Introduction
Colorectal cancer (CRC) is the second most prevalent
type of cancer and the third leading cause of cancer-associated
mortality worldwide (1). Despite
significant advances in the diagnosis and treatment of CRC, the
overall survival rate of CRC remains unsatisfactory (2,3).
Therefore, it is necessary to identify the molecular mechanisms
underlying the occurrence and development of CRC, in order to
develop more effective diagnostic and therapeutic methods.
Genomic studies have confirmed that only 2% of human
gene transcripts encode proteins, whereas numerous transcripts
encode non-coding ribonucleic acids (ncRNAs) (4). Long ncRNAs (lncRNAs) are a subtype of
ncRNAs >200 nucleotides long, which lack protein-coding ability.
Although the exact functions of lncRNAs remain unclear, they appear
to regulate numerous biological behaviors through epigenetic
regulation, transcription and post-transcriptional processing
(5). Accumulating evidence has
indicated that the aberrant expression of lncRNAs may regulate
cancer cell proliferation, migration, invasion, apoptosis and
metastasis (6,7).
p63 is a member of the p53 family, which is highly
homologous and structurally similar to p53 (8). p63 has two isoforms, TAp63 and ΔNp63,
which differ at the N-terminal. TAp63 contains an N-terminal
transactivation domain, whereas ΔNp63 lacks this domain (9). These two isoforms perform different
functions during tumorigenesis. TAp63 acts as a tumor suppressor,
similar to p53, and induces cell cycle arrest and cell apoptosis
(10). Conversely, ΔNp63 acts as an
oncogene, facilitating proliferation, survival, invasion,
metastasis and chemoresistance, and reducing apoptosis of various
cancer cells (11–13). Numerous studies have revealed that
lncRNAs mediate the biological functions of CRC cells through
modulating p53 expression (14–16).
However, the number of studies on lncRNAs regulating CRC
tumorigenesis by targeting p63 is limited. The present study aimed
to determine whether there is a newly identified lncRNA targeting
ΔNp63, and to investigate the biological functions of this newly
identified lncRNA in the proliferation, cell cycle progression,
invasion, apoptosis and pyroptosis of CRC cells, in the aim of
providing a novel target for the treatment of patients with
CRC.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of The Third Xiangya Hospital of Central South University
(Changsha, China). The study protocol conformed to the principles
outlined in the Helsinki Declaration and to local legislation.
Informed consent was obtained from all of the participants.
Patient specimens
A total of 34 pairs of human primary CRC tissues and
corresponding non-cancerous tissues (NCTs) were collected from
patients who had undergone tumor resection at the Department of
Gastrointestinal Surgery, The Third Xiangya Hospital of Central
South University between October 2014 and June 2015. The specimens
were immediately frozen in liquid nitrogen following surgical
resection and maintained at −80°C. The clinical characteristics of
all patients, including age, tumor site, tumor differentiation,
stage, lymph node metastasis and distant metastasis, are summarized
in Table I. Cancer staging was
performed according to the 7th TNM classification by the American
Joint Committee on Cancer (17).
None of the patients underwent radiotherapy or chemotherapy prior
to surgery.
 | Table I.Association of clinical and
pathological characteristics with lncRNA RP1-85F18.6, ΔNp63 and
GSDMD mRNA expression. |
Table I.
Association of clinical and
pathological characteristics with lncRNA RP1-85F18.6, ΔNp63 and
GSDMD mRNA expression.
|
|
| lncRNA
RP1-85F18.6 | ΔNp63 | GSDMD |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases | Lowa (n=17) | Higha (n=17) | P-value | Lowa (n=17) | Higha (n=17) | P-value | Lowa (n=17) | Higha (n=17) | P-value |
|---|
| Age (years) | 34 | 56.36±15.17 | 58.55±10.31 | 0.905 | 58.11±12.10 | 56.27±13.16 | 0.918 | 57.68±11.25 | 56.89±12.39 | 0.962 |
| Tumor site |
| Left
colon | 9 | 5 | 4 | 0.774 | 4 | 5 | 0.257 | 5 | 4 | 0.856 |
| Right
colon | 5 | 3 | 2 |
| 1 | 4 |
| 2 | 3 |
|
|
Rectum | 20 | 9 | 11 |
| 12 | 8 |
| 10 | 10 |
|
| Tumor
differentiation |
|
Well | 6 | 2 | 4 | 0.634 | 4 | 2 | 0.533 | 2 | 4 | 0.285 |
|
Moderate | 23 | 12 | 11 |
| 10 | 13 |
| 11 | 12 |
|
|
Poor | 5 | 3 | 2 |
| 3 | 2 |
| 4 | 1 |
|
| Stage |
| I | 5 | 2 | 3 | 0.726 | 3 | 2 | 0.362 | 3 | 2 | 0.905 |
| II | 9 | 4 | 5 |
| 3 | 6 |
| 5 | 4 |
|
|
III | 16 | 8 | 8 |
| 10 | 6 |
| 7 | 9 |
|
| IV | 4 | 3 | 1 |
| 1 | 3 |
| 2 | 2 |
|
| Lymph node
metastasis |
| N0 | 16 | 11 | 5 | 0.034b | 12 | 4 | 0.022b | 3 | 13 | 0.003b |
| N1 | 10 | 5 | 5 |
| 3 | 7 |
| 8 | 2 |
|
| N2 | 8 | 1 | 7 |
| 2 | 6 |
| 6 | 2 |
|
| Distant
metastasis |
| M0 | 30 | 17 | 13 | 0.033b | 17 | 13 | 0.033b | 13 | 17 | 0.033b |
| M1 | 4 | 0 | 4 |
| 0 | 4 |
| 4 | 0 |
|
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
RNA was extracted from CRC tissues/NCTs, or fresh
cultured cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. RT of 3 µg total RNA was performed using
RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RT-qPCR was performed
using the SYBR-Green Master Mix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and was run using a thermal cycler (Bio-Rad
Laboratories, Inc.). The thermal cycling conditions were as
follows: 95°C for 2 min, followed by 40 cycles at 95°C for 30 sec,
50°C for 30 sec and 72°C for 30 sec, and a final extension step at
72°C for 10 min. GAPDH was used for normalization. Relative gene
expression was calculated using the 2−ΔΔCq method
(18). The primers used in the
present study are listed in Table
II.
 | Table II.Reverse transcription-quantitative
polymerase chain reaction primers used in this study. |
Table II.
Reverse transcription-quantitative
polymerase chain reaction primers used in this study.
| Gene name | Primer sequence
(5′-3′) |
|---|
| lncRNA | Forward:
GGCTCTTTGCTCACATCG |
| RP1-85F18.6 |
|
|
| Reverse:
AAGGAAACCACAGGCTCA |
| ΔNp63 | Forward:
GAAGAAAGGACAGCAGCATTGA |
|
| Reverse:
GGGACTGGTGGACGAGGAG |
| TAp63 | Forward:
TGTATCCGCATGCAGGACT |
|
| Reverse:
CTGTGTTATAGGGACTGGTGGAC |
| GSDMD | Forward:
GTGTGTCAACCTGTCTATCAAGG |
|
| Reverse:
CATGGCATCGTAGAAGTGGAAG |
| GAPDH | Forward:
ACCACAGTCCATGCCATCAC |
|
| Reverse:
TCCACCACCCTGTTGCTGTA |
Cell culture
The NCM460 human colorectal epithelial cell line,
and the SW480, SW620 and HCT116 CRC cell lines were obtained from
Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China). The cells were
cultured in Dulbecco's modified Eagle's medium containing 10% fetal
bovine serum (FBS) (HyClone; GE Healthcare, Logan, UT, USA) at 37°C
in a humidified atmosphere containing 5% CO2.
