary glioma into a secondary glioblastoma,
the transition from a proneural subtype of the tumor to a
mesenchymal one, the creation of more aggressive clones of CSCs and
the generation of new CSC populations (12).
Transforming growth factor (TGF)-β1 serves a key
role in EMT-inducing mechanisms. This cytokine has been
demonstrated to trigger EMT in certain cases of breast cancer
(13), renal carcinoma (14), intestinal tumors (15–17),
pancreatic cancer (18) and
glioblastoma (19). Previous
research (20) demonstrated that
stimulation of U87 MG cell lines with TGF-β1 significantly
increases the production of proteins associated with invasion,
proliferation, migration, DNA regeneration, stemness, and
resistance to drugs and radiation. The resulting pool of
glioblastoma cells has a molecular phenotype that is very similar
to that of CSCs (21). It was
reported that in human glioblastoma tumor cells, only two signaling
pathways (the integrin and focal adhesion pathways) are available
for regulatory effects on gene expression processes in the CSC
nucleus (20).
It is likely that the upregulation of the protein
components of these signaling pathways due to EMT determines the
interactions of the CSCs with the microenvironment and
extracellular matrix. Theoretically, normal СD45+
CD34+ stem cells are able to regulate these processes
during their interaction with CSCs, which may become a key point in
cancer therapy and for overcoming therapeutic resistance. Bone
marrow cells represent 30–50% of non-neoplastic glioblastoma cells.
The aim of the present study was to investigate patterns of
interaction between hematopoietic stem cells and glioblastoma cells
stimulated by TGF-β1 in vitro.
Materials and methods
Cancer cells
The current study used human glioblastoma U-87 MG
cells (American Type Culture Collection® HTB-14™;
Manassas, VA, USA). This cell line is not the original U87 line
established at the University of Uppsala (Uppsala, Sweden),
although it is likely to be derived from a glioblastoma of unknown
origin (21). However, as
demonstrated in a previous study, cells of this line expressing the
epitope prominin-1 (CD133) have high similarity of proteasomal
profiles with neural CD133+ human stem cells and
significant proteomic differences compared with normal mesenchymal
stem cells of the human bone marrow (20). In a previous study (19), stimulation of U87 glioblastoma cells
with TGF-β1 led to a significant increase in the expression of
proteins associated with EMT, which markedly increased their
invasiveness. These arguments served as the basis for choosing this
line of tumor cells for studying the processes of cell-cell
interaction in vitro.
Human bone marrow cells
A leukoconcentrate of CD45+
mononucleocytes mobilized from the peripheral blood following an
injection of a granulocyte colony-stimulating factor (filgrastim)
was used, according to a previously described method (22). The cells were provided freely by the
CJSC NeuroVita Clinic of Restorative and Interventional Neurology
and Therapy (Moscow, Russia), with the donors' permission.
According to the supporting documents, the sample contained 4.5%
cells with a СD34+CD45+ immunophenotype and
1.8% cells with integrin-β1+, CD44 antigen
(CD44)+, 5′-nucleotidase+ and Thy-1 membrane
glycoprotein+ markers. An immunosorting method was used
for isolating CD45+ CD34+ cells from the
leukoconcentrate (21), and these
cells were used for the present study. The use of human samples in
this study was approved by the Ethical Committee of the School of
Biomedicine, The Far Eastern Federal University (Vladivostock,
Russia; minute no. 1 of February 2nd 2017) and the Academic Council
of the School of Biomedicine.
The cells were frozen and cultivated at 37°C and 5%
CO2, in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
10,000 U/ml penicillin/streptomycin, with 25 mg/ml fungizone. Cells
were stained with fluorescent dyes and used for subsequent
experiments.
Cell staining with fluorescent
dyes
The cells were stained with CellTracker™ Red CMTPX
Dye (cat. no. C34552; Molecular Probes; Thermo Fisher Scientific,
Inc.; λ, 546 nm; 15 µM in DMEM; 25 min; 37°С) and
Vybrant® CFDA SE (cat. no. V12883; Molecular Probes;
Thermo Fisher Scientific, Inc.; λ, 488 nm; 25 µM in PBS; 25 min;
37°С), according to the manufacturer's protocols. Fluorescence was
analyzed using a Carl Zeiss LSM 710 confocal laser-scanning
microscope (Carl Zeiss AG, Oberkochen, Germany) with a standard set
of filters. Lenses with magnification ×10, ×20 and ×40 were used
for observation.
Experimental design
The effects of recombinant human TGF-β1
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) on glioblastoma
cells were studied in 24-well plates; each well was seeded with
3×104 cancer cells stained with CMTPX. TGF-1β, at a
concentration of 10, 20 or 30 ng/ml, was placed into the culture
medium, while the control wells contained cells without TGF-1β. The
experiment was conducted with the computer-aided Cell IQ system (CM
Technologies GmbH, Elmshorn, Germany).
To study the migration of CD45+
CD34+ cells, 24-well plates with preset cell culture
inserts were used, each with a pore size of 8 µm (SPL Life
Sciences, Pocheon, Korea). The cell culture inserts were seeded
with 3×104 cancer cells stained with CMTPX dye; the
wells contained 6×104 HSCs stained with CFDA dye, and
certain wells contained cultures with 10 ng/ml TGF-1β. In the
control group, HSCs were also introduced into the wells, although
the inserts had the culture medium without any cells or additives.
Analysis was conducted with a flow cytofluorometer.
The process of exchanging a fluorescent tag was
examined by co-culturing the cells at a ratio of 1:1, specifically
2.5×104 cancer cells stained with red CMTPX with
2.5×104 monocytes stained with green CFDA; certain wells
contained 10 ng/ml TGF-1β. For monitoring, a confocal laser
microscope was used for 96 h. Lenses with magnification ×10, ×20
and ×40 were used for observation. The samples were placed in a
chamber filled with 5% CO2. Images were captured every 3
h. HSC adhesion to cancer cells was also studied using an electron
microscope. Cells were cultured in DMEM with 10% FBS and 0.5%
penicillin/streptomycin in Petri dishes for 72 h, following which
the cells were extracted, washed and centrifuged, (120 × g; 5 min;
37°C), fixed and embedded for further analysis.
To study the proteomes of the glioblastoma cells, a
combination of high-performance liquid chromatography and mass
spectrometry was used. The label-free method (21) was applied for evaluating the protein
expression levels. The purposes of the study required the primary
emphasis of the bioinformatic analysis to be placed on marker
proteins of EMT and proteins of signaling pathways determining the
interactions between cancer cells and the extracellular matrix.
Electron microscopy
The cells were fixed in 2.5% glutaraldehyde for 1 h,
and washed in PBS twice (30 min/wash; 25°С). Post-fixation was
conducted in osmium for 1 h, following which the cells were
dehydrated in 30, 50, 70 and 80% ethanol, and twice in 96% ethanol,
for 10–15 min in each solution (25°С). Subsequently, the samples
were treated with ethanol-acetone at ratios of 3:1, 1:1 and 1:3 for
15 min in each solution (25°C). The samples were embedded in resin
(Epon-812, 4.4 ml; dodecenylsuccinic anhydride, 3.7 ml; methyl
nadic anhydride, 1.9 ml) by infiltrating them into a graded series
of acetone and resin mixture with a 3:1 ratio for 30 min, 1:1 for 2
h, 1:3 for 30 min, and resin alone for 12 h (25°С). The embedding
was finalized as follows: Two drops of accelerator per 1 ml of
resin; and polymerization lasting for 72 h at 60°C. Sections of
70-nm thickness were prepared using Ultracat E Reichert (Leica
Microsystems, Inc., Buffalo Grove, IL, USA). The sections were
stained with the following solutions: 1% uranyl acetate for 6 min;
and lead citrate for 1.5 min. The transmission electron microscope
(Libra 120; Carl Zeiss AG) was used for visualization of the
results.
Flow cytofluorometry
The exchange of a fluorescent tag was additionally
studied with a flow cytofluorometer. The cells stained with
fluorescent tracers were extracted using trypsin and washed by
double centrifuging in PBS (120 × g; 3 min; 37°С). The precipitate
was resuspended in 400 µl PBS, and analyzed with a BD
Accuri® C6 flow cytometer (BD Biosciences, Franklin
Lakes, NJ, USA). At least 10,000 single cells were analyzed in each
sample. To distinguish single cells from aggregated ones, and to
exclude the aggregated cells from the analysis, the following
methods were used: A combination of signals from forward (value
proportional to the size of the cells) and side (value
characterizing the cell structure) light scattering; and the
combination of the intensity of the peak signal against the
intensity of integrated forward or side scattering signals. The
results obtained were analyzed with Kaluza 1.3 software (Beckman
Coulter, Inc., Brea, CA, USA).
Comparative analysis of cellular
proteomes
Cancer cells (50,000) were seeded into 6-well plates
and cultivated at 37°C with 5% СО2 to reach 30%
confluence, in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.)
medium containing 10% FBS, 1% 200 mM L-glutamine and 20 mM HEPES.
Subsequently, the cells were washed with Hanks' Balanced Salt
Solution and transferred into DMEM/F12 serum-free medium with the
addition of 5 ng/ml TGF-β1 for 72 h. The cells of the monolayer
culture were extracted, the precipitate was centrifuged out (120 ×
g; 10 min; 25°С), supernatant was removed, and the cells were
resuspended in 3 ml PBS (рН 7.4). PBS washing was repeated twice.
