Introduction
Gastric cancer (GC) is one of the most common
malignant tumors and a major cause of cancer-associated mortality
worldwide (1). Early diagnosis and
effective treatment for GC are difficult to achieve as the
underlying molecular mechanisms are unclear (2). As such, increasing our understanding
of the mechanisms responsible for GC is crucial to allow for early
diagnosis, effective therapy and survival prediction.
Long noncoding RNAs (lncRNAs) are RNAs >200 bp in
length that are not directly translated into protein (3,4).
LncRNAs account for only 4–9% of the mammalian transcriptome
(5); however, a number of reports
indicate that lncRNAs serve key roles in gene expression, gene
imprinting, DNA recombination, chromatin modification, cell cycle
regulation and encoding peptides (6–9). It
has been reported that lncRNAs interact with genomic DNA and
chromatin in the following ways: i) Forming a DNA-RNA heteroduplex
by binding to regions of single-stranded DNA; ii) forming
DNA-DNA-RNA triplexes by Hoogsteen or reverse Hoogsteen base
pairing; iii) tethering to chromatin through association with RNA
polymerase II and thereby acting as allele-specific signatures for
a specific locus; iv) indirectly binding to chromatin by binding to
chromatin-associated proteins or transcription factors; and v)
directly forming sense chromatin structures (10). LncRNA is able to exert a wide range
of regulatory functions, including on cell growth, apoptosis,
migration, invasion and autophagy (11).
A previous study by our research group revealed that
lncRNA STCAT3 overexpression promoted the proliferation and
invasion of GC cells and that STCAT3 interference significantly
inhibited GC (12). However, the
potential pathological mechanisms of STCAT3 and its prognostic
significance in patients with GC remain uncertain. The aim of the
present study was to screen and identify proteins that interact
with STCAT3 by using RNA pull-down technology and mass
spectrometry. Reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and western blotting were used to measure mRNA
and protein expression, respectively. RNA binding protein
immunoprecipitation (RIP) experiments were also performed. The
prognostic value of STCAT3 and its binding protein in GC was
evaluated using univariate Cox proportional hazards regression
models.
Materials and methods
Patients and clinical samples
The present study was approved by the institutional
review board of the Affiliated Hospital of Nantong University
(Nantong, China) and performed according to the Declaration of
Helsinki. Written informed consent was obtained from all patients
prior to the experiments. A total of 98 patients (32–84 years old;
male: female, 63: 35) with GC were recruited at the Department of
General Surgery, the Affiliated Hospital of Nantong University
between May 2014 and December 2016. GC tissues and pair-matched
normal gastric tissues (5 cm adjacent to carcinoma tissue) were
harvested. No patients had previously undergone chemotherapy,
radiotherapy or treatment with long-term anti-inflammatory
nonsteroidal drugs or corticosteroids prior to surgery. Cancer
classification and pathological typing was performed by two
experienced pathologists in a double-blind manner.
Cell culture and transfection
The human GC cell lines AGS, MGC-803, BGC-823,
MKN-28, MKN-45 and SGC-7901 and the normal human gastric mucosa
cell line GES-1 were purchased from the Shanghai Institutes for
Biological Sciences (Chinese Academy of Sciences, Shanghai, China).
Cells were cultured in RPMI-1640 culture medium (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal
bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., Hangzhou,
China), 100 µg/ml streptomycin, and 100 U/ml penicillin at 37°C in
an atmosphere containing 5% CO2. STCAT3-overexpression
plasmid (STCAT3-pCDNA3.1), negative control vector (pCDNA3.1),
STCAT3 knockdown plasmid [pGPU6/GFP/Neo-STCAT3-short hairpin
(sh)RNA, sh-STCAT3], and negative control vector (sh-NC) were
constructed and synthesized by Shanghai GenePharma Co., Ltd
(Shanghai, China). Cells in the log phase were selected and
transfected with 4.0 µg/well plasmid DNA in 24-well plates
(5×105 cells/well) at 85% confluence using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cells were dissociated using a
solution containing 0.25% trypsin EDTA (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Primer sequences used for plasmid construction
are provided in Table I. Subsequent
experiments were performed 72 h post-transfection.
 | Table I.Primers used for shRNA plasmid
construction. |
Table I.
Primers used for shRNA plasmid
construction.
| shRNA | Direction | Sequence
(5′-3′) |
|---|
| STCAT3 shRNA | Sense |
CACCGTACGCTTCACAAGGTTCTCATTCAAGAGATGAGAACCTTGTGAAGCGTACTTTTTTG |
|
| Antisense |
GATCCAAAAAAGTACGCTTCACAAGGTTCTCATCTCTTGAATGAGAACCTTGTGAAGCGTAC |
| Control shRNA | Sense |
CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG |
|
| Antisense |
GATCCAAAAAAGTTCTCCGAACGTGTCACGTAATCTCTTGACGTGACACGTTCGGAGAAC |
In vitro transcription assays and RNA
pull down assays
Primers were designed and synthesized by Wuhan
GeneCreate Biological Engineering Co., Ltd. (Wuhan, China). The
STCAT3 gene primer sequences were as follows: Sense chain, forward
5′-TAATACGACTCACTATAGGGGTCTATTTGATTCTTCTCTC-3′ and reverse
5′-GGAGCTGAAAAACACAGC-3′; antisense chain, forward
5′-TAATACGACTCACTATAGGGGGAGCTGAAAAACACAGCAT-3′ and reverse
5′-GTCTATTTGATTCTTCTCTC-3′. In vitro amplification of the
sense and antisense chains was performed to obtain DNA templates;
in vitro transcription reactions were then performed using a
MAXIscript® T7 Transcription kit (no. AM1312, Ambion;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. STCAT3 RNAs were labeled by desthiobiotinylation using a
Pierce™ RNA 3′ End Desthiobiotinylation kit (no. 20163; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA pulldown assays were then performed using a Pierce™ Magnetic
RNA-Protein Pull-Down kit (no. 20164; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol.