Cell transfection
LncRNA RP1-85F18.6 small interfering (si)RNA, ΔNp63
siRNA and negative control (NC) siRNA were purchased from Guangzhou
RiboBio Co., Ltd. (Guangzhou, China). The sequences were as
follows: lncRNA RP1-85F18.6 siRNA, 5′-GACTCCGCCGTGAACCCTTCA-3′;
ΔNp63 siRNA, 5′-ACAAUGCCCAGACUCAAUUUU-3′. A scramble siRNA
(siN05815122147) was used as the NC siRNA. Once the SW620 cells
reached 70% confluence, the cells were transfected for 48 h at 37°C
with lncRNA RP1-85F18.6 siRNA, ΔNp63 siRNA or NC siRNA using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
siRNAs were diluted to a final concentration of 60 nM for
transfection. In addition, the entire sequence of human lncRNA
RP1-85F18.6 was amplified from SW620 cells using PCR and cloned
into the pcDNA3.1 vector. The negative control empty vector, which
was purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China),
and the lncRNA RP1-85F18.6 plasmid were transfected into SW620
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
The plasmid was diluted to a final concentration of 2 µg/ml for
transfection.
Cell proliferation assay
Transfected SW620 cells were cultured in 96-well
plates and incubated for 24, 48 and 72 h. Optical density values
were measured using the MTT Cell Proliferation and Cytotoxicity
Assay kit (Beyotime Institute of Biotechnology, Shanghai, China),
according to the manufacturer's protocol. Cell proliferation was
calculated using the absorbance values, which were measured at 490
nm at each time point.
Flow cytometric analysis of
apoptosis
Cell apoptosis was estimated using flow cytometric
analysis with the Apoptosis Detection kit (Nanjing KeyGen BioTech
Co., Ltd.), according to the manufacturer's protocol. After
transfection for 48 h, the cells were washed with ice-cold PBS and
resuspended with binding buffer. Subsequently, the cells were
incubated with propidium iodide (PI) and Annexin V at room
temperature for 15 min in the dark. The cells were then resuspended
with PBS and analyzed by flow cytometry (BD Biosciences, Franklin
Lakes, NJ, USA) and FlowJo software (v7.6.2, FlowJo; LLC, Ashland,
OR, USA).
Cell cycle analysis
Cell cycle analysis was performed as previously
described (19). Briefly, after
transfection for 48 h, the cells were washed with PBS and fixed
with ice-cold 70% ethanol at 4°C overnight. Fixed cells were washed
with PBS and incubated with PI and RNase, which were obtained from
the Cell Cycle Detection kit (Nanjing KeyGen BioTech Co., Ltd.),
for 30 min at room temperature in the dark. Subsequently, the
incubated cells were analyzed by flow cytometry (BD Biosciences)
and FlowJo software (v7.6.2, FlowJo; LLC).
Transwell assay
Cellular invasion was evaluated using Transwell
migration chambers precoated with a layer of Matrigel®.
This assay was performed as previously described at 48 h
post-transfection (20). Briefly,
cells suspended in 100 µl medium without FBS were seeded in the
upper chamber (104 cells/well). To the lower chamber,
600 µl medium supplemented with 20% FBS was added. After 24 h
incubation at 37°C, cells on the top of the membrane were removed
with cotton swabs. The cells that invaded through the membrane were
washed with PBS, fixed in 4% methanol for 20 min and stained with
0.1% crystal violet for 10 min at room temperature. The number of
invasive cells was counted in five randomly selected fields under a
light microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Protein extraction and western
blotting
Protein extraction and western blotting were
performed as previously described (21). Briefly, proteins were extracted from
tissues and cells using radioimmunoprecipitation assay lysis buffer
containing 1% 100 mM phenylmethylsulfonyl fluoride (both from
Beyotime Institute of Biotechnology). Protein concentration was
examined using the bicinchoninic acid method (Nanjing KeyGen
Biotech Co., Ltd.). Subsequently, proteins (20 µg) were separated
by 10% SDS-PAGE and were transferred to polyvinylidene fluoride
membranes. The membranes were blocked in 5% non-fat milk for 2 h at
room temperature, and were incubated with the following primary
antibodies at 4°C overnight: Anti-β-actin (1:2,000, cat. no.
20536-1-AP) and anti-gasdermin D (GSDMD; 1:1,000, cat. no.
20770-1-AP) (both from Wuhan Sanying Biotechnology, Wuhan, China);
anti-ΔNp63 (1:500, cat. no. 619001) and anti-TAp63 (1:500, cat. no.
618901) (both from BioLegend, Inc., San Diego, CA, USA). After
washing with PBS-0.1% Tween, the membranes were incubated with
anti-rabbit immunoglobulin G secondary antibody (1:5,000, cat. no.
SA00001-2; Wuhan Sanying Biotechnology) for 1 h at room
temperature. The images were obtained using Advansta WesternBright
enhanced chemiluminescence (Advansta lnc., Menlo Park, CA, USA) and
the ChemiDoc™ MP Imaging system (Bio-Rad Laboratories, Inc.).
Lactate dehydrogenase (LDH) release
assay
After transfection for 48 h, the supernatants of
transfected cells were collected to measure LDH release using LDH
Cytotoxicity Assay kit, according to the manufacturer's protocol
(Beyotime Institute of Biotechnology). Data were collected based on
the absorbance at 490 nm, which was measured using a microplate
reader (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were repeated three times and data
are expressed as the means ± standard deviation. All statistical
calculations were performed using GraphPad Prism version 6.01
(GraphPad Software, Inc., La Jolla, CA, USA) and SPSS (PASW
Statistics) version 20 (IBM Corporation, Armonk, NY, USA). The
Mann-Whitney U test was used to compare differences between two
groups. One-way analysis of variance followed by the
Student-Newman-Keul's test was used to compare multiple groups. The
correlation between two factors was determined using the Spearman's
rank correlation test. The sensitivity and specificity in CRC
tissues were calculated using the Youden's index. The area under
the receiver operating characteristic (ROC) curve (AUC) was also
estimated. P<0.05 was considered to indicate a statistically
significant difference.
Results
p63-associated lncRNA
identification
To identify novel lncRNAs targeting p63, a
microarray analysis was performed in our previous study, and
thousands of lncRNAs and mRNAs were differentially expressed
between CRC tissues and NCTs (22).
Subsequently, LncTar, which is an efficient tool for predicting RNA
targets of lncRNAs that is provided by Cui Lab from the Department
of Biomedical Informatics, Peking University Health Science Center
(http://www.cuilab.cn/lnctar) (23), was used. The Gibbs chemical bond
free energy between tumor protein 63 (TP63; NM_003722.4) and
candidate lncRNAs was determined. Using a value of <0.05 as the
threshold, lncRNAs below that threshold were considered to have a
lower Gibbs chemical bond free energy. A total of 24 lncRNAs were
predicted to target TP63; among those, six were upregulated and 18
were downregulated in a CRC lncRNA microarray. The top five
upregulated TP63-associated lncRNAs are listed in Table III.
 | Table III.Top five upregulated tumor protein
63-associated long non-coding RNAs. |
Table III.