The cells were lysed with a Mammalian Cell Lysis kit
(Sigma-Aldrich; Merck KGaA). The received samples were purified
from low-molecular components using Agilent 5K MWCO Spin
Concentrators for Proteins (Agilent Technologies GmbH, Waldbronn,
Germany). Tryptic cleavage: Lysates were added to
2.2.2-trifluoroethanol (Reagent Plus; Sigma-Aldrich; Merck KGaA),
NH4HCO3 (Ultra; Fluka Chemie AG, Buchs,
Switzerland) and trichloroethylphosphate (Fluka Chemie AG) (1 h;
600°C). Aqueous iodoacetomide (Sigma-Aldrich; Merck KGaA) was added
(30 min; 250°C); subsequently, NH4HCO3
solution, water and trypsin solution (porcine pancreatic;
proteomics grade; demethylated; Sigma-Aldrich; Merck KGaA) in 1 mM
hydrochloric acid (Purum; Chimmed, Moscow, Russia) (18 h; 370°C).
Solutions were analyzed by mass spectrometry for trypsinolysis
completeness, which was controlled by the peaks of tryptic peptides
and by the areas of peaks with m/z 842.51 Da and 421.76 Da.
Mass-spectrometry: Tryptic peptides were analyzed in the nanoflow
chromatograph Dionex Ultimate 3000 in combination with mass
spectrometer LTQ Orbitrap XL (Thermo Fisher Scientific, Inc.) with
a nanospray ionization ion source. Mass spectra were obtained using
the positive ion mode in the m/z 300–2,000 Da range, a needle
voltage of 1.7 kB, a source temperature of 200°С, a voltage to
capillary of 43 В, and to lens 165 В. Mass spectra were registered
in the orbital trap in Fourier transform mode; tandem spectra were
obtained by ionization induced by collisions in a linear trap
(enhanced scanning mode).
To identify proteins, the mass spectra were
converted into.mgf files using Proteome Discoverer 1.0 software
(Thermo Fisher Scientific, Inc.); pattern WF_Spectrum_Export_MGF
was set as the default except for the mass range (between 300 and
10,000 Da) and retention time (0–180 min). Protein searches were
performed on the local server with Mascot Server 2.3.02 software
(Matrix Science, Ltd., London, UK). Search parameter were as
follows: Database, National Center for Biotechnology Information
non-redundant GenBank (version, January 25th 2012); species,
Homo sapiens; enzyme, trypsin; number of missed cleavages,
2; accuracy of parent ion mass, 10 ppm; fragments, 0.8 Da; ion
trap. Identified proteins were sorted by MudPIT score (Mascot
Server 2.3.02 software; Matrix Science, Ltd.; http://www.matrixscience.com/server.html), presenting