Mass spectrometry
Proteomic detection of the purified RNA-binding
proteins was achieved using mass spectrometry. Briefly, 10 mM
dithiothreitol (DTT) and 55 mM iodoacetamide (IAM) were added into
the polyacrylamide gel electrophoresis (PAGE) gel along with a
trypsin enzyme (Beijing Biodee Diagnostics, Beijing, China) to
enzymatically hydrolyze overnight. The resulting peptides were
treated with a C18 column (YMC Co., Ltd., Shanghai, China) and then
dissolved in 15 µl loading buffer (0.1% formic acid and 3%
acetonitrile). Peptides were investigated using LC-MS/MS (ekspert™
nanoLC, TripleTOF 5600-plus; SCIEX, Framingham, MA, USA). Data were
analyzed using ProteinPilot™ Software version 4.5 (SCIEX) with
Triple TOF 5600-plus for database retrieval (UniProt, June 2016;
http://www.uniprot.org/proteomes/UP000005640) to
obtain the possible interacting proteins.
RNA immunoprecipitation analysis
(RIP)
RIP experiments were performed using a Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore,
Billerica, MA, USA) according to the manufacturer's protocol. The
dermcidin antibody used for RIP was from Abcam (1:20; cat. no.
ab52138, Cambridge, UK). An aliquot of mouse IgG (1:20; cat. no.
ab190475, Abcam) served as the input control. Messenger RNAs of
cells were isolated, then samples RNA enrichment was determined by
RT-qPCR as described in the following section and normalized to the
input control. Data was from triplicate experiments.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cDNA was obtained using a
PrimeScript™ RT Reagent kit (cat. no. RR014A; Takara Biotechnology,
Co., Ltd., Dalian, China) according to the manufacturer's protocol.
The temperature protocol was as follows: 42°C for 60 min and 70°C
for 5 min. qPCR was performed using SYBR-Green I (Takara
Biotechnology, Co., Ltd.) and an Applied Biosystems 7500 Real-Time
PCR System (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The primers were synthesized by GeneCreate
(Wuhan, China) and are presented in Table II. Relative gene expression was
calculated with β-actin as a control. The PCR system comprised the
following: 25 µl SYBR Premix Ex Taq™ (2X), 2 µl PCR forward primer,
2 µl PCR reverse primer, 4 µl PCR template (cDNA solution), 1 µl
ROX Reference Dye (50X) and 16 µl sterile double steamed water.
Thermocycling conditions were as follows: 95°C for 30 sec, 95°C for
5 sec and 60°C for 30 sec. Data were quantified using the
2−ΔΔCq method (13).
Each RT-qPCR experiment was performed in triplicate. Tumor tissues
were assigned to the high expression group when STCAT3 expression
was two fold higher compared with adjacent normal tissues.
 | Table II.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| STCAT3 |
CCACTGTTTGTCTGATGGGC |
AGCAAGACAAGCCAGCATTC |
| Dermcidin |
TCTTTGGGGCTCCTGTGAATC |
CTGCTGCTCCTGGGTATCATT |
| Suprabasin |
AAATAGCAGCGTGGCTTCCC |
AGTAGCAGAAGGAGGGAGCA |
| Cathepsin D |
TGCTCAAGAACTACATGGACGC |
CGAAGACGACTGTGAAGCACT |
| Calmodulin |
TACGAGGAGTTCGCGAGGAT |
AGAGTCCCAGCACAAAAGCA |
| Annexin A2 |
TGCCTTCGCCTACCAGAGAA |
GCCCAAAATCACCGTCTCC |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC |
| β-actin |
AGCGAGCATCCCCCAAAGTT |
GGGCACGAAGGCTCATCATT |
Western blotting
Western blotting was performed to assess the
expression of dermcidin, GAPDH, calmodulin, cathepsin D,
suprabasin, Annexin A2 and β-actin proteins as described previously
(14). Briefly, cells were rapidly
homogenized in a buffer containing 10 µg/ml leupeptin, 1% Triton
X-100, 10% sodium dodecyl sulfate (SDS), 1 M Tris-HCl (pH 7.5),
0.5% sodium deoxycholate, 10 µg/ml aprotinin, 1% Nonidet™ P-40, 1
mM phenylmethylsulfonyl fluoride and 0.5 M
ethylenediaminetetraacetic acid, following which they were
centrifuged at 10,000 × g for 30 min at 4°C and collect the
supernatant. Protein concentrations were determined using a Bio-Rad
protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Equal amounts of protein (40 µg/lane) were separated by 15%
SDS-PAGE and transferred to polyvinylidene difluoride membranes.
The membranes were blocked with 5% dried skimmed milk in
Tris-buffered saline and Tween-20 containing 0.05% Tween-20, 20 mM
Tris, and 150 mM NaCl for 2 h at room temperature. Membranes were
incubated at 4°C overnight with the following monoclonal primary
antibodies: Rabbit anti-dermcidin (cat. no. ab52138), rabbit
anti-GAPDH (cat. no. ab181602), rabbit anti-calmodulin (cat. no.
ab45689), rabbit anti-cathepsin D (cat. no. ab75852), rabbit
anti-suprabasin (cat. no. ab232771) and rabbit anti-Annexin A2
(cat. no. ab178677; all 1:2,000; all Abcam). Mouse anti-β-actin
(cat. no. ab8226; 1:2,000; Abcam) monoclonal antibody was used as
an internal control. Horseradish peroxidase (HRP)-conjugated goat
anti-rabbit IgG H&L (1:10,000; cat. no. ab97051; Abcam) and
rabbit anti-mouse IgG H&L cat. no. ab6728; 1:5,000; Abcam) were
used as secondary antibodies for 2 h at room temperature. The
proteins were visualized using an enhanced chemiluminescent
detection system (GE Healthcare Life Sciences, Little Chalfont,
UK), according to the manufacturer's protocol. Following
chemiluminescence, bands were exposed to X-ray films (Kodak,
Rochester, NY, USA) and ImageJ 1.37 software (National Institutes
of Health, Bethesda, MD, USA) was used for grey value analysis.
Relative amounts of proteins were quantified by absorbance analysis
and normalized to β-actin. The experiments were repeated three
times.
Immunohistochemistry
Tumor and adjacent normal tissues were fixed in 10%
buffered formalin for 24 h at room temperature, embedded in
paraffin and cut into sections (4 µm). The sections were
deparaffinized in xylene, rehydrated in graded ethanol solutions
and treated with 0.1% hydrogen peroxide in methanol for 20 min to
remove endogenous peroxidase activity. Antigen retrieval was
performed by microwaving the sections immersed in 10 mM citric acid
buffer for 10 min, followed by 1-h incubation at room temperature.