Top five upregulated tumor protein
63-associated long non-coding RNAs.
| Sequence name | ndG | Fold change |
|---|
|
ENSG00000232754|ENST00000415054 | −0.0889 | 13.3513184 |
|
ENSG00000257453|ENST00000552367 | −0.0823 | 4.7515723 |
|
ENSG00000259933|ENST00000563284 | −0.0746 | 6.7542724 |
|
ENSG00000259479|ENST00000564140 | −0.0712 | 5.0080102 |
|
ENSG00000204787|ENST00000447469 | −0.0698 | 10.0279087 |
Expression of a newly identified
lncRNA is upregulated in CRC tissues
To further verify the lncRNAs that are associated
with p63 in tumor samples, RT-qPCR was performed to assess the
expression levels of upregulated lncRNAs in 10 pairs of matched CRC
tissues and NCTs. Among these lncRNAs, lncRNA RP1-85F18.6
(ENST00000415054) was the most markedly increased in CRC tissues
(data not shown). Therefore, lncRNA RP1-85F18.6 was selected for
subsequent experiments. Compared with the NCTs, lncRNA RP1-85F18.6
expression was increased in 24 tumor tissues (70.6%, P<0.05),
out of the 34 pairs of matched CRC tissues and NCTs (Fig. 1A). These results provided further
evidence to suggest that lncRNA RP1-85F18.6 was increased in CRC
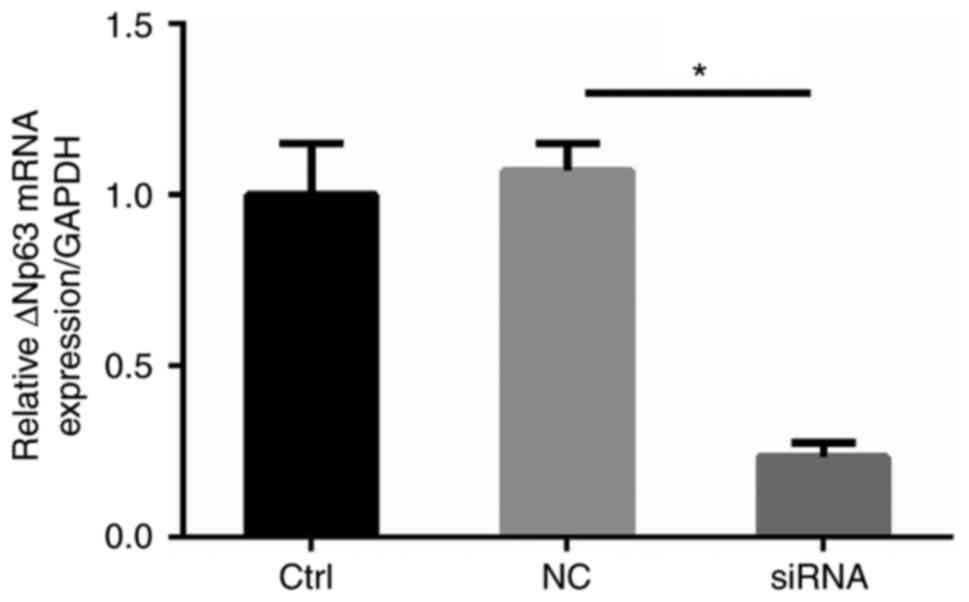
tissues. The results of RT-qPCR also demonstrated that ΔNp63
expression was increased in 22 tumor tissues (64.8%, P<0.05),
and TAp63 expression was reduced in 22 tumor tissues (64.8%,
P<0.05) (Fig. 1B and C).
Similarly, the protein expression levels of ΔNp63 were obviously
increased in CRC tissues (Fig. 1D),
whereas the protein expression levels of TAp63 were reduced
(Fig. 1D). These results were
consistent with the findings of previous studies (13,21).
Furthermore, lncRNA RP1-85F18.6 was positively correlated with
ΔNp63 expression (r=0.678, P<0.001), whereas the correlation
with TAp63 expression was negative (r=−0.371, P=0.0308) (Fig. 1E and F). These findings indicated
that lncRNA RP1-85F18.6 may be correlated with TP63, and may
facilitate the progression of CRC.
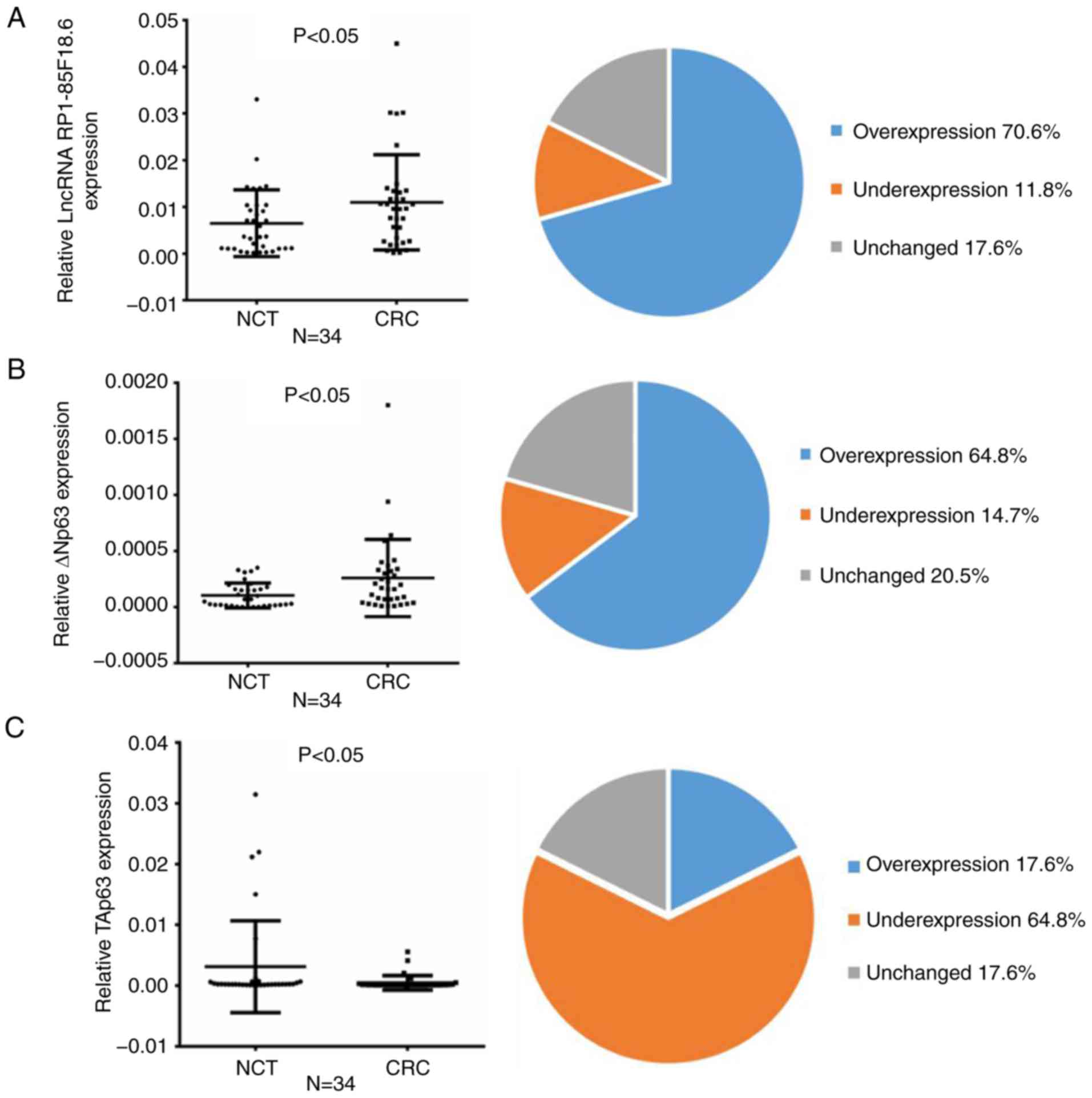 | Figure 1.Expression of lncRNA RP1-85F18.6,
ΔNp63 and TAp63 in CRC tissues and cell lines. (A-C) Expression of
lncRNA RP1-85F18.6, ΔNp63 and TAp63 in CRC tissues, as assessed by
RT-qPCR (n=34, P<0.05). The mean expression levels are shown in
the left scatter diagram and the expression distributions are
summarized in the right pie chart. GAPDH was used for
normalization. (D) Representative results of increased ΔNp63
expression and reduced TAp63 expression in CRC tissues, as assessed
by western blotting (N, NCT; C, CRC). (E and F) lncRNA RP1-85F18.6
was positively correlated with ΔNp63 and negatively correlated with
TAp63 in CRC samples. (G and H) Expression levels of lncRNA
RP1-85F18.6, ΔNp63 and TAp63 in various cell lines were evaluated
by RT-qPCR and western blotting; n=3, *P<0.05. CRC, colorectal
cancer; lncRNA, long non-coding RNA; NCT, non-cancerous tissue;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
Expression of lncRNA RP1-85F18.6,
ΔNp63 and TAp63 in CRC cells
To assess the expression of lncRNA RP1-85F18.6,
ΔNp63 and TAp63 in CRC cells, their expression was examined in
various CRC cell lines (SW480, SW620 and HCT116) compared with in
the NCM460 normal human colorectal epithelial cell line, using
RT-qPCR and western blotting. Consistent with the results of CRC
tumor samples, the mRNA expression levels of lncRNA RP1-85F18.6 and
ΔNp63 were markedly upregulated, whereas those of TAp63 were
downregulated in the SW480 and SW620 cell lines (Fig. 1G); the protein expression of ΔNp63
and TAp63 was altered accordingly (Fig.