peptides at a significance level of P<0.05. Received lists of
identified proteins and mass spectra were uploaded to Skyline
1.2.0.3303 (University of Washington, Seattle, WA, USA; http://skyline.gs.washington.edu/labkey/project/home/software/Skyline),
and peak peptide areas were received for every probe. The areas of
all the peaks of every identified peptide were summed and
normalized according to total area of all identified peaks in the
probe. Biological processes, molecular functions, cellular
localization and protein signaling pathways were annotated using
the PubMed (https://www.ncbi.nlm.nih.gov/pubmed), PANTHER
(http://www.pantherdb.org), Gene Ontology
(http://geneontology.org) and Kyoto Encyclopedia
of Genes and Genomes databases (https://www.genome.jp/kegg/kegg1.html).
Statistical analysis
The data were processed using GraphPad Prism 4.0
software (GraphPad Software, Inc., La Jolla, CA, USA). Results of
the migration and electron microscopy study are presented as
boxplots, indicating quartiles 1 and 3, the median, and the minimum
and maximum data values (whiskers). Statistical analysis was
performed using the Mann-Whitney U test. The results were
considered to be statistically significant at
U<Ucrit=93 (n=19) for α=0.01; and U<Ucrit=113
(n=19) for α=0.05.
The results of the study of the effects of TGF-β1 on
glioblastoma cells are presented as the mean ± standard error of
the mean. The analysis of variance F-test was used for data
analysis (n=75; m=25) followed by a post hoc t-test with Bonferroni
correction. Results were considered to be statistically significant
at values of family-wise error rate <P<α.
Results
Stimulation of control cells with
TGF-β1
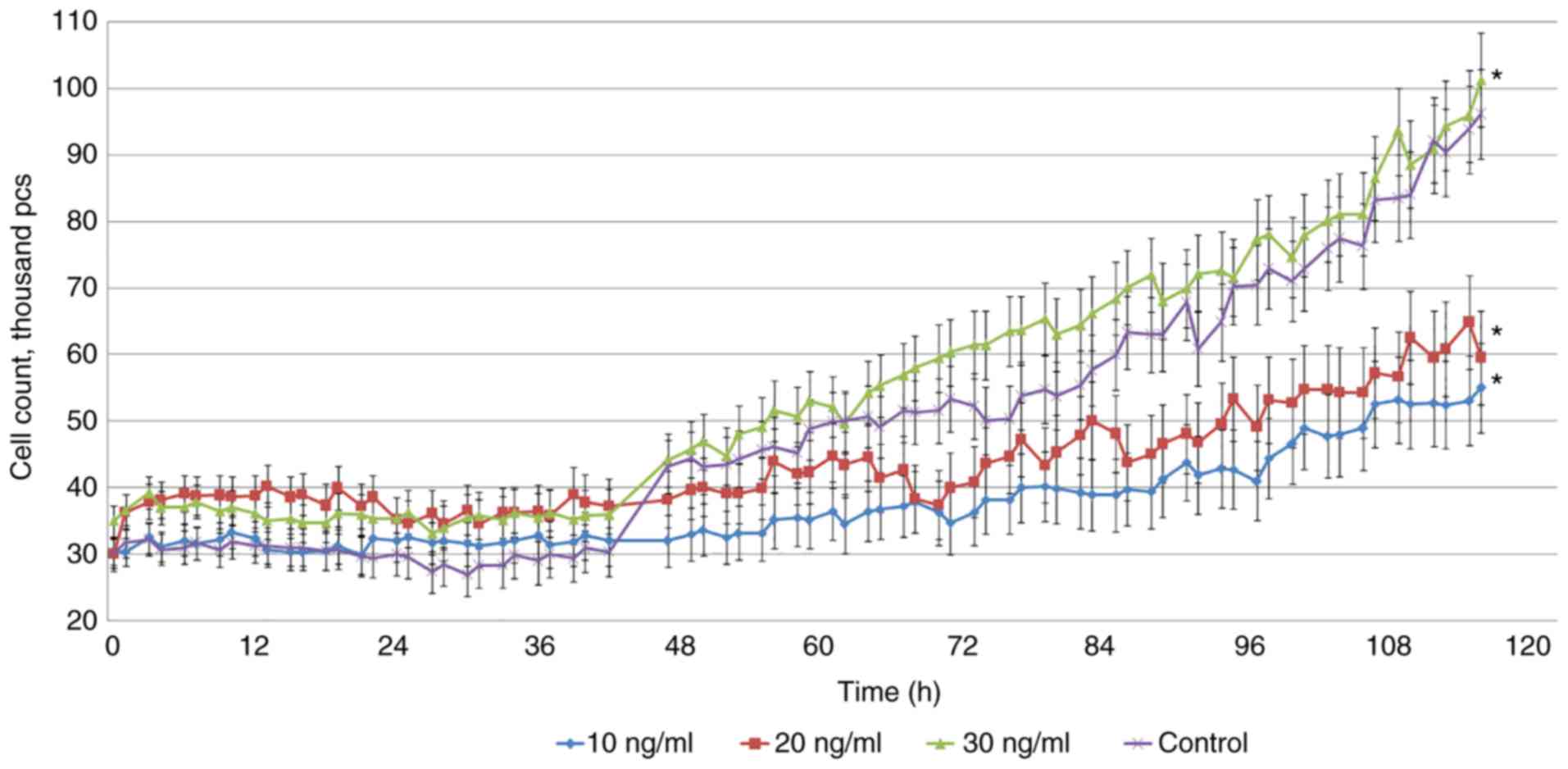
TGF-β1 (30 ng/ml) did not have a pronounced effect
on the proliferation of U87 glioblastoma cells. The gradual
reduction of the TGF-β1 concentration to 20 and 10 ng/ml (Fig. 1) slowed the cell proliferation and
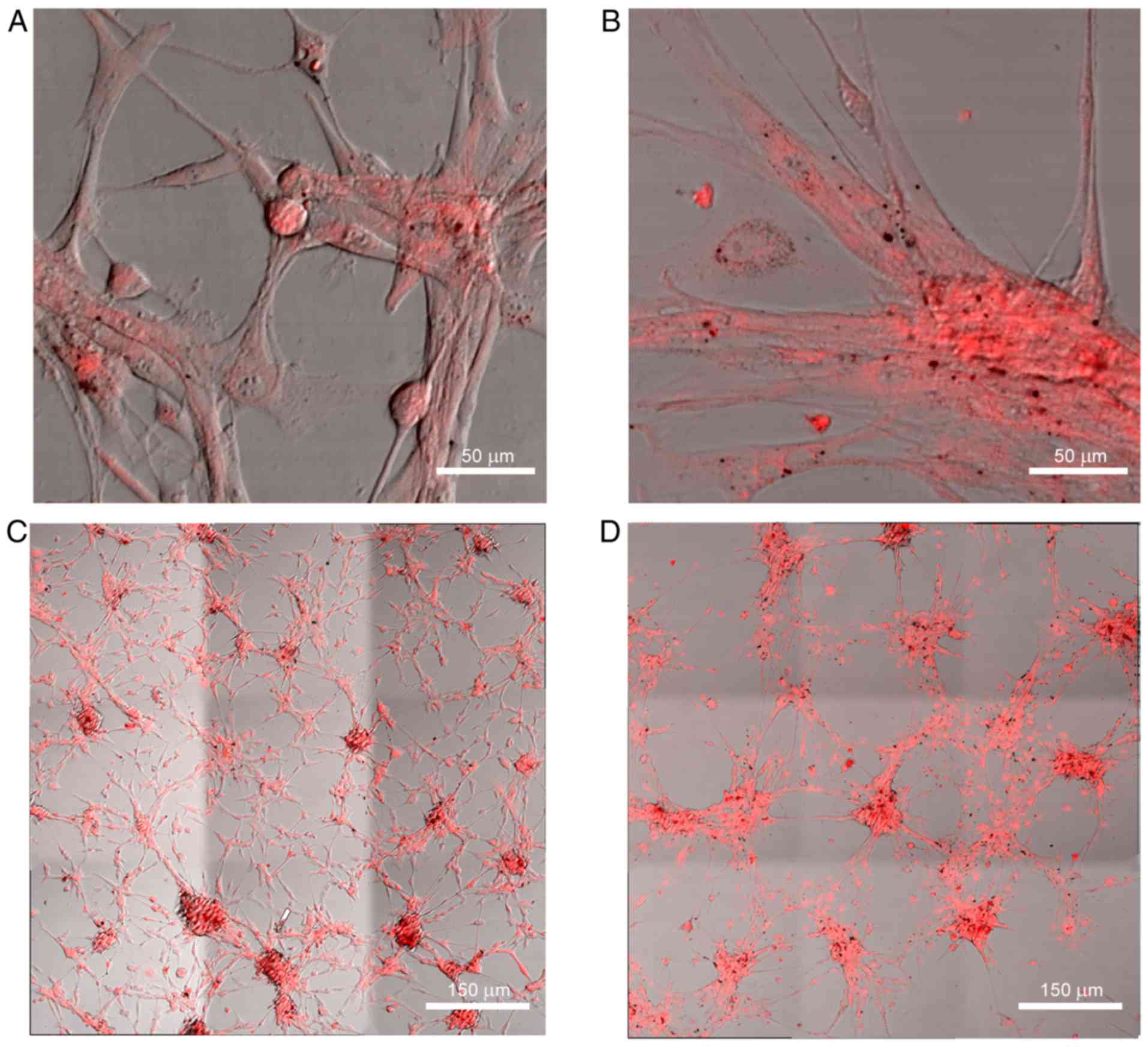
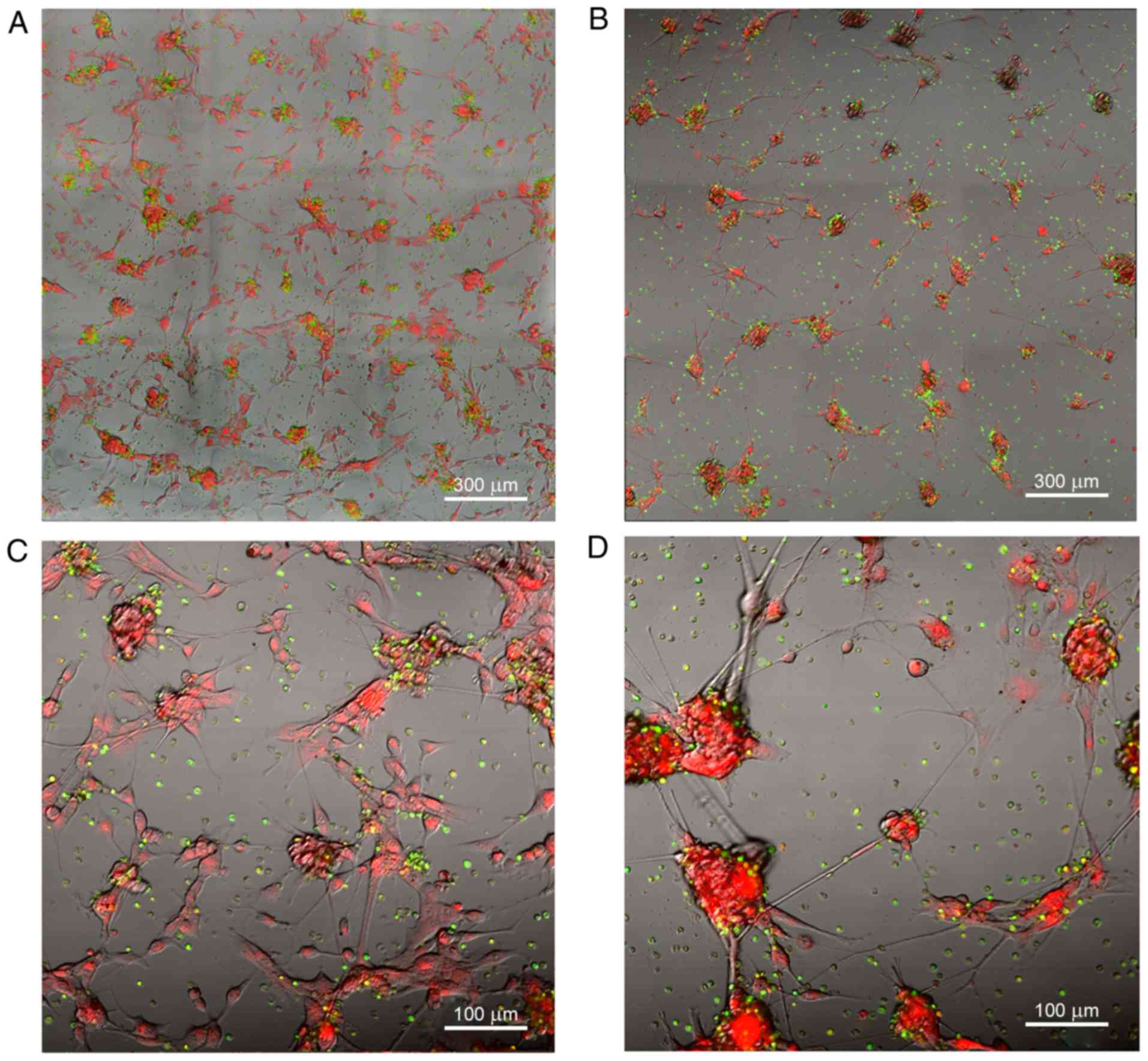
led to notable morphological alterations (Fig. 2A and B). Glioblastoma cells were
dispersed over the surface of the well and created conglomerates
with an evenly distributed fluorescent tag. These were connected
with the microtubes of dispersed cells, which was particularly
noticeable in the panoramic view (Fig.
2С and D). This phenomenon was absent in the control group.