To block non-specific protein binding, sections were incubated with
5% bovine serum albumin (cat. no. LLBB-1000-01; SurModics, Inc.,
Eden Prairie, USA) for 10 min at 37°C. Sections were incubated at
room temperature for 1 h with monoclonal antibody against dermcidin
(cat. no. ab52138; 1:50; Abcam) and again overnight at 4°C.
Sections were washed with PBS and incubated with HRP-conjugated
polyclonal goat anti-rabbit IgG (H&L) biotin secondary
antibodies (cat. no. E030110-01; 1:500; EarthOx, LLC, San
Francisco, CA, USA) for 1 h at room temperature, followed by 0.1%
hydrogen peroxide and 0.6 mM 3,3′-diaminobenzidine (DAB) in PBS for
8 min at room temperature. Omission of the primary antibody served
as a negative control. All sections were counterstained with
hematoxylin for 2 min at room temperature and examined using a
light microscope (BX51; Olympus Corporation, Tokyo, Japan;
magnification, ×100 and ×400). Two consultant pathologists assessed
slides in a blinded fashion. Staining was evaluated as previously
described (14).
Statistical analysis
All statistical analyses were performed using SPSS
17.0 (SPSS, Inc., Chicago, IL, USA), and graphs were generated
using GraphPad Prism software version 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). Differences between groups were analyzed using
Student's t-test, one-way analysis of variance, the χ2
test, Spearman's rank correlation coefficient or Pearson's
correlation coefficient, as appropriate. Student-Newman-Keuls post
hoc test was performed following ANOVA. Variables associated with
overall survival (OS) and time to recurrence (TTR) were assessed
using univariate Cox proportional hazard regression models.
Kaplan-Meier plots (log-rank tests) were used to describe OS and
TTR. P<0.05 was considered to indicate a statistically
significant difference.
Results
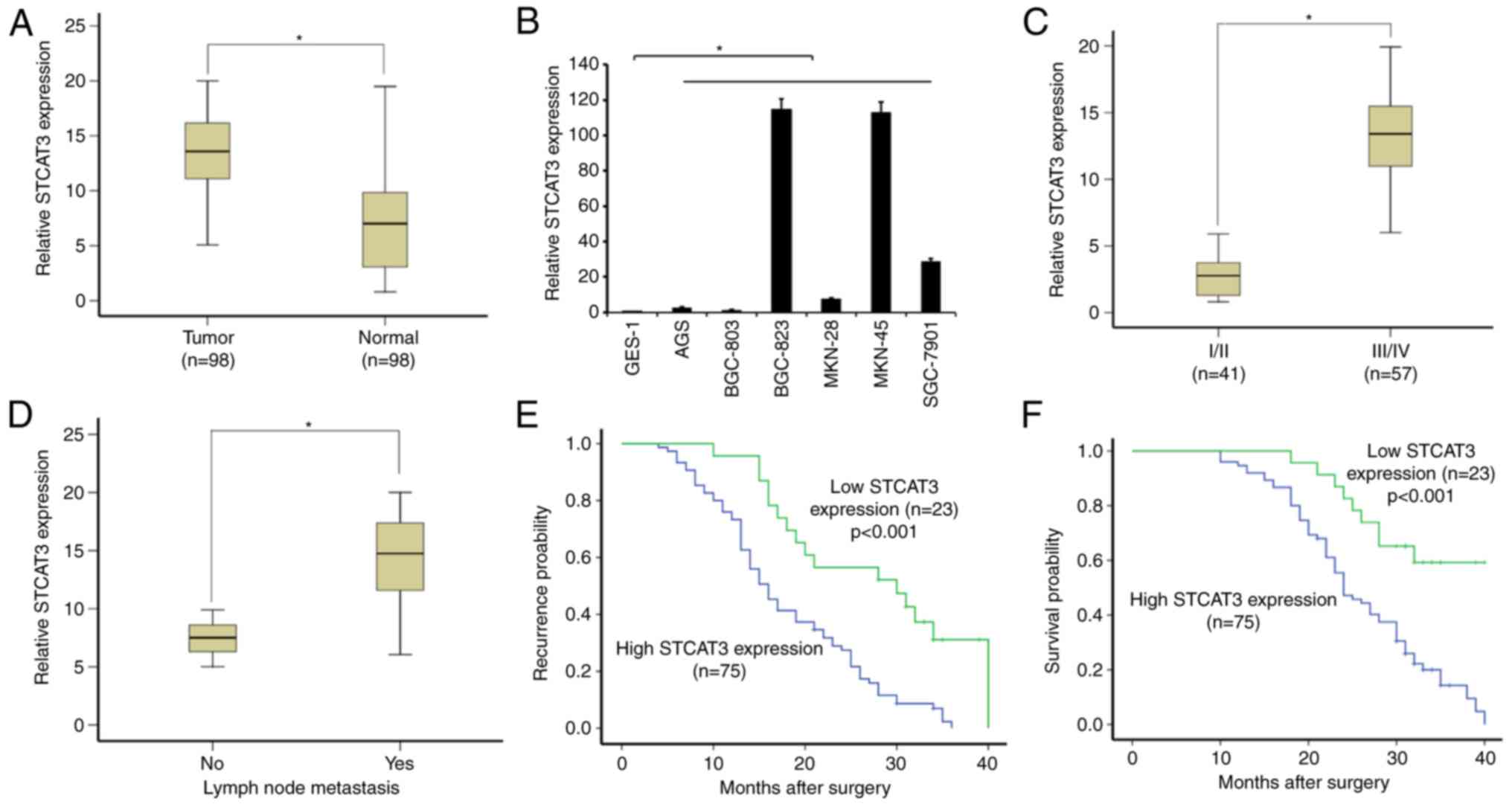
STCAT3 is highly expressed in GC
tissues
The expression of STCAT3 was detected in 98 paired
GC tissues and adjacent normal tissues using RT-qPCR normalized to
β-actin. STCAT3 expression in GC tissues was significantly higher
compared with in the adjacent normal gastric tissues (P<0.05;
Fig. 1A). The expression of STCAT3
varied dramatically between GC cell lines, however it was
upregulated in all GC cell lines compared with the normal gastric
mucosa cell line GES-1 (all P<0.05). In particular, STCAT3
expression was much higher in BGC-823, MKN-45 and SGC-7901 compared
with the GES-1 cell line (Fig.
1B).