1H). These results further confirmed that lncRNA RP1-85F18.6
and TP63 may serve key roles in triggering the tumorigenic process
in CRC. In addition, the differences in the expression of lncRNA
RP1-85F18.6, ΔNp63 and TAp63 were most significant in the SW620
cell line; therefore, SW620 cells were selected for subsequent
experimentation.
lncRNA RP1-85F18.6 acts as an oncogene
in CRC cells
To evaluate the biological role of lncRNA
RP1-85F18.6 in CRC tumorigenesis, lncRNA RP1-85F18.6 was silenced
by transfecting SW620 cells with lncRNA RP1-85F18.6 siRNA or NC
siRNA (Fig. 2A). The
Matrigel-coated Transwell assay revealed that silencing lncRNA
RP1-85F18.6 inhibited invasion of CRC cells compared with in the NC
group (215 vs. 272, P<0.05) (Fig.
2B). This finding suggested that lncRNA RP1-85F18.6 may promote
the invasive phenotype of CRC cells. To investigate whether lncRNA
RP1-85F18.6 regulated the proliferation of CRC cells, the MTT assay
was performed. As shown in Fig. 2C,
the proliferative ability of lncRNA RP1-85F18.6 siRNA-transfected
SW620 cells was significantly reduced. Subsequently, cell cycle
analysis revealed that SW620 cells exhibited a shortened S phase
(11.9 vs. 22.0%, P<0.05) and were arrested at the G2
phase (17.8 vs. 7.6%, P<0.05) following transfection with lncRNA
RP1-85F18.6 siRNA compared with in the NC group (Fig. 2D). Furthermore, the apoptotic rate
of SW620 cells was upregulated after silencing lncRNA RP1-85F18.6
(13.38 vs. 4.81%, P<0.05) compared with in the NC group
(Fig. 2E). These results suggested
that the reduced cell proliferation may be attributed to disrupted
cell cycle progression and increased apoptosis.
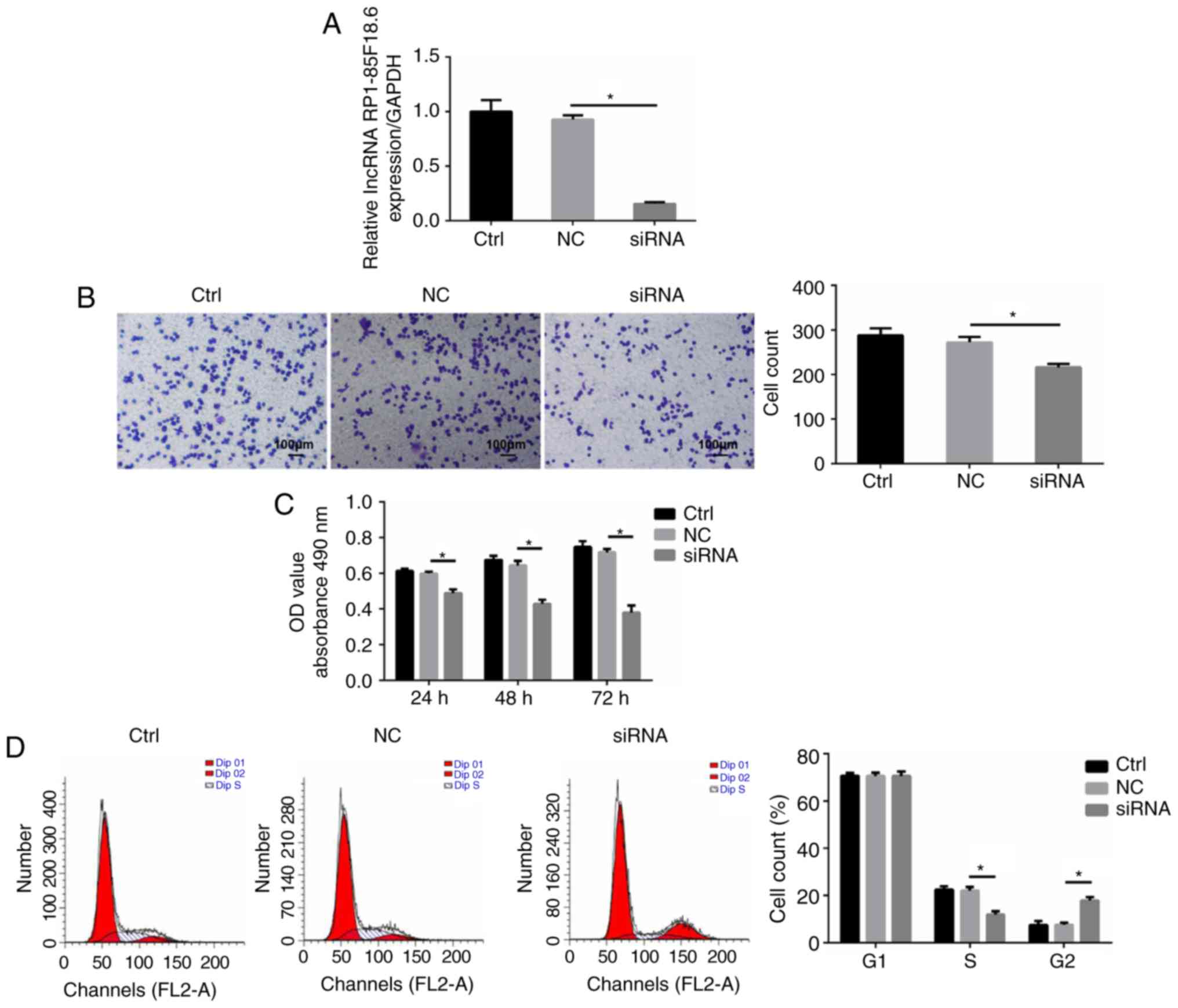 | Figure 2.Biological functions of lncRNA
RP1-85F18.6 in CRC cells. The CRC cells were transfected with
lncRNA RP1-85F18.6 siRNA or NC siRNA. (A) Post-transfection with
lncRNA RP1-85F18.6 siRNA, the expression levels of lncRNA
RP1-85F18.6 were assessed by RT-qPCR. (B) Matrigel-coated Transwell
assay was used to determine the invasive ability of cells;
magnification, ×200. (C) Proliferative ability was assessed by the
MTT assay. (D and E) Cell cycle distribution and apoptosis were
determined by flow cytometric analysis. (F) LDH release was
measured by colorimetric assay. (G and H) mRNA and protein
expression levels of GSDMD in various cell lines were evaluated by
RT-qPCR and western blotting, respectively. n=3, *P<0.05. CRC,
colorectal cancer; GSDMD, gasdermin D; LDH, lactate dehydrogenase;
lncRNA, long non-coding RNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; siRNA, small
interfering RNA. |
Furthermore, LDH release was markedly elevated
following inhibition of lncRNA RP1-85F18.6, thus indicating rupture
of the plasma membrane and pyroptosis (Fig. 2F). This result suggested that the
reduced proliferation may also be caused by cell pyroptosis. GSDMD
is a member of the gasdermin family, which has been confirmed to be
the main effector molecule for pyroptosis (24,25).