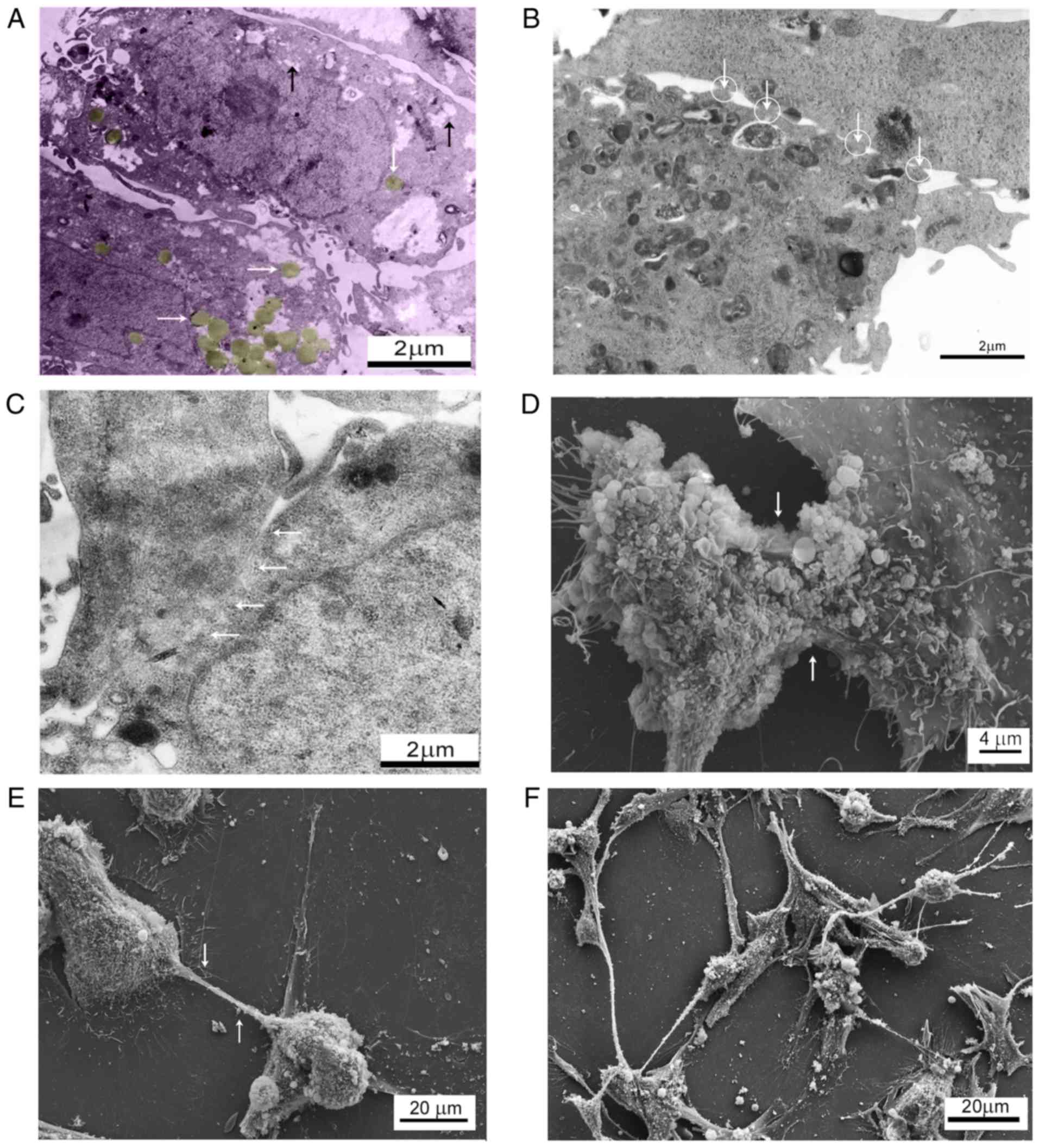
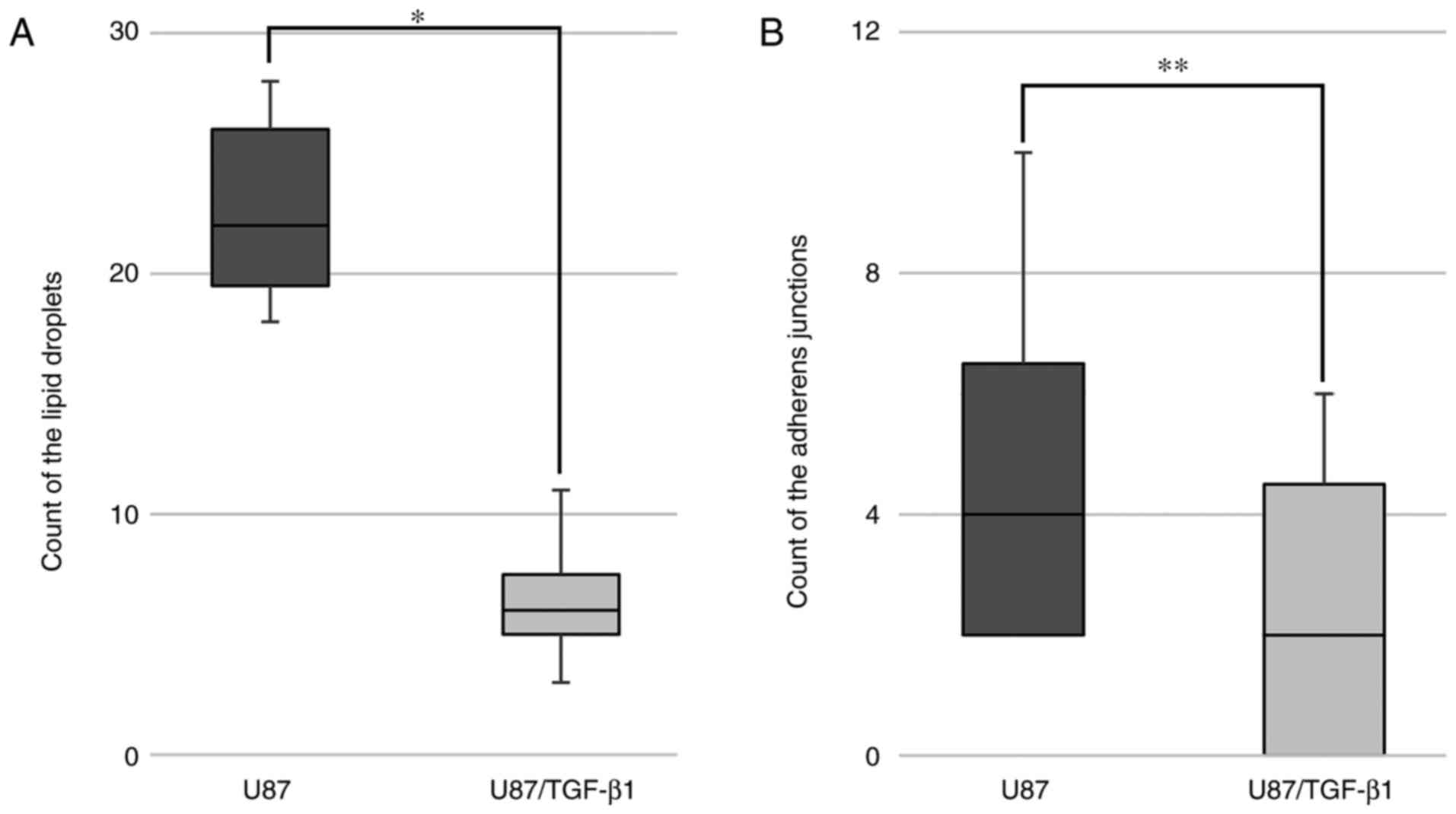
The analysis of electronograms revealed cells with
large nuclei and irregular shapes, dispersed chromatin and
well-defined nucleoli. The cytoplasm of the control cells contained
larger areas of lower electron density and dispersed distribution
of components resembling carbohydrate inclusions, among which there
were round objects resembling lipid droplets (Fig. 3A). Touching the cell surface, they
created adhesion and contact bands with a spongy amorphous matrix
spread among them (Fig. 3B). In
places, there were large areas of cell adhesion with
interdigitation represented by adjacent cellular membranes that had
their surface dissolved in sections, thus creating zones of
cytoplasmic fusion with homogenous content (Fig. 3C). The scanning microscopy revealed
certain cells with fused membranes (Fig. 3D), and cells joined by cytoplasm
tube-like extensions that allowed single cells (Fig. 3E) and cell conglomerates (Fig. 3F) to interact with each other.
Stimulation of glioblastoma cells with TGF-β1 was accompanied by a
decrease in the number of intercellular contacts and the almost
complete disappearance of specific inclusions in the cytoplasm
(Fig. 4). The presence of numerous
filopodia indicated an increased rate of cancer cell mobility.
Interaction of HSCs and cancer cells
in vitro
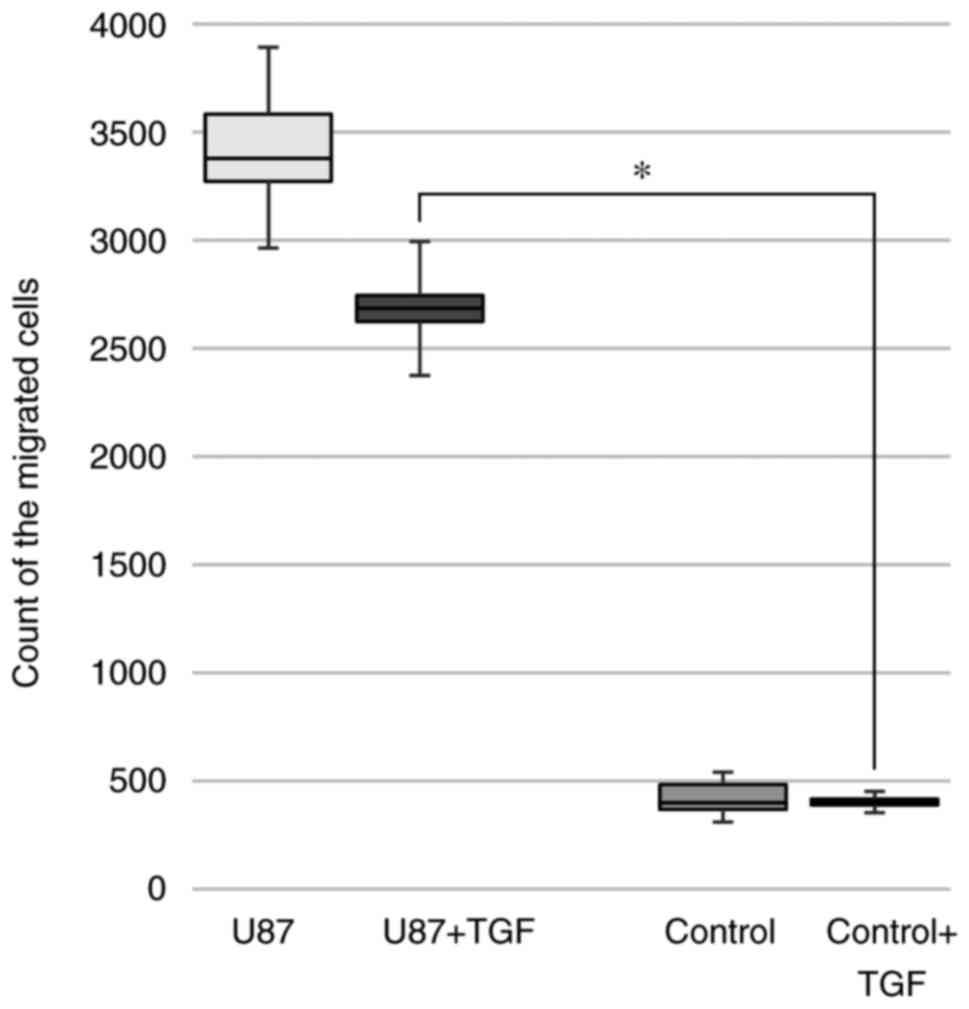
According to the automated monitoring, the migration
of HSCs to the inserts with glioblastoma cells in the
non-stimulated experimental group was more dynamic compared with
the group stimulated with TGF-β1 (Fig.