STCAT3 is and is associated with
advanced clinical stage and poor survival
The clinical features of patients with GC are
presented in Table III. STCAT3
expression in GC tissues was positively correlated with TNM stage
and lymph node metastasis; patients with GC of TNM stages III/IV
had significantly higher STCAT3 expression compared with those with
GC at stages I/II (P<0.05; Fig.
1C). Furthermore, patients with positive lymph node metastasis
had significantly higher STCAT3 expression compared with patients
with no lymph node metastasis (P<0.05; Fig. 1D). Other clinical characteristics,
including histological grade, invasion depth, sex and age, were not
correlated with STCAT3 expression.
 | Table III.The association between STCAT3
expression and clinical characteristics. |
Table III.
The association between STCAT3
expression and clinical characteristics.
|
|
| STCAT3
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | N | Low | Higha | P-value |
|---|
| Age (years) |
|
|
| 0.478 |
|
<60 | 41 | 8 | 33 |
|
|
≥60 | 57 | 15 | 42 |
|
| Sex |
|
|
| 0.804 |
|
Male | 63 | 14 | 49 |
|
|
Female | 35 | 9 | 26 |
|
| Tumor size
(cm) |
|
|
| 0.456 |
|
<5 | 65 | 17 | 48 |
|
| ≥5 | 33 | 6 | 27 |
|
| Invasion depth |
|
|
| 0.096 |
|
T1/T2 | 44 | 14 | 30 |
|
|
T3/T4 | 54 | 9 | 45 |
|
| TNM phase |
|
|
| 0.001 |
|
I/II | 41 | 17 | 24 |
|
|
III/IV | 57 | 6 | 51 |
|
| Differentiation
degree |
|
|
| 1 |
| High or
mid | 48 | 11 | 37 |
|
|
Low | 50 | 12 | 38 |
|
| Lymph node
metastasis |
|
|
| 0.025 |
| No | 35 | 13 | 22 |
|
|
Yes | 63 | 10 | 53 |
|
Kaplan-Meier analysis and log-rank test were
performed to determine the association between the STCAT3
expression in GC tissues and patient prognosis. The median time
until postoperative recurrence in the patients with high STCAT3
expression levels was significantly shorter compared with the low
STCAT3 group (P<0.05; Fig. 1D).
Furthermore, the median survival time in the low STCAT3 expression
group was significantly longer compared with the high expression
group (P<0.05; Fig. 1E).
Screening and identification of
dermcidin as a novel binding protein of lncRNA STCAT3
STCAT3 was used to capture its interacting protein
using the BGC-823 cell line. A notable difference was observed
between the sense and antisense strands of STCAT3 following
electrophoresis silver staining. The protein of band 2 was
decomposed and analyzed using mass spectrometry (Fig. 2A). Species database retrieval and
bioinformatics analysis were performed and 56 proteins were