In various CRC cells, the expression of GSDMD was revealed to be
downregulated, both at the mRNA and protein levels (Fig. 2G and H). Furthermore, it was
demonstrated that, although silencing lncRNA RP1-85F18.6 did not
alter the full-length protein expression of GSDMD, it increased
GSDMD-N domain cleavage, as demonstrated by western blotting
(Fig. 3A). Following cleavage from
the full-length GSDMD, the GSDMD-N domain acts as the executioner
of pyroptosis, perforating the cell membrane, and causing cell
swelling and lysis (26).
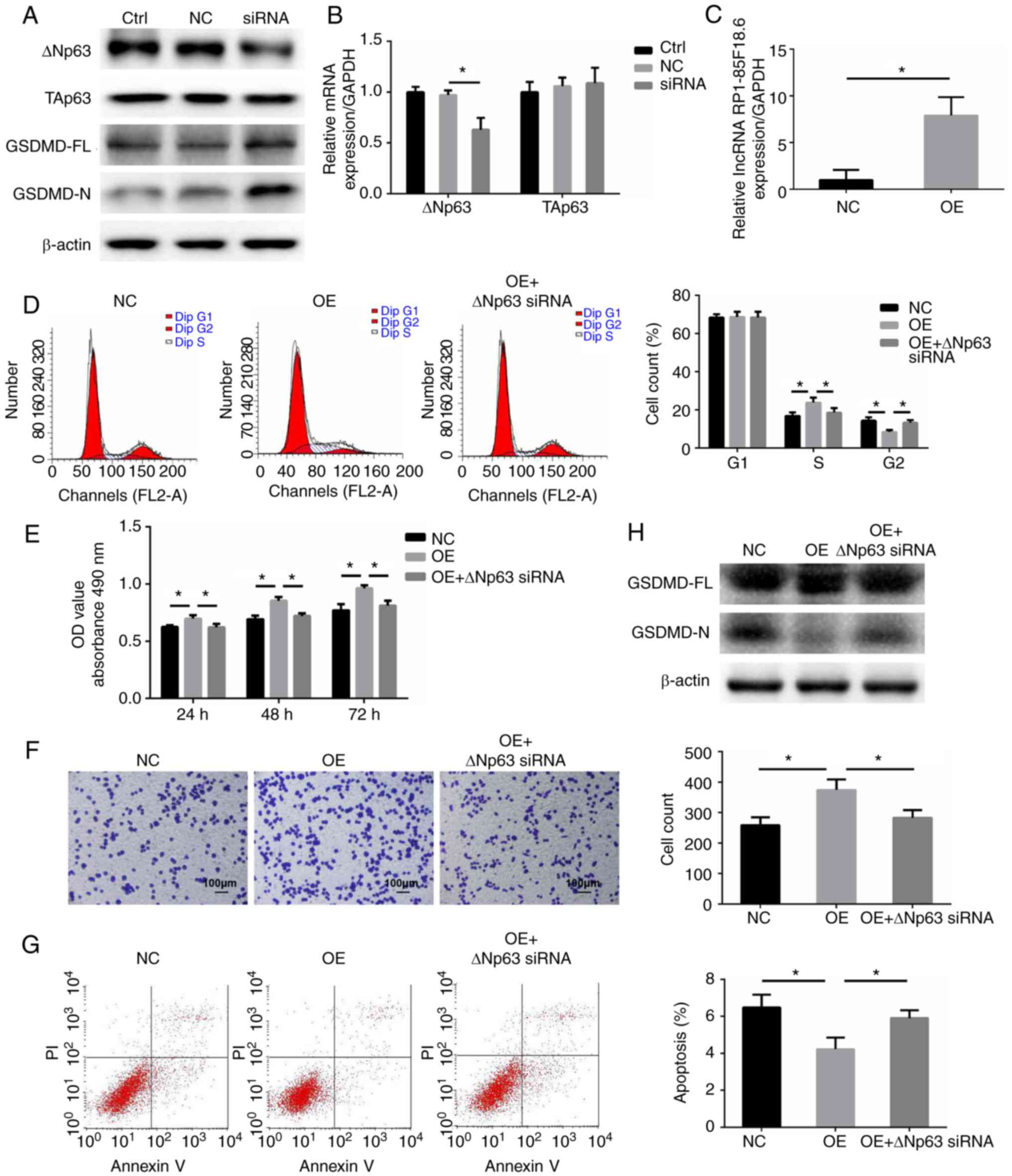 | Figure 3.lncRNA RP1-85F18.6 mediates
biological functions of CRC cells through regulating ΔNp63. (A)
Western blotting was performed to evaluate ΔNp63, TAp63, GSDMD-FL
and GSDMD-N expression after knockdown of lncRNA RP1-85F18.6. (B)
Following transfection with lncRNA RP1-85F18.6 siRNA, the
expression levels of ΔNp63 and TAp63 were assessed by RT-qPCR. CRC
cells were transfected with empty vector or lncRNA
RP1-85F18.6-expressing vector, or were co-transfected with lncRNA
RP1-85F18.6-expressing vector and ΔNp63 siRNA. (C) Following
transfection with the lncRNA RP1-85F18.6-expressing vector, the
expression of lncRNA RP1-85F18.6 was assessed by RT-qPCR. (D) Cell
cycle distribution was determined by flow cytometry. (E)
Proliferative ability of CRC cells was evaluated by the MTT assay.