5). According to the monitoring data, the increase in the
number of HSCs migrating to U87 cells enhanced the ability of the
tumor cells to detach from the substrate and move across the well
surface.
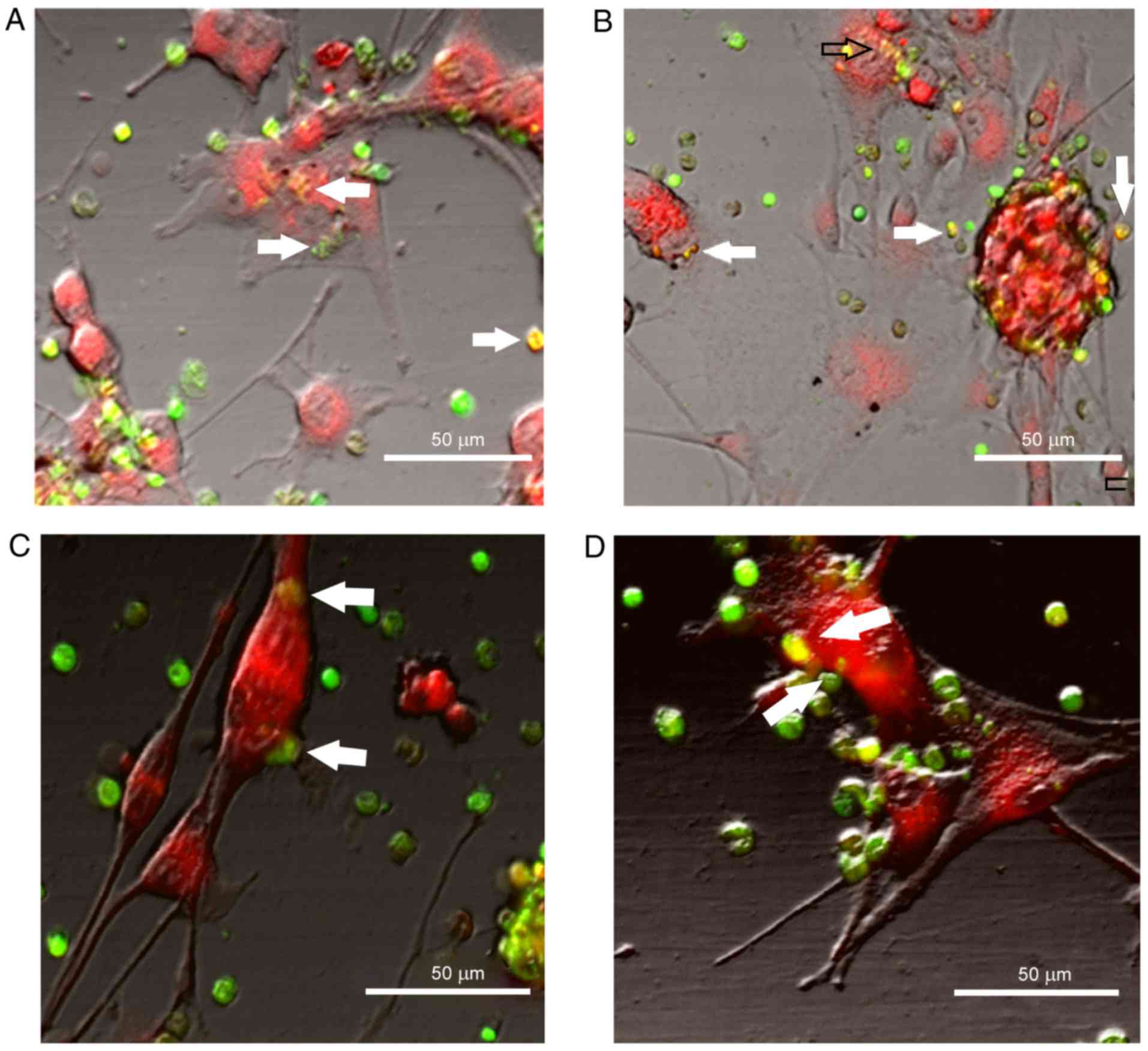
After 48 h, cancer cells in the control group and
the experimental co-culture with adhesive HSCs were arranged in
grape-like clusters (Fig. 6). After
12 h, in addition to the fluorescent cells stained with CMTPX RED
(λ, 546 nm; red tag) and adherent HSCs stained with CFDA (λ, 488
nm; green tag), green fluorescent objects of a smaller size began
to appear on the surface of and inside glioblastoma cells (Fig. 7). This phenomenon was also present
in the cell culture stimulated by TGF-β1; however, by the end of
the experiment there were notably fewer HSCs in the well with
co-culture compared with the control group.
The exchange of the fluorescent tag was not
one-sided. As the observation progressed, the fluorescence of the
stem cells adherent to glioblastoma cells exhibited dominant yellow
tones (Fig. 7) that were attributed
to a spectral intersection of fluorescent signals from СFDA (λ, 488
nm) and CMTPX Red (λ, 546 nm) dyes. This was likely to have been
caused by the fluorescent tag carried by the cancer cells. In the
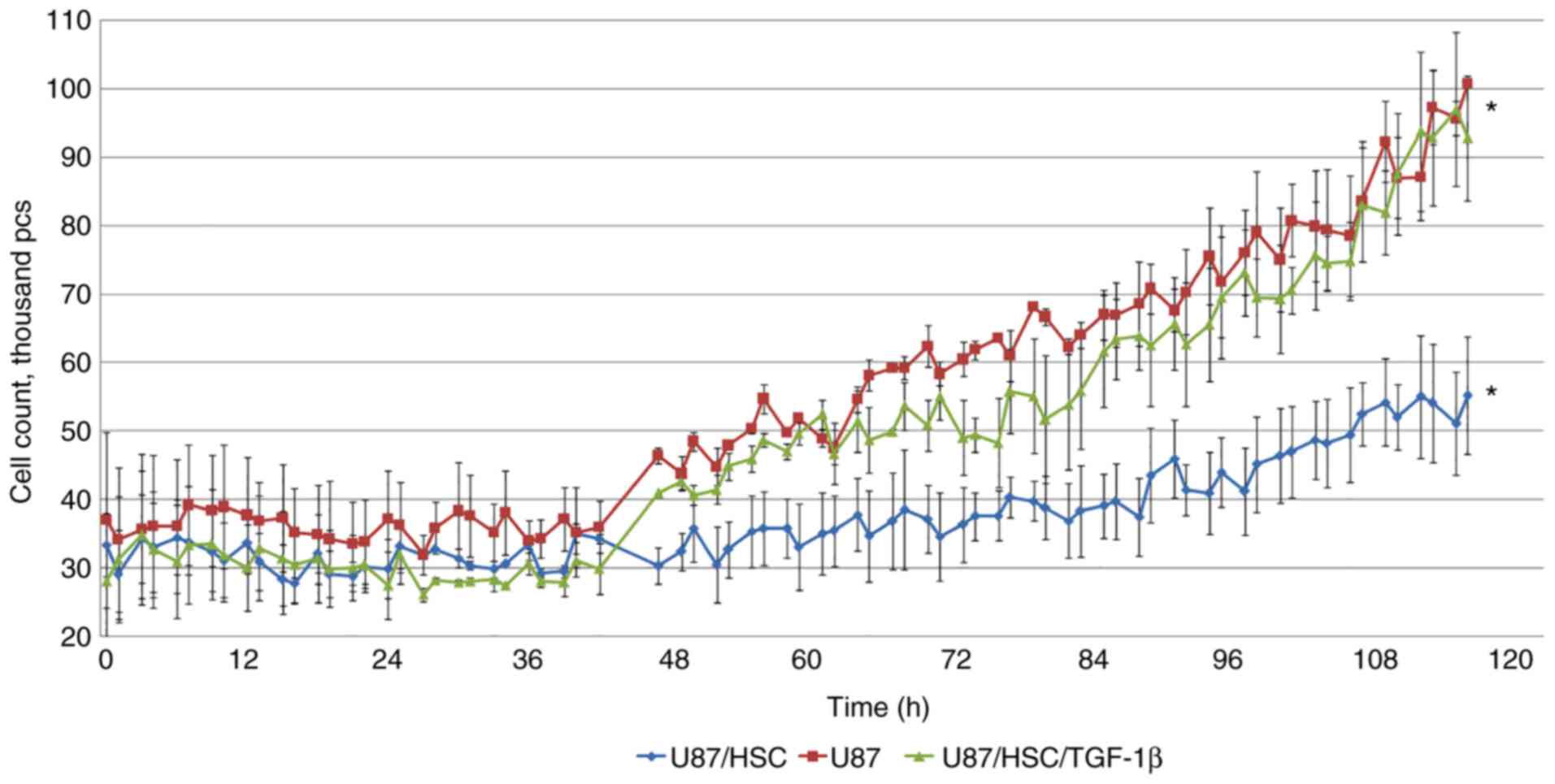
non-stimulated group these results were similar. Interaction with
HSCs had a pronounced effect on glioblastoma cells, which was
indicated by the proliferation rate returning to a level similar to
that of the control group (Fig.
8).
Comparative analysis of cancer cell
proteomes
This phase of the experiment involved analyzing the
impact of TGF-β1 on the molecular phenotype of glioblastoma cells.