different regarding statistics or function (data not shown).
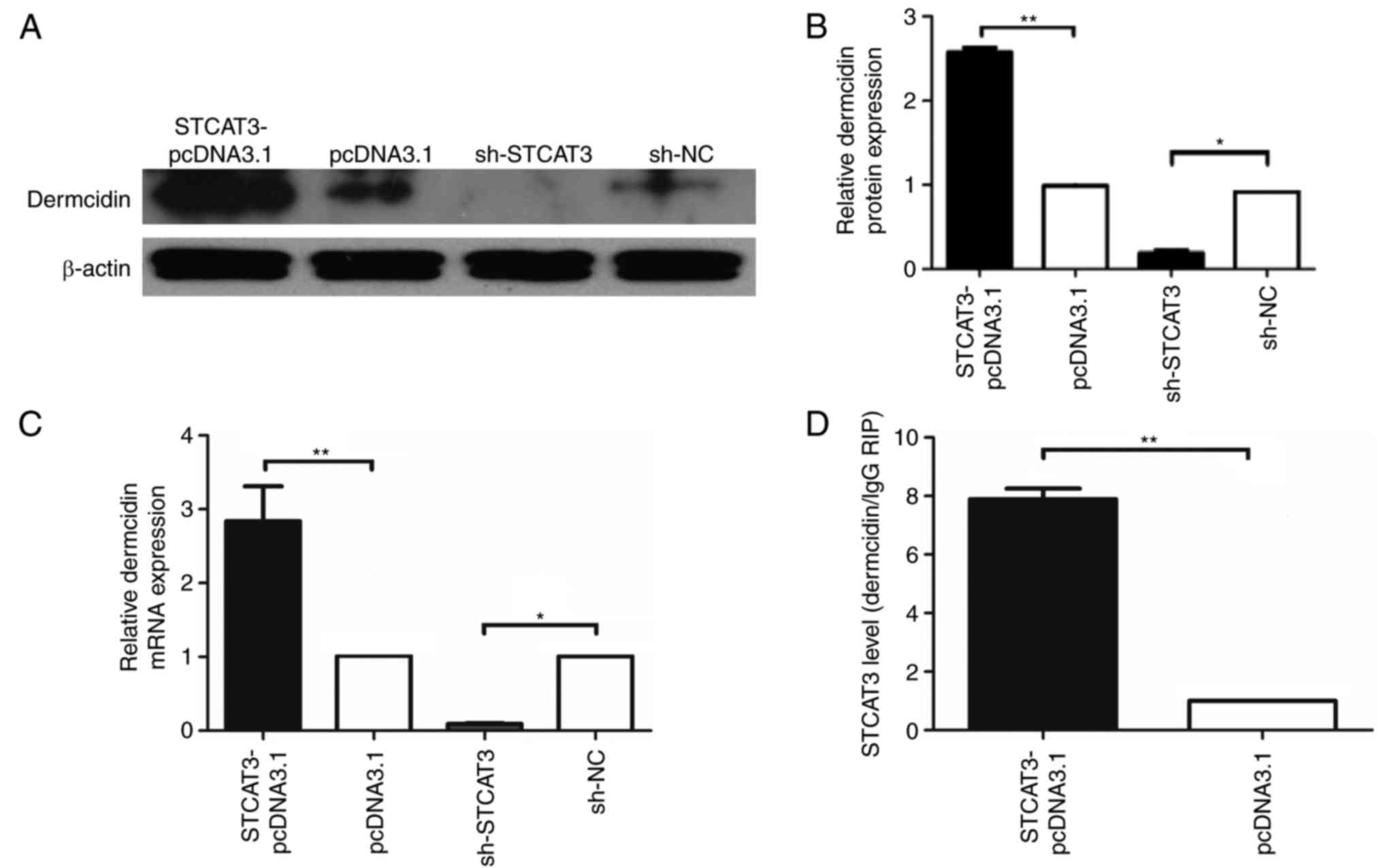
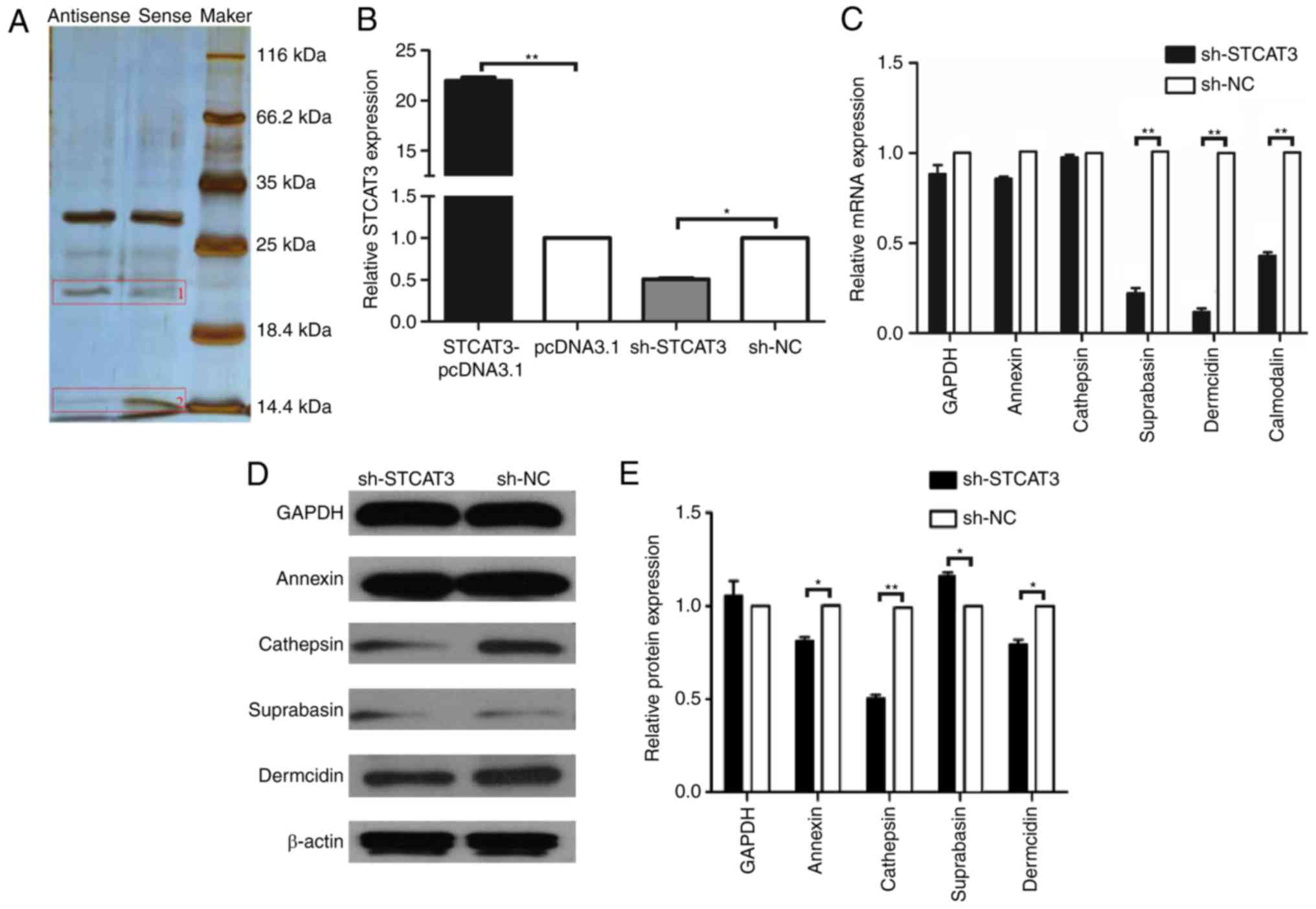 | Figure 2.Screening and identification of
candidate lncRNA STCAT3 binding proteins. (A) Silver
electrophoresis staining with the sense and antisense RNA of lncRNA
STCAT3. Reverse transcription-quantitative polymerase chain
reaction was used to assess (B) STCAT3 expression in the
STCAT3-pCDNA3.1, pCDNA3.1, STCAT3-shRNA and sh-NC groups,
respectively, and (C) the expression of annexin, cathepsin-D,
suprabasin, dermcidin, and calmodulin mRNA in BGC-823 cells
following transfection. (D and E) Western blotting was uses to
measure the expression of annexin, cathepsin-D, suprabasin,
dermcidin, and calmodulin-like proteins in the sh-STCAT3 and sh-NC
groups. β-actin was used as a reference control.
**P<0.001 and *P<0.05. lncRNA, long non-coding
RNA; STCAT3, stomach cancer-associated transcript 3; sh, short
hairpin RNA; NC, negative control. |
Mounting studies have reported that dermcidin
(15–17), GAPDH (18), annexin (19,20),
calmodulin-like protein (21,22),
cathepsin-D (23) and suprabasin
(24) are associated with malignant
tumors. Based on this, RNA-protein interaction prediction was used
to assess whether these proteins were binding candidates for
STCAT3. The interaction probabilities for all proteins were
>0.5, suggesting that these proteins may be candidate binding
proteins for STCAT3.
To identify the binding protein for STCAT3, the
plasmid vectors STCAT3-pCDNA3.1, pCDNA3.1, sh-STCAT3 and sh-NC were
transfected into BGC-823 cells. STCAT3 expression in the
STCAT3-pCDNA3.1 group was significantly higher compared with the
pCDNA3.1 group (P<0.001; Fig.
2B). In contrast, STCAT3 expression in the sh-STCAT3 group was
decreased compared with the sh-NC group (P<0.001; Fig. 2B). RT-qPCR was used to measure the
mRNA expression of the potential binding proteins in the sh-STCAT3
and sh-NC groups. Dermcidin, suprabasin and calmodulin-like
proteins were significantly downregulated in the sh-STCAT3 group
compared with the sh-NC group (P<0.05; Fig. 2C). However, the expression of
annexin, cathepsin-D and GAPDH was not significantly different
between groups. Furthermore, dermcidin, suprabasin and
calmodulin-like protein expression was positively correlated with
lncRNA STCAT3 expression (Pearson r=0.985, 0.932 and 0.941;
P<0.05), suggesting that they were candidate STCAT3 binding
proteins.