(F) Matrigel-coated Transwell assay was used to determine invasive
ability of cells; magnification, ×200. (G) Flow cytometric analysis
was applied to evaluate cell apoptosis. (H) Protein expression
levels of GSDMD-FL and GSDMD-N were determined by western blotting.
n=3, *P<0.05. CRC, colorectal cancer; GSDMD, gasdermin D;
GSDMD-FL, GSDMD-full length; GSDMD-N, GSDMD-N domain; lncRNA, long
non-coding RNA; NC, negative control; OD, optical density; OE,
overexpression of lncRNA RP1-85F18.6; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; siRNA, small
interfering RNA. |
lncRNA RP1-85F18.6 acts as an oncogene
in CRC cells through regulating ΔNp63
To further investigate the underlying mechanisms by
which lncRNA RP1-85F18.6 mediates the biological functions of CRC
cells, the present study examined whether lncRNA RP1-85F18.6
regulates the expression of ΔNp63 and TAp63. lncRNA RP1-85F18.6 was
silenced in SW620 cells, and the RT-qPCR and western blotting
results revealed that the expression of ΔNp63 was reduced at both
the mRNA and protein levels (Fig. 3A
and B). Unexpectedly, there were no significant alterations in
the expression of TAp63 at the mRNA or protein levels (Fig. 3A and B).
Subsequently, SW620 cells were transfected with
lncRNA RP1-85F18.6-expressing vector, or were co-transfected with
lncRNA RP1-85F18.6-expressing vector and the ΔNp63 siRNA (Fig. 3C-G), which was revealed to decrease
ΔNp63 expression (Fig. 4). The
expression of lncRNA RP1-85F18.6 was markedly upregulated following
transfection with lncRNA RP1-85F18.6-expressing vector (Fig. 3C). Compared with the NC group,
overexpression of lncRNA RP1-85F18.6 increased the percentage of
cells in S phase and decreased the percentage of cells in
G2 phase (Fig. 3D). In
addition, increased lncRNA RP1-85F18.6 expression promoted cell
proliferation (Fig. 3E) and
invasion (Fig. 3F), and inhibited
apoptosis (Fig. 3G) and cleavage of
the GSDMD-N domain (Fig. 3H) in
SW620 cells. However, co-transfection with lncRNA
RP1-85F18.6-expressing vector and ΔNp63 siRNA reversed these
tumor-promoting effects. These data suggested that lncRNA
RP1-85F18.6 may induce the proliferation, invasion and cell cycle
disruption, and inhibit apoptosis and pyroptosis of CRC cells
through regulating ΔNp63.
Association of lncRNA RP1-85F18.6,
ΔNp63 and GSDMD expression with clinicopathological parameters
To establish the association of GSDMD with CRC, the
mRNA and protein expression levels of GSDMD were detected in CRC
tissues. The mRNA expression levels of GSDMD were reduced in 26
tumor tissues (76.4%, P<0.05) (Fig.
5A). Consistently, the protein expression levels of GSDMD were
markedly decreased in CRC tissues (Fig.
5B). Furthermore, the association of lncRNA RP1-85F18.6, ΔNp63
and GSDMD expression with the clinicopathological characteristics
of patients with CRC was evaluated. As shown in Table I, lncRNA RP1-85F18.6 and ΔNp63
expression was positively correlated with lymph node and distant
metastases (P<0.05). Conversely, an inverse association was
determined between GSDMD expression and lymph node and distant
metastases (P<0.05). In addition, there were no statistically
significant associations between lncRNA RP1-85F18.6, ΔNp63 or GSDMD
expression, and other clinicopathological parameters
(P>0.05).
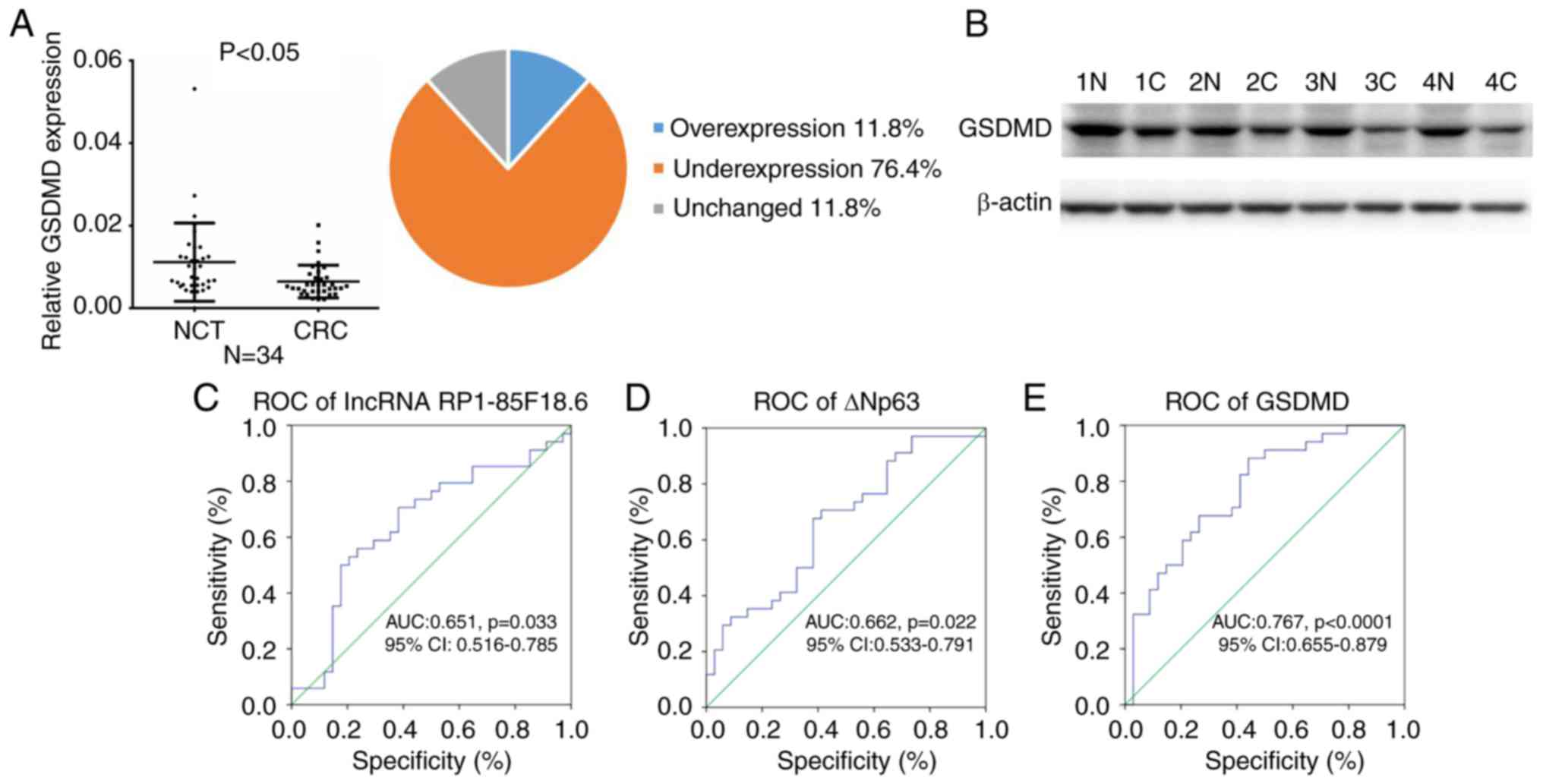 | Figure 5.Association between lncRNA
RP1-85F18.6, ΔNp63 and GSDMD expression, and clinicopathological
characteristics of patients with CRC. (A) Expression of GSDMD in
CRC tissues, as assessed by reverse transcription-quantitative
polymerase chain reaction (n=34, P<0.05). The mean expression of
GSDMD is shown in the left scatter diagram and the expression
distribution is summarized in the right pie chart. GAPDH was used
for normalization. (B) Representative results of reduced expression
of GSDMD in CRC tissues from four cases, as assessed by western
blotting (N, NCT; C, CRC). (C-E) ROC curve analysis of the
expression levels of lncRNA RP1-85F18.6, ΔNp63 and GSDMD for
discriminating CRC from NCT samples. AUC, area under the ROC curve;
CRC, colorectal cancer; GSDMD, gasdermin D; lncRNA, long non-coding
RNA; NCT, non-cancerous tissue; ROC, receiver operating
characteristic. |
The present study also demonstrated that lncRNA
RP1-85F18.6, ΔNp63 and GSDMD were suitable predictors of CRC
through a ROC curve analysis. The AUC of lncRNA RP1-85F18.6 for CRC
was 0.651 [95% confidence interval (CI): 0.516–0.785; P=0.033],
with 55.9% sensitivity and 76.5% specificity (Fig. 5C). The AUC of ΔNp63 for CRC was
0.662 [95% CI: 0.533–0.791; P=0.022], with 70.6% sensitivity and
58.8% specificity (Fig. 5D). The
AUC of GSDMD for CRC was 0.767 [95% CI: 0.655–0.879; P<0.0001],
with 88.2% sensitivity and 55.9% specificity (Fig. 5E). These results suggested that
lncRNA RP1-85F18.6, ΔNp63 and GSDMD may be implicated in CRC
tumorigenesis and metastasis, and may serve as potential prognostic
biomarkers of CRC in the future.