During the incubation of the U87 line cells in the medium with
TGF-β1, 637 proteins exhibited a significant alteration in
expression; 513 proteins had an increase in expression, while 124
proteins had an expression decrease (P<0.01). Bioinformatics
analysis of proteins with significant alterations in their
expression was based on their status as EMT markers (Table I) and their involvement in signaling
mechanisms regulating cellular interactions with the
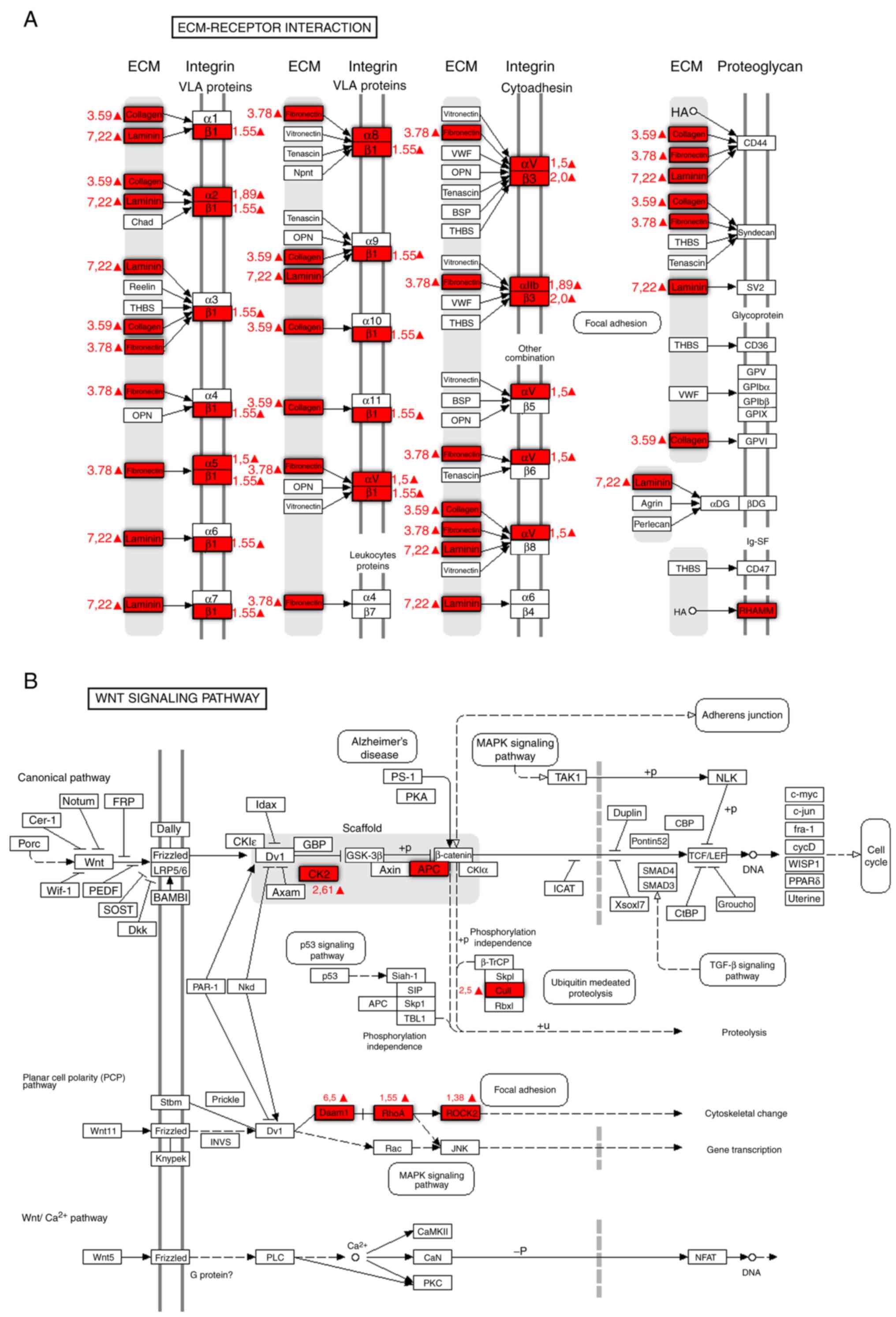
microsurroundings and extracellular matrix (Table II). Fig. 9 presents a schematic representation
of protein upregulation under the influence of TGF-β1. These
proteins are components of the interaction between the
extracellular matrix and the cell membrane. There are also a number
of proteins that are simultaneously adjusted with the components of
the ECM in this schematic. The synthesized components of the
extracellular matrix allow cells to invade surrounding tissues. The
components of the integrin signaling pathway, following stimulation
with TGF-β1, also undergo upregulation, which transforms cells
stimulated by TGF-β1 into cancer stem cells (Table II; Fig.
9)
 | Table I.Alteration in the expression of
proteins involved in epithelial-mesenchymal transition in cells of
the U87 human glioblastoma line, following stimulation with
TGF-β1. |
Table I.
Alteration in the expression of
proteins involved in epithelial-mesenchymal transition in cells of
the U87 human glioblastoma line, following stimulation with
TGF-β1.
| Abbreviation | Protein name | Ratio,
TGF-β1/control |
|---|
| CDH1 | E-cadherin | ↓ |
| OCLN | Occludin | ↓ |
| CLDN1 | Claudin1 | 0.65 |
| VIM | Vimentin | 2.81 |
| FN1 | Fibronektin1 | 4.10 |
| FNDC3B | Fibronectin type
III domain containing 3B | 3.32 |
| Actl6a | Actin-like 6A | ↑ |
| Actn1 | Actinin, α1 | 2.04 |
| ARPC3 | Actin related
protein 2/3 complex subunit 3 | 2.31 |
| MYBPC3 | Myosin binding
protein C, cardiac | 5.71 |
| Myo1C | Myosin IC | 2.82 |
| MYO5A | Myosin VA (heavy
chain 12, myoxin) | ↑ |
| Myo7a | Myosin VIIA | ↑ |
| MMP2 | Matrix
metallopeptidase 2 | 2.85 |
| MMP9 | Matrix
metallopeptidase 9 | 2.10 |
| MMP14 | Matrix
metallopeptidase 14 | ↑ |
| ADAMTS1 | ADAM
metallopeptidase with thrombospondin type 1 motif, 1 | ↑ |
 | Table II.Proteins of the integrin and focal
adhesion signaling pathways with enhanced expression in
glioblastoma cells of the U87 line after stimulation of TGF-β1. |
Table II.
Proteins of the integrin and focal
adhesion signaling pathways with enhanced expression in
glioblastoma cells of the U87 line after stimulation of TGF-β1.
| Abbreviation | Protein name | Ratio,
TGF-β1/control |
|---|
| ITGA2 | Integrin, α2 | 1.91 |
| ITGA8 | Integrin, α8 | ↑a |
| ITGAX | Integrin, αX | 1.61 |
| ITGA3 | Integrin, α3 | 0.81 |
| ITGAV | Integrin, αV | 1.51 |
| ITGA5 | Integrin, α5 | 1.55 |
| ITGB2 | Integrin, β2 | 1.21 |
| ITGB3 | Integrin, β3 | 2.01 |
| ITGB1 | Integrin, β1 | 1.52 |
| Memo1 | Mediator of cell
motility 1 | ↑ |
| LAMB1 | Laminin, β1 | 7.22 |
| Col15a1 | Collagen, type XV,
α1 | 2.42 |
| COL1A2 | Collagen, type I,
α2 | 3.59 |
| COL6A1 | Collagen, type VI,
α1 | 2.34 |
| COL7A1 | Collagen, type VII,
α1 | 4.58 |
| RHAMM | Hyaluronan-mediated
motility receptor | ↑ |
| CDC42 | Cdc42
GTPase-activating protein | ↑ |
| RhoA | Ras homolog gene
family, member A | 1.45 |
| RHOC | Ras homolog gene
family, member C | 1.21 |
| ROCK2 | Rho-associated,
coiled-coil containing protein kinase 2 | 1.38 |
| CK2A2 | Casein kinase 2,
alpha prime polypeptide | 2.61 |
| CTNND1 | Catenin
(cadherin-associated protein), δ1 | 2.60 |
| Cul1 | Cullin 1 | 2.53 |
| DAAM1 | Dishevelled
associated activator of morphogenesis 1 | 6.54 |
| APC | Adenomatous
poliposis coli | ↑ |
Discussion
The role of TGF-β1 in the process of carcinogenesis
of glioblastoma multiforme is extremely complex. According to a
previous study, when TGF-β1 functions under normal conditions it
inhibits cell proliferation, arrests the cell cycle, and initiates
differentiation or apoptosis (22).
Following neoplastic transformation, certain parts of the TGF-β1
signaling pathway are altered, and TGF-β1 no longer exerts its
effects on the cell (22). This may
explain the present observation of high proliferation rates in
glioblastoma cells treated with 30 ng/mg TGF-β1. A high
concentration of TGF-β1 may be essential for neoplastic growth. In
normal in vivo conditions, TGF-β1 arises from the cancer
cells themselves, fibroblasts, cells of cancerous
microglia/macrophages, and possibly from immunocytes recruited by
the tumor (22).
It is likely that the intensive growth of cancer
cells creates competition for oxygen and other sources of
nutrition, which leads to inhibition of cell metabolism and a lower
level of TGF-β1 synthesis. Therefore, this transformation may
initiate cell migration from hypoxic areas to other areas with a
better blood supply, where the local microenvironment may be more
favorable. This hypothesis is supported by the gradual decrease in
replicative activity among cancer cells in the present study, when
the TGF-β1 concentration was reduced to 20 and 10 ng/ml. Other
studies also support this hypothesis (18,19).