In contrast, the results of western blotting
(Fig. 2D) revealed that dermcidin,
annexin and cathepsin D protein expression in the sh-STCAT3 group
was significantly lower compared with the sh-NC group (P<0.05;
Fig. 2E). These protein levels were
positively correlated with STCAT3 expression (Pearson r=0.967,
0.868, 0.971; P<0.05). Suprabasin protein expression was
significantly increased in the sh-STCAT3 group compared with the
sh-NC group and was negatively correlated with STCAT3 (Pearson
r=−0.951; P<0.05; Fig. 2E). As
such, in terms of protein expression, dermcidin, annexin and
cathepsin D were identified as potential binding proteins of
STCAT3. Based on these above results, dermcidin was identified as a
novel binding protein that can interact with the lncRNA STCAT3.
Validation of dermcidin as a novel
binding protein of lncRNA STCAT3
To confirm dermcidin as a novel binding protein of
STCAT3, dermcidin expression was measured in a number of GC cell
lines. RT-qPCR and western blotting revealed that dermcidin
expression was significantly increased in the STCAT3-pCDNA3.1 group
compared with the pCDNA3.1 group (P<0.001; Fig. 3A-C). However, dermcidin expression
was significantly lower in the sh-STCAT3 group compared with the
sh-NC group (P<0.01; Fig.
3A-C).
To confirm direct binding between lncRNA STCAT3 and
dermcidin, RIP-qPCR was performed to measure STCAT3 expression in
the STCAT3-pCDNA3.1 and pCDNA3.1 groups. LncRNA STCAT3
co-immunoprecipitated by the protein dermcidin was significantly
higher in the STCAT3-pCDNA3.1 group compared with the pCDNA3.1
group (P<0.001; Fig. 3D). These
data suggest that dermcidin is the direct-binding protein of lncRNA
STCAT3.
Dermcidin expression is positively
correlated with STCAT3 and poor survival in patients with GC
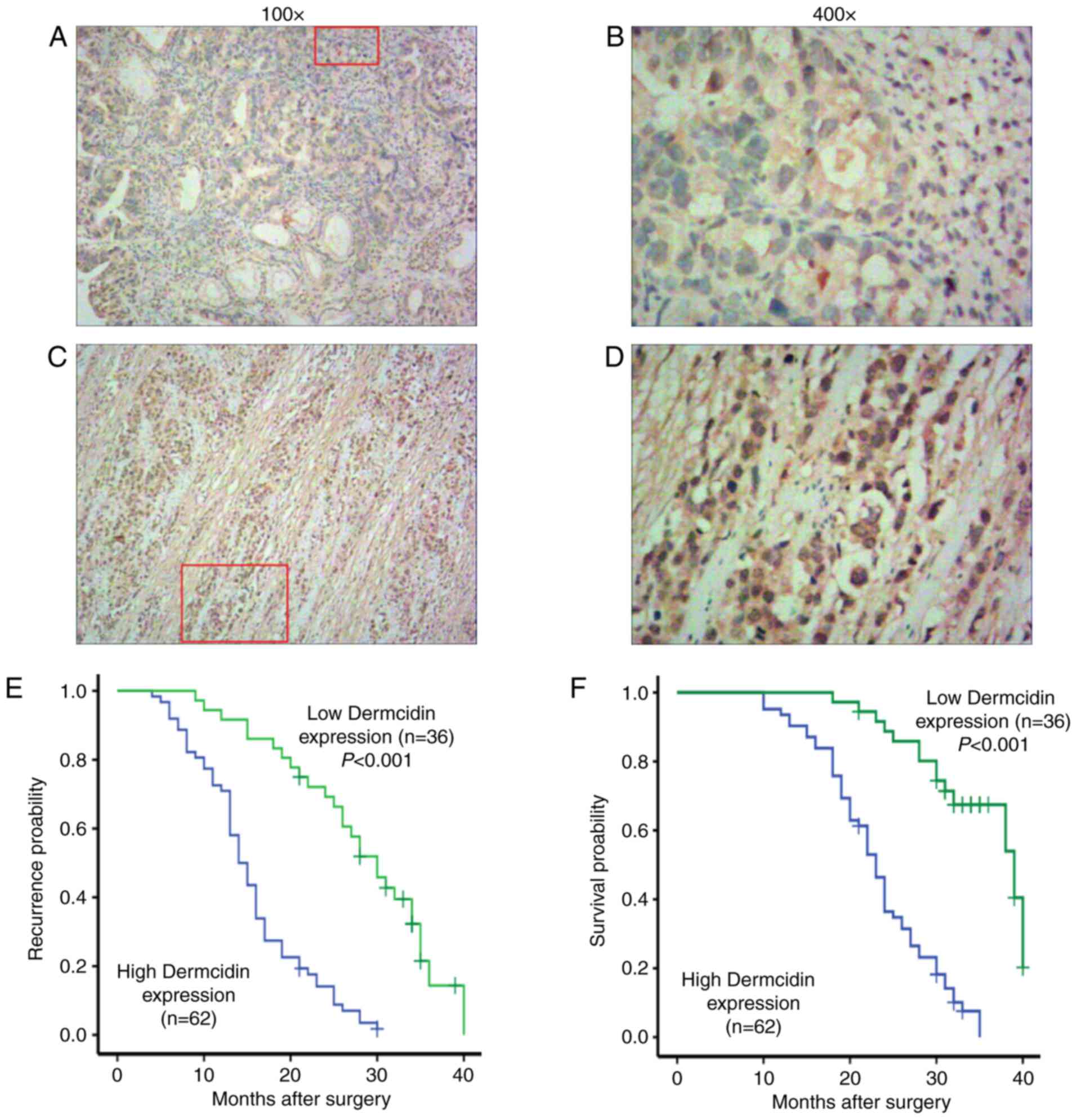
The expression of dermcidin was measured in GC
tissues from 98 patients with GC using immunohistochemistry.
Positive staining for dermcidin was localized to the cell nucleus
and cytoplasm in GC tissues (Fig.
4A-D). The rate of dermcidin expression was 63.26% (62/98) in
GC tissues, whereas it was weakly expressed in normal adjacent
tissues. Dermcidin expression was also demonstrated to be
significantly associated with clinical TNM stage, tumor
differentiation and lymph node metastasis (all P<0.05; Table IV).
 | Table IV.Association between Dermcidin
expression and clinical characteristics. |
Table IV.