Discussion
Initially, lncRNAs were considered to be
‘transcriptional noise’; however, over the past few years, research
has indicated that lncRNAs may act as oncogenes or tumor suppressor
genes, and exert a multitude of biological effects on various types
of cancer, including CRC (20,27,28).
Accumulating evidence has indicated that lncRNAs regulate the
proliferation, migration, invasion, apoptosis, pyroptosis and
metastasis of cancer cells during tumorigenesis (6,7).
In the present study, the expression levels of the
newly identified lncRNA RP1-85F18.6 were upregulated in CRC tissues
and cell lines, thus indicating that it may have a key role in CRC
tumorigenesis. Subsequently, the biological role of lncRNA
RP1-85F18.6 was evaluated in CRC cells, and the underlying
molecular mechanisms were explored. The results demonstrated that
lncRNA RP1-85F18.6 triggered the proliferation, invasion and cell
cycle disruption, and suppressed apoptosis and pyroptosis of CRC
cells. In addition, it was revealed that knockdown of lncRNA
RP1-85F18.6 decreased ΔNp63 expression without affecting TAp63
expression. Furthermore, the results demonstrated that the
tumor-promoting effects of lncRNA RP1-85F18.6 overexpression were
reversed by knockdown of ΔNp63. It is well known that p53 serves as
a tumor suppressor in CRC cells through the regulation of various
target genes (29). ΔNp63, which is
a member of the p53 family, acts as an oncogene (30,31);
specifically, ΔNp63 acts as a transcriptional repressor and
oncoprotein through opposing the activity of p53 (32,33).
Previous studies have reported that ΔNp63 facilitates transition
through the cell cycle, proliferation, migration, invasion and
metastasis of cancer cells, whereas it inhibits apoptosis, through
stimulating the expression of several target genes (12,13,34).
These biological functions of ΔNp63 are in accordance with those of
lncRNA RP1-85F18.6, which further supports the present evidence.
These findings indicated that the tumor-promoting effects of lncRNA
RP1-85F18.6 on CRC cells are partially mediated through the ΔNp63
signaling pathway.
Based on the results obtained over the last few
years, lncRNAs appear to regulate gene expression at epigenetic,
transcriptional or post-transcriptional levels (5). In the present study, knockdown of
lncRNA RP1-85F18.6 reduced ΔNp63 expression at both the mRNA and
protein levels, thus suggesting that lncRNA RP1-85F18.6 regulates
ΔNp63 partly through transcriptional regulation. Previous research
revealed that lncRNA PURPL regulates p53 expression and stability
through association with MYB Binding Protein 1a, a protein that
binds to and stabilizes p53 (16).
It may be hypothesized that lncRNA RP1-85F18.6 regulates ΔNp63 by a
certain gene that promotes ΔNp63 stability; we hope to elucidate
this in future studies.
In the present study, silencing lncRNA RP1-85F18.6
also induced pyroptosis of CRC cells through cleavage of GSDMD. To
the best of our knowledge, there are few studies focusing on the
role of non-protein-coding genes in pyroptosis (7). In addition to apoptosis, pyroptosis is
another type of programmed cell death, which is characterized by
plasma membrane rupture. Recently, Shi et al redefined
pyroptosis as gasdermin-mediated programmed necrosis (26). Furthermore, Wang et al
revealed that chemotherapeutic drugs induce pyroptosis of cancer
cells through cleavage of gasdermin E (35). In the present study, cleavage of
GSDMD, which was induced by silencing lncRNA RP1-85F18.6, promoted
the pyroptosis of CRC cells, thus suggesting that GSDMD may
represent a novel focus in CRC research.
Carcinoembryonic antigen (CEA) is the most widely
used molecular marker of CRC, which has been proven to be a
valuable tool for the diagnosis of CRC (36). Previous research demonstrated that
the sensitivity of CEA is 46.59% and its specificity is 80%
(37). In the present study, the
sensitivity and specificity values of lncRNA RP1-85F18.6 were 55.9
and 76.5%, respectively, which are comparable with those of CEA.
These findings suggested that lncRNA RP1-85F18.6 may be a valuable
prognostic and diagnostic biomarker for CRC in the future.
In conclusion, to the best of our knowledge, the
present study is the first to report that lncRNA RP1-85F18.6
expression may be increased in CRC tissues and cell lines. This
lncRNA was revealed to promote proliferation, invasion and cell
cycle disruption, whereas it inhibited the apoptosis and pyroptosis
of CRC cells through inducing ΔNp63. In addition, the present
findings indicated that lncRNA RP1-85F18.6, ΔNp63 and GSDMD may
prove to be valuable prognostic and diagnostic biomarkers for
early-stage CRC in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the New Xiangya
Talent Project of The Third Xiangya Hospital of Central South
University (grant no. JY201508).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
YM performed the experiments and wrote the
manuscript; YC statistically analyzed the data; CL collected the
tissues and analyzed the clinical characteristics of all patients;
GH designed the experiments, analyzed the ata, supervised the
experiments and gave final approval of the version to be published.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Third Xiangya Hospital of Central South University
(No. 2014-S009), and patients provided written informed
consent.
Patient consent for publication
The patients provided consent for publication.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
CRC
|
colorectal cancer
|
|
GSDMD
|
gasdermin D
|
|
ncRNA
|
non-protein-coding ribonucleic
acid
|
|
NCT
|
non-cancerous tissue
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
LDH
|
lactate dehydrogenase
|
|
ROC curve
|
receiver operating characteristic
curve
|
|
AUC
|
area under the ROC curve
|
|
CEA
|
carcinoembryonic antigen
|
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RG, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
burden of disease study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Y, Junjie P, Sanjun C and Ma Y: Long
non-coding RNAs in colorectal cancer: Progression and future
directions. J Cancer. 8:3212–3225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng H, Wang JM, Li M, Tang R, Tang K, Su
Y, Hou Y and Zhang J: Long non-coding RNAs: New biomarkers for
prognosis and diagnosis of colon cancer. Tumour Biol.