On the one hand, this mechanism hinders the progress
of the neoplastic process; on the other hand, it ensures the
selection of hypoxia-resistant cellular elements that make a tumor
more aggressive. Switching from a proliferation to a migration
program is reflected by more active interaction with the surface of
the culture plate. TGF-β1 stimulation leads to an intensification
of exocrine function in cancer cells, causing a decrease in the
number of intracellular inclusions and intercellular contacts, and
creating multiple exocyte bubbles and actively releasing cell
contents (22). The synthesis of
extracellular matrix components combined with the production of
proteolytic enzymes is an important part of a complex invasive
growth program (23). By secreting
components of the extracellular matrix and interacting with them, a
cancer cell may penetrate the surrounding tissues. The ability of
cancer cells to synthesize components of the extracellular matrix
may be considered to be a crucial mechanism in shaping the
aggressive nature of cancer (24).
The production of matrix proteins and molecules involved with
cellular adhesion and migration explains the marked alteration in
the shape of cells and the appearance of multiple filopodia
(25).
However, the present study suggested that these
changes do not exclude a possibility of coordinated interaction
among glioblastoma cells due to a complex system of intercellular
communication creating a unified system of cells.
Cross-talk between cells in living organisms is
based on the exchange of information. With the help of
intercellular interactions, the coordinated regulation of
metabolism, differentiation and cell proliferation occurs in
different tissues. The complex system of microtubes joining
glioblastoma cells merits consideration. Certain studies have
suggested that there is a cancer cell communication network
(24–27). This network is thought to be
responsible for transporting proteins that confer chemoresistance
and radiation resistance, proteins responsible for DNA repair,
microRNAs (miRNAs) disrupting the processes of epigenetic control
over oncogene expression, the hierarchical development of
glioblastoma cells (6), and the
creation of CSC niches (21).
It is known that the development of an invasive
phenotype in cancer cells following stimulation by TGF-β1, as
described by the authors of the present study (20) and others (23), is not limited by their localization.
Appearing as a response to the local conditions, a transformed
resistant and invasive molecular phenotype is transmitted to other
cells through adhesive contacts, multiple connective tubes, the
fusion of cancer cells and the production of microvesicles. To an
extent, this system of communication may explain the dynamic nature
of CSC populations, and the presence of cancer/stem progenitor
cells, tumor-inducing cells and other neoplastic elements with
properties that are not typical for ordinary glioblastoma cells
(6,13,14).
The production of microvesicles is one of the
less-studied types of communication between neoplastic cancer cells
(24–26). This type of communication is used
for long-distance transportation of materials or to protect
materials from an aggressive microenvironment. In addition to DNA
and RNA, microvesicles may transport CD44, CD133+
mitogen activated protein kinase, epidermal growth factor vIII
receptor, disintegrin and metalloproteinase domain-containing
protein 10, Annexin A2 and certain pro-metastatic molecules
(28–30). It is possible to transfer drug
resistance between invasive glioma cells through exosomes (31). Therefore, it is possible make a
justified assumption that microvesicle synthesis is a
self-sufficient mechanism of tumor aggression, which renders it
possible to transfer an invasive phenotype to other cells and
tissues.
Normal CD45+ CD34+ HSCs are
able to migrate to cells of different types, although they have
increased mobility towards cancer cells. In animals with implanted
glial brain tumors, intravenously injected HSCs migrate to the
tumor nidus and accumulate in areas of invasion and necrosis
(32). A previous study reported
that hematopoietic CD34+ CD45+ stem cells
migrate towards glioblastoma cells and interact with them,
indicative of a strong association between these cell types
(32). It is possible that by
recruiting bone marrow cells, the tumor creates its own
microenvironment, allowing it to optimize resources and escape the
innate immune system and other defense mechanisms of the body
(32). As demonstrated by
fluorescent microscopy, HSCs attach themselves to cancer cells in
‘piggy-back fashion’, as described by Aboody et al (7). The method of electron microscopy
failed to reveal specific intercellular contacts between the
mononuclear cells and cancer cells. This may be associated with the
disappearance of mononuclear cells, caused by the toxic products of
glioblastoma cellular metabolism.
According to the results of the present study,
TGF-β1 stimulation did not reduce the ability of glioblastoma cells
to attract HSCs. The cells actively attached themselves to
glioblastoma cells and exchanged fluorescent tags.
Exchanging a fluorescent tag during the interaction
of normal stem cells and cancer cells suggests the possibility of
transferring cellular proteins and other cytoplasm components that
the dye adheres to. The results of the flow cytometry indicated the
presence of cancer cells with a double fluorescent tag, which
represented cells with new properties. It is noteworthy that the
exchange of a fluorescent tag goes in both directions. The most
likely mechanisms of these interactions are gap junction contacts,
the exchange of biological information through microvesicles or the
direct receipt of biological material secreted by the membrane of
neoplastic cells (31).
Gap junction contacts (33) do not allow for the transfer of large
protein molecules between cells, although they do allow cells to
exchange miRNAs. Previously, miRNAs that inhibit [the Let-7 family,
miRNA (miR)34, miR31, miR451, miR145, miR200/141, the miR14/15
family, miR23b, miR223 and miR224] (34) and stimulate (the miR17-92 cluster,
miR21, H19, MALAT and HOTAIR/miR-10b) cellular transition to a
pro-tumor phenotype have been described (35–37,28).
Together with cellular proteins, these miRNAs may be transported to
cancerous and non-cancerous cells through microvesicles, or may be
transferred during cell fusion or contact. However, the possibility
of direct absorption by non-cancer cells may not be ignored, and
was partially demonstrated by the results of the fluorescence
microscopy in the present study.
It is notable that during cell interactions, the
proliferation speed of glioblastoma cells that were treated with
TGF-β1 became similar to that of the control group. The intensified
proliferation rates of glioblastoma cells stimulated by TGF-β1
while interacting with HSCs indicated the ability of normal
CD45+ CD34+ stem cells to switch
proliferation and migration programs, which is likely to be a focal
point of HSC antitumor potential. This has significant theoretical
importance. The triggering of EMT stimulates invasion and
migration, accompanied by intensified interaction with the
extracellular matrix and a decreased rate of cancer cell
replication (13). The decline in
replication activity to the control values suggested a decreasing
interaction with the extracellular matrix and an inhibition of
invasion. In this respect, it may be proposed that the combination
of a stem cell-based medication with cytostatic chemotherapy and
radiation may be essential for the destruction of neoplastic
cells.
However, the present experimental data are not
sufficient for the successful implementation of cellular
technologies in practice. It is necessary to have a clear
understanding of what specific molecular mechanisms are activated
in glioblastoma cell TGF-β1 stimulation, and how exactly HSCs are
able to regulate them. The answers to these questions are not
trivial. TGF-β1 stimulation of cancer cells is accompanied by
substantial modification of their molecular phenotype, rendering
them similar to cancer stem cells.
The integrin and focal adhesion signaling pathways
are the most accessible pathways for regulatory influence in normal
neural stem cells and stem cells of the bone marrow, and in cancer
stem cells of human glioblastoma. The present study demonstrated
that TGF-β1 stimulation of cancer cells was accompanied by a
significant alteration in E-cadherin, occluding and claudin-1
production, and intensified synthesis of vimentin, actin and other
EMT markers. However, the maximum upregulation was achieved for
proteins of integrin and focal adhesion signaling pathways. This
was not unexpected; increased expression of proteins regulating the
interaction of cancer cells with the extracellular matrix during
EMT is a logical outcome of TGF- β1 stimulation.
The fact that the focal adhesion proteins with
increased expression (Ras homolog gene family, member A, Ras
homolog gene family, member C, Rho-associated, coiled-coil
containing protein kinase 2, dishevelled associated activator of
morphogenesis 1 and cullin 1) were components of the WNT signaling
pathway was noteworthy. Key components of canonical WNT/β-catenin
pathway (CK2A2 and APC) appeared de novo as a reaction to
stimulation. This signaling pathway is a strategically significant
mechanism of stemness, indicating a possibility of developing CSC
properties in these cells in response to invasion programs.
Proliferation intensification in cancer cells with EMT upon
interacting with HSCs indicates the ability of normal stem cells to
regulate this process, and requires further study.
The following conclusions may be drawn from the
present study. U87 glioblastoma cells have a complex system of
communication, including adhesive intercellular contacts, areas of
interdigitation with dissolution of the cytoplasm, cell fusion,
communication microtubes and microvesicles. The effect of TGF-β1 on
glioblastoma cell proliferation was inversely proportional to its
concentration in the medium. When the concentration of TGF-β1
reached 10 ng/ml, it resulted in the modification of cell shape and
the intensification of exocrine functioning. HSCs migrated to
glioblastoma cells, interacted with them and exchanged fluorescent
tags. Stimulation of cancer cells with 10 ng/ml TGF-β1 weakened
their ability to attract HSCs and exchange a fluorescent tag. The
proliferation rate of glioblastoma cells treated with TGF-β1
increased during intercellular interactions. TGF-β1 triggered the
mechanisms of EMT in glioblastoma cells, which was accompanied by
an alteration in the production of E-cadherin, occluding and
claudin-1, and enhanced synthesis of vimentin and actin.
Upregulation of the proteins of the integrin and focal adhesion
signaling pathways was accompanied by an increase in the expression
of proteins of the WNT signaling pathway. These processes indicated
a direct association between the initiation of EMT and the stemness
of the cancer cells phenotype. It is apparent that HSCs may be able
to regulate this process, which requires continued research.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Ministry of
Education and Science of the Russian Federation (grant no.