Association between Dermcidin
expression and clinical characteristics.
|
|
| Dermcidin
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | N | − | + | P-value |
|---|
| Age (years) |
|
|
| 0.21 |
|
<60 | 41 | 12 | 29 |
|
|
≥60 | 57 | 24 | 33 |
|
| Sex |
|
|
| 0.194 |
|
Male | 63 | 20 | 43 |
|
|
Female | 35 | 16 | 19 |
|
| Tumor size
(cm) |
|
|
| 1 |
|
<5 | 65 | 24 | 41 |
|
| ≥5 | 33 | 12 | 21 |
|
| Invasion depth |
|
|
| 0.058 |
|
T1/T2 | 44 | 21 | 23 |
|
|
T3/T4 | 54 | 15 | 39 |
|
| TNM stage |
|
|
| 0.005 |
|
I/II | 41 | 22 | 19 |
|
|
III/IV | 57 | 14 | 43 |
|
| Differentiation
degree |
|
|
| 0.012 |
| High or
mid | 48 | 24 | 24 |
|
|
Low | 50 | 12 | 38 |
|
| Lymph node
metastasis |
|
|
| 0.002 |
| No | 35 | 20 | 15 |
|
|
Yes | 63 | 16 | 47 |
|
Spearman's rank correlation between STCAT3 mRNA and
dermcidin protein expressions in human GC tissue samples was
calculated and a positive correlation was observed between STCAT3
and dermcidin protein expression in GC tissues (r=0.5270;
P<0.001; Table V).
 | Table V.The correlation between STCAT3 and
Dermcidin protein expression in patients with GC. |
Table V.
The correlation between STCAT3 and
Dermcidin protein expression in patients with GC.
|
| Dermcidin |
|
|---|
|
|
|
|
|---|
| STCAT3 | + | − | Sum |
|---|
| + | 58 | 17 | 75 |
| − | 4 | 19 | 23 |
| Sum | 62 | 36 | 98 |
To investigate the association between dermcidin
expression and GC prognosis, Kaplan-Meier analysis and the log-rank
test were performed. The median time until postoperative recurrence
and median survival duration were significant higher in the low
dermcidin expression group compared with the high expression group
(P<0.001; Fig. 4E and F),
indicating that dermcidin overexpression is a predictor of poor
prognosis in patients with GC.
To confirm the prognostic value of STCAT3 and
dermcidin expression in patients with GC, univariate and
multivariate survival analyses (Cox proportional hazards regression
model) were performed. The univariate analysis included ten
prognostic factors: Age, sex, tumor size, invasion depth, TNM
stage, differentiation grade, lymph node metastasis, STCAT3
expression and dermcidin expression. The results revealed that
STCAT3 and dermcidin expression are independent predictors of OS
(P<0.05) and TNM stage (P=0.014) in patients with GC (Table VI).
 | Table VI.Univariate and multivariate analysis
of clinical factors associated with survival in patients with
GC. |
Table VI.
Univariate and multivariate analysis
of clinical factors associated with survival in patients with
GC.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Risk factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age | 0.764 | 0.237–1.599 | 0.709 | 0.517 | 0.227–1.412 | 0.211 |
| Sex | 0.892 | 0.434–2.211 | 0.843 | 1.478 | 0.461–3.473 | 0.474 |
| Tumor size | 1.18 | 0.499–2.865 | 0.714 | 1.167 | 0.414–3.062 | 0.922 |
| Invasion depth | 2.238 | 0.974–4.234 | 0.062 | 0.617 | 0.213–2.612 | 0.439 |
| TNM stage
(I/II/III/IV) | 6.835 | 2.338–16.427 | 0.001 | 5.558 | 1.240–25.513 | 0.014 |
| Differentiation
degree (high/middle/low) | 3.495 | 1.438–8.438 | 0.097 | 3.502 | 1.226–10.316 | 0.129 |
| Lymph node
metastasis (no/yes) | 4.33 | 1.874–12.208 | 0.027 | 3.243 | 1.127–9.247 | 0.145 |
| STCAT3
expression | 13.828 | 3.847–37.180 | 0.001 | 10.61 | 2.079–47.012 | 0.006 |
| Dermcidin
expression | 7.24 | 2.151–24.913 | 0.002 | 6.395 | 1.424–26.969 | 0.019 |
Discussion
Despite advances in surgery, chemotherapy and
targeted molecular therapies, GC remains a leading contributor to
cancer-associated mortality worldwide (1,25).
Developing a better understanding of the molecular mechanisms
underlying the development and progression of GC may allow for
early diagnostic and treatment methods to be developed, improving
the, overall prognosis.
A number of studies have reported that lncRNAs serve
vital roles in a range of human diseases, including GC and other
cancers (26,27), functioning as potential oncogenes or
tumor suppressors (28–31). For example, HOTAIR expression is
upregulated in GC tissues and is an independent predictor of poor
prognosis and HOTAIR knockdown in GC cells suppresses the migration
and invasion of GC cells; this suggests that HOTAIR functions as a
novel oncogene in GC (32,33). LncRNA gastric carcinoma high
expressed transcript 1 overexpression is associated with gastric
tumor growth and invasion, promoting the proliferation of GC cells
by enhancing the interaction between c-Myc mRNA and insulin-like
growth factor 2 binding protein 1 (34). Additionally, GHET1 overexpression
promotes the development of multidrug resistance, which is
associated the expressions B-cell lymphoma 2 (Bcl-2),
Bcl-2-associated X, MDR, and multidrug resistance-associated
protein 1 genes in GC cells (35).
LncRNA urothelial cancer-associated 1 increases the metastatic
ability of GC cells by regulating GRK2 protein stability via
Cbl-c-mediated GRK2 ubiquitination and degradation (36).
The authors of the present study have previously
reported that novel lncRNA STCAT3 was highly expressed in GC
tissues, was able to promote the proliferation, migration and
invasion of GC cells in vitro and promote the growth of
transplanted tumors in vivo (12). The results of the present study
support these findings. It was also demonstrated that STCAT3
expression was significantly upregulated in GC compared with paired
adjacent tissues. STCAT3 upregulation in patients with GC was
positively correlated with advanced TNM stage and lymph node
metastasis. Furthermore, high STCAT3 expression in GC tissues was
associated with poor prognosis. These results suggest that STCAT3
may function as an oncogene in GC.