39:10104283177063322017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Liu X, Bai X, Lin Y, Li Z, Fu J,
Li M, Zhao T, Yang H, Xu R, et al: Melatonin prevents endothelial
cell pyroptosis via regulation of long noncoding RNA
MEG3/miR-223/NLRP3 axis. J Pineal Res. 64:doi: 10.1111/jpi.12449.
2018. View Article : Google Scholar :
|
|
8
|
Chen Y, Peng Y, Fan S, Li Y, Xiao ZX and
Li C: A double dealing tale of p63: An oncogene or a tumor
suppressor. Cell Mol Life Sci. 75:965–973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gressner O, Schilling T, Lorenz K,
Schleithoff Schulze E, Koch A, Schulze-Bergkamen H, Lena AM, Candi
E, Terrinoni A, Catani MV, et al: TAp63alpha induces apoptosis by
activating signaling via death receptors and mitochondria. EMBO J.
24:2458–2471. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu G, Nomoto S, Hoque MO, Dracheva T,
Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, et al:
DeltaNp63alpha and TAp63alpha regulate transcription of genes with
distinct biological functions in cancer and development. Cancer
Res. 63:2351–2357. 2003.PubMed/NCBI
|
|
11
|
Srivastava K, Pickard A, McDade S and
McCance DJ: p63 drives invasion in keratinocytes expressing HPV16
E6/E7 genes through regulation of Src-FAK signalling. Oncotarget.
8:16202–16219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He YF, Tian DY, Yi ZJ, Yin ZK, Luo CL,
Tang W and Wu XH: Upregulation of cell adhesion through delta Np63
silencing in human 5637 bladder cancer cells. Asian J Androl.
14:788–792. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Compagnone M, Gatti V, Presutti D, Ruberti
G, Fierro C, Markert EK, Vousden KH, Zhou H, Mauriello A, Anemone
L, et al: ΔNp63-mediated regulation of hyaluronic acid metabolism
and signaling supports HNSCC tumorigenesis. Proc Natl Acad Sci USA.
114:13254–13259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Jiang X and Niu X: Long non-coding
RNA reprogramming (ROR) promotes cell proliferation in colorectal
cancer via affecting P53. Med Sci Monit. 23:919–928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thorenoor N, Faltejskova-Vychytilova P,
Hombach S, Mlcochova J, Kretz M, Svoboda M and Slaby O: Long
non-coding RNA ZFAS1 interacts with CDK1 and is involved in
p53-dependent cell cycle control and apoptosis in colorectal
cancer. Oncotarget. 7:622–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li XL, Subramanian M, Jones MF, Chaudhary
R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, et al:
Long noncoding RNA PURPL suppresses basal p53 levels and promotes
tumorigenicity in colorectal cancer. Cell Rep. 20:2408–2423. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edge SB and Compton CC: The American Joint
Committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yun DP, Wang YQ, Meng DL, Ji YY, Chen JX,
Chen HY and Lu DR: Actin-capping protein CapG is associated with
prognosis, proliferation and metastasis in human glioma. Oncol Rep.
39:1011–1022. 2018.PubMed/NCBI
|
|
20
|
Xie S, Ge Q, Wang X, Sun X and Kang Y:
Long non-coding RNA ZFAS1 sponges miR-484 to promote cell
proliferation and invasion in colorectal cancer. Cell Cycle.
17:154–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Zhang Y, He J, Fu Y, Lin C and Li
X: MicroRNA-133b is regulated by TAp63 while no gene mutation is
present in colorectal cancer. Oncol Rep. 37:1646–1652. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H, Wu R, Chen M, Li D, Dai J, Zhang Y,
Gao K, Yu J, Hu G, Guo Y, et al: Comprehensive analysis of
differentially expressed profiles of lncRNAs and construction of
miR-133b mediated ceRNA network in colorectal cancer. Oncotarget.
8:21095–21105. 2017.PubMed/NCBI
|
|
23
|
Li J, Ma W, Zeng P, Wang J, Geng B, Yang J
and Cui Q: LncTar: A tool for predicting the RNA targets of long
noncoding RNAs. Brief Bioinform. 16:806–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi J, Zhao Y, Wang K, Shi X, Wang Y,
Huang H, Zhuang Y, Cai T, Wang F and Shao F: Cleavage of GSDMD by
inflammatory caspases determines pyroptotic cell death. Nature.
526:660–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding J, Wang K, Liu W, She Y, Sun Q, Shi
J, Sun H, Wang DC and Shao F: Pore-forming activity and structural
autoinhibition of the gasdermin family. Nature. 535:111–116. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi J, Gao W and Shao F: Pyroptosis:
Gasdermin-mediated programmed necrotic cell death. Trends Biochem
Sci. 42:245–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F,
Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al: Long
non-coding RNA UICLM promotes colorectal cancer liver metastasis by
acting as a ceRNA for microRNA-215 to regulate ZEB2 expression.
Theranostics. 7:4836–4849. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the interleukin-6/STAT3 signaling
pathway. Hepatology. 67:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Q, Deng J, Fan D, Duan Z, Zhu C, Fu R
and Wang S: Ginsenoside Rh4 induces apoptosis and autophagic cell
death through activation of the ROS/JNK/p53 pathway in colorectal
cancer cells. Biochem Pharmacol. 148:64–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu K, Yao H, Lei S, Xiong L, Qi H, Qian
K, Liu J, Wang P and Zhao H: The miR-124-p63 feedback loop
modulates colorectal cancer growth. Oncotarget. 8:29101–29115.
2017.PubMed/NCBI
|
|
31
|
Nayak KB, Kuila N, Das Mohapatra A, Panda
AK and Chakraborty S: EVI1 targets ΔNp63 and upregulates the cyclin
dependent kinase inhibitor p21 independent of p53 to delay cell
cycle progression and cell proliferation in colon cancer cells. Int
J Biochem Cell Biol. 45:1568–1576. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Balboni AL, Cherukuri P, Ung M, DeCastro
AJ, Cheng C and DiRenzo J: p53 and ΔNp63α coregulate the
transcriptional and cellular response to TGFβ and BMP signals. Mol
Cancer Res. 13:732–742. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He Y, Wu X, Tang W, Tian D, Luo C, Yin Z
and Du H: Impaired delta NP63 expression is associated with poor
tumor development in transitional cell carcinoma of the bladder. J
Korean Med Sci. 23:825–832. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giacobbe A, Compagnone M,
Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH,
Annicchiarico-Petruzzelli M, Melino G and Peschiaroli A: p63
controls cell migration and invasion by transcriptional regulation
of MTSS1. Oncogene. 35:1602–1608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Gao W, Shi X, Ding J, Liu W, He H,
Wang K and Shao F: Chemotherapy drugs induce pyroptosis through
caspase-3 cleavage of a gasdermin. Nature. 547:99–103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thomas DS, Fourkala EO, Apostolidou S,
Gunu R, Ryan A, Jacobs I, Menon U, Alderton W, Gentry-Maharaj A and
Timms JF: Evaluation of serum CEA, CYFRA21-1 and CA125 for the
early detection of colorectal cancer using longitudinal preclinical
samples. Br J Cancer. 113:268–274. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao Y, Wang J, Zhou Y, Sheng S, Qian SY
and Huo X: Evaluation of serum CEA, CA19-9, CA72-4, CA125 and
ferritin as diagnostic markers and factors of clinical parameters
for colorectal cancer. Sci Rep. 8:27322018. View Article : Google Scholar : PubMed/NCBI
|