14.584.21.0027; ID, RFMEFI58417X0027).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EM was involved in the experiments stimulating
control cells with TGF-β1, and the interactions between HSCs and
cancer cells in vitro, in addition to working on the text of
the article. IB worked on the text of the article, proposed the
study idea, developed the design, offered methodological support,
organized the scientific team and provided scientific guidance on
the experimental part of the study. AP undertook experiments on the
interactions between HSCs and cancer cells in vitro. MK
conducted the flow cytofluorometry. IL performed the work with the
CellIQ system. YZ worked on the bioinformatics experiments and the
text of the article. SZ performed the statistical analysis. PM
performed experiments stimulating control cells with TGF-β1. ME
conducted the electron microscopy. YK was involved in the
experimental design and proteomic analysis, and worked on the text
of the article. VS performed the proteomic analysis. HS consulted
on the neuropathology aspects of this work, and worked on the text
of the article. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The use of human samples for this study was approved
by the Ethical Committee of the School of Biomedicine, The Far
Eastern Federal University (Vladivostock, Russia; minute no. 1 of
February 2nd 2017) and the Academic Council of the School of
Biomedicine. Consent was received.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hambardzumyan D and Bergers G:
Glioblastoma: Defining tumor niches. Trends Cancer. 1:252–265.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Toms SA and Kesari S: Treatment
for patients with newly diagnosed glioblastoma-reply. JAMA.
315:2348–2349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brown DV, Daniel PM, D'Abaco GM, Gogos A,
Ng W, Morokoff AP and Mantamadiotis T: Coexpression analysis of
CD133 and CD44 identifies proneural and mesenchymal subtypes of
glioblastoma multiforme. Oncotarget. 6:6267–6280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bradshaw A, Wickremsekera A, Tan ST, Peng
L, Davis PF and Itinteang T: Cancer stem cell hierarchy in
glioblastoma multiforme. Front Surg. 3:212016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aboody KS, Brown A, Rainov NG, Bower KA,
Liu S, Yang W, Small JE, Herrlinger U, Ourednik V, Black PM, et al:
Neural stem cells display extensive tropism for pathology in adult
brain: Evidence from intracranial gliomas. Proc Natl Acad Sci USA.
97:12846–12851. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bryukhovetskiy I, Bryukhovetskiy A,
Khotimchenko Y and Mischenko P: Novel cellular and post-genomic
technologies in the treatment of glioblastoma multiforme (Review).
Oncol Rep. 35:639–648. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garber K: Epithelial-to-mesenchymal
transition is important to metastasis, but questions remain. J Natl
Cancer Inst. 100:232–239. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weng J, Zhang H, Wang C, Liang J, Chen G,
Li W, Tang H and Hou J: miR-373-3p targets DKK1 to promote
EMT-induced metastasis via the Wnt/β-catenin pathway in tongue
squamous cell carcinoma. Biomed Res Int. 2017:60109262017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Huang S, Zhu X, Zhang W and Zhang X:
FOXK1 promotes glioblastoma proliferation and metastasis through
activation of Snail transcription. Exp Ther Med. 15:3108–3116.
2018.PubMed/NCBI
|
|
12
|
de Almeida Sassi F, Brunetto Lunardi A,
Schwartsmann G, Roesler R and Abujamra AL: Glioma revisited: From
neurogenesis and cancer stem cells to the epigenetic regulation of
the niche. J Oncol. 2012:5378612012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang N, Hong B, Zhou C, Du X, Chen S,
Deng X, Duoerkun S, Li Q, Yang Y and Gong K: Cobalt
chloride-induced hypoxia induces epithelial-mesenchymal transition
in renal carcinoma cell lines. Ann Clin Lab Sci. 47:40–46.
2017.PubMed/NCBI
|
|
15
|
Yang SW, Zhang ZG, Hao YX, Zhao YL, Qian
F, Shi Y, Li PA, Liu CY and Yu PW: HIF-1α induces the
epithelial-mesenchymal transition in gastric cancer stem cells
through the Snail pathway. Oncotarget. 8:9535–9545. 2017.PubMed/NCBI
|
|
16
|
Sun LL, Song Z, Li WZ and Tang SY: Hypoxia
facilitates epithelial-mesenchymal transition-mediated rectal
cancer progress. Genet Mol Res. 15:gmr150489362016.doi:
10.4238/gmr15048936. View Article : Google Scholar
|
|
17
|
Lehmann S, TeBoekhorst V, Odenthal J,
Bianchi R, van Helvert S, Ikenberg K, Ilina O, Stoma S, Xandry J,
Jiang L, et al: Hypoxia induces a HIF-1-dependent transition from
collective-to-amoeboid dissemination in epithelial cancer cells.
Curr Biol. 27:392–400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Qu C, Ning Z, Wang H, Zang K, Zhuang
L, Chen L, Wang P and Meng Z: Radiation promotes
epithelial-to-mesenchymal transition and invasion of pancreatic
cancer cell by activating carcinoma-associated fibroblasts. Am J
Cancer Res. 6:2192–2206. 2016.PubMed/NCBI
|
|
19
|
Huang W, Zhang C, Cui M, Niu J and Ding W:
Inhibition of bevacizumab-induced epithelial-mesenchymal transition
by BATF2 overexpression involves the suppression of Wnt/β-catenin
signaling in glioblastoma cells. Anticancer Res. 37:4285–4294.
2017.PubMed/NCBI
|
|
20
|
Bryukhovetskiy I and Shevchenko V:
Molecular mechanisms of the effect of TGF-β1 on U87 human
glioblastoma cells. Oncol Lett. 12:1581–1590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bryukhovetskiy A, Shevchenko V, Kovalev S,
et al: To the novel paradigm of proteome-based cell therapy of
tumors: Through comparative proteome mapping of tumor stem cells
and tissue-specific stem cells of humans. Cell Transplant. 1 Suppl
23:S151–S170. 2014. View Article : Google Scholar
|
|
22
|
Massagué J: TGFbeta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Santos JC, Ribeiro ML, Sarian LO, Ortega
MM and Derchain SF: Exosomes-mediate microRNAs transfer in breast
cancer chemoresistance regulation. Am J Cancer Res. 6:2129–2139.
2016.PubMed/NCBI
|
|
24
|
Osswald M, Jung E, Sahm F, Solecki G,
Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M,
et al: Brain tumour cells interconnect to a functional and
resistant network. Nature. 528:93–98. 2015.PubMed/NCBI
|
|
25
|
Weil S, Osswald M, Solecki G, Grosch J,
Jung E, Lemke D, Ratliff M, Hänggi D, Wick W and Winkler F: Tumor
microtubes convey resistance to surgical lesions and chemotherapy
in gliomas. Neuro Oncol. 19:1316–1326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sontheimer H: Brain cancer: Tumour cells
on neighbourhood watch. Nature. 528:49–50. 2015.PubMed/NCBI
|
|
27
|
Murphy SF, Varghese RT, Lamouille S, Guo
S, Pridham KJ, Kanabur P, Osimani AM, Sharma S, Jourdan J, Rodgers
CM, et al: Connexin 43 inhibition sensitizes chemoresistant
glioblastoma cells to temozolomide. Cancer Res. 76:139–149. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reza AM, Choi YJ, Yasuda H and Kim JH:
Human adipose mesenchymal stem cell-derived exosomal-miRNAs are
critical factors for inducing anti-proliferation signaling to A2780
and SKOV-3 ovarian cancer cells. Sci Rep. 6:384982016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Katakowski M, Buller B, Zheng X, Lu Y,
Rogers T, Osobamiro O, Shu W, Jiang F and Chopp M: Exosomes from
marrow stromal cells expressing miR-146b inhibit glioma growth.
Cancer Lett. 335:201–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gopal SK, Greening DW, Rai A, Chen M, Xu
R, Shafiq A, Mathias RA, Zhu HJ and Simpson RJ: Extracellular
vesicles: Their role in cancer biology and epithelial-mesenchymal
transition. Biochem J. 474:21–45. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Richter N, Wendt S, Georgieva PB,
Hambardzumyan D, Nolte C and Kettenmann H: Glioma-associated
microglia and macrophages/monocytes display distinct
electrophysiological properties and do not communicate via gap
junctions. Neurosci Lett. 583:130–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bryukhovetskiy IS, Dyuizen IV, Shevchenko
VE, Bryukhovetskiy AS, Mischenko PV, Milkina EV and Khotimchenko
YS: Hematopoietic stem cells as a tool for the treatment of
glioblastoma multiforme. Mol Med Rep. 14:4511–4520. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rappa G, Mercapide J, Anzanello F, Pope RM
and Lorico A: Biochemical and biological characterization of
exosomes containing prominin-1/CD133. Mol Cancer. 12:622013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baumann P, Cremers N, Kroese F, Orend G,
Chiquet-Ehrismann R, Uede T, Yagita H and Sleeman JP: CD24
expression causes the acquisition of multiple cellular properties
associated with tumor growth and metastasis. Cancer Res.
65:10783–10793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Katakowski M and Chopp M: Exosomes as
tools to suppress primary brain tumor. Cell Mol Neurobiol.
36:343–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou RJ, Xu XY, Liu BX, Dai WZ, Cai MQ,
Bai CF, Zhang XF, Wang LM, Lin L, Jia SZ, et al: Growth-inhibitory
and chemosensitizing effects of microRNA-31 in human glioblastoma
multiforme cells. Int J Mol Med. 36:1159–1164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yarygin VN, Zaitsev AYu, Bryukhovetskiy
AS, Mentkevich GL and Mheidze DM: Autologic hemopoietic stem cells
preparation and method for producing, cryogenic preserving and
applying it in treating traumatic nervous system disease cases.
Patent RU 2283119. Filed March 29, 2005; issued September 10.
2006.
|