To further identify the molecular mechanisms and
binding partners of STCAT3, mass spectrometry was performed in
BGC-823 cells followed by an RNA pull-down assay. The results
demonstrated that STCAT3 interacts with dermcidin, which was
further verified by RT-qPCR and western blotting. The results were
also verified using an RIP-qPCR assay and it was demonstrated that
STCAT3 could be enriched in dermcidin precipitates, indicating that
dermcidin is a novel binding protein of lncRNA STCAT3.
A number of studies have reported that dermcidin
serves a role in the carcinogenesis of a number of tumor types,
including lung cancer (16,37), melanoma (17,38),
breast cancer (39–41), hepatocellular carcinoma (HCC)
(42,43), prostatic carcinoma (44) and pancreatic cancer (15). Dermcidin may also affect breast
cancer development through the ERBB signaling pathway (41).
Immunohistochemical results revealed that, compared
with pair-matched normal gastric tissues, dermcidin was
overexpressed in GC. Furthermore, the dermcidin expression was
significantly correlated with clinical TNM stage, tumor
differentiation and lymph node metastasis. Importantly, a positive
correlation was observed between the expression of STCAT3 and
dermcidin in GC tissues. The median time until postoperative
recurrence was increased in the low dermcidin expression group, as
was the OS duration. These results indicate that dermcidin
overexpression is a predictor of poor prognosis in patients with
GC.
The present study is not without limitations.
Although it was determined that dermcidin is the interaction
protein of lncRNA STAT3, the mechanism that leads to the formation
of lncRNA STCAT3 remain to be elucidated. Furthermore, the sample
size used in the present study was small. These limitations should
be addressed in future studies.
In summary, the results of the present study
indicate that dermcidin is a novel binding protein of lncRNA
STCTA3, which serves an important role in cancer progression and
patient clinical outcomes in GC. The results of the present study
provide a theoretical basis for follow-up studies investigating the
regulatory mechanism of lncRNA STCAT3 in the occurrence and
development of GC and its potential as a molecular therapeutic
target.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Youth Foundation
of Affiliated Hospital of Nantong University (grant no. TDFY0332),
the Hospital Talent Training Foundation (grant no. 2015-68), the
Youth Foundation of Nantong Municipal Commission of Health and
Family Planning (grant no. WQ2016079) and Jiangsu Province's Key
Young Medicine Talents Program of Ding (grant no. QNRC2016688),
China Postdoctoral Science Foundation (grant no. 2018M630592).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JZ, WD, ZM and ZW conceived and designed the study.
JZ, WD, XK, YJ and ZZ performed the experiments. JZ and WD wrote
the manuscript. ZM and ZW reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in this study involving
human participants were in accordance with the 1964 Declaration of
Helsinki and its later amendments or comparable ethical standards.
The ethical approval was obtained from the independent ethics
committees at Affiliated Hospital of Nantong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang
Y, Zhou J, Pan K, Sun L, Fang J, et al: A global burden of gastric
cancer: The major impact of China. Expert Rev Gastroenterol
Hepatol. 11:651–661. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nam JW, Choi SW and You BH: Incredible
RNA: Dual functions of coding and noncoding. Mol Cells. 39:367–374.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang JZ, Chen M, Chen, Gao XC, Zhu S,
Huang H, Hu M, Zhu H and Yan GR: A peptide encoded by a putative
lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell.
68(171–184): e62017.
|
|
9
|
Matsumoto A, Pasut A, Matsumoto M,
Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI,
Clohessy JG and Pandolfi PP: mTORC1 and muscle regeneration are
regulated by the LINC00961-encoded SPAR polypeptide. Nature.
541:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roberts TC, Morris KV and Weinberg MS:
Perspectives on the mechanism of transcriptional regulation by long
non-coding RNAs. Epigenetics. 9:13–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang JF, Sun ZS, Zhang QF, Ding WF, Wu XH
and Mao ZB: Expression of long noncoding RNA STCAT3 in gastric
cancer tissues and its effect on malignant phenotype of gastric
cancer cells. Zhonghua Yi Xue Za Zhi. 96:3735–3740. 2016.(In
Chinese). PubMed/NCBI
|
|
13
|
Zhang JF, Qu LS, Qian XF, Xia BL, Mao ZB
and Chen WC: Nuclear transcription factor CDX2 inhibits gastric
cancer-cell growth and reverses epithelial-to-mesenchymal
transition in vitro and in vivo. Mol Med Rep.
12:5231–5238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guan C, Zhang J, Zhang J, Shi H and Ni R:
Enhanced expression of early mitotic inhibitor-1 predicts a poor
prognosis in esophageal squamous cell carcinoma patients. Oncol
Lett. 12:114–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stewart GD, Skipworth RJ, Pennington CJ,
Lowrie AG, Deans DA, Edwards DR, Habib FK, Riddick AC, Fearon KC
and Ross JA: Variation in dermcidin expression in a range of
primary human tumours and in hypoxic/oxidatively stressed human
cell lines. Br J Cancer. 99:126–132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang WC, Huang MS, Yang CJ, Wang WY, Lai
TC, Hsiao M and Chen CH: Dermcidin identification from exhaled air
for lung cancer diagnosis. Eur Respir J. 35:1182–1185. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ortega-Martínez I, Gardeazabal J,
Erramuzpe A, Sanchez-Diez A, Cortés J, García-Vázquez MD,
Pérez-Yarza G, Izu R, Luís Díaz-Ramón J, de la Fuente IM, et al:
Vitronectin and dermcidin serum levels predict the metastatic
progression of AJCC I–II early-stage melanoma. Int J Cancer.
139:1598–1607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yun J, Mullarky E, Lu C, Bosch KN,
Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et
al: Vitamin C selectively kills KRAS and BRAF mutant
colorectal cancer cells by targeting GAPDH. Science. 350:1391–1396.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang CY, Chen CL, Tseng YL, Fang YT, Lin
YS, Su WC, Chen CC, Chang KC, Wang YC and Lin CF: Annexin A2
silencing induces G2 arrest of non-small cell lung
cancer cells through p53-dependent and -independent mechanisms. J
Biol Chem. 287:32512–32524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng X, Liu H, Zhang Z, Gu Y, Qiu H and He
Z: Annexin A2 contributes to cisplatin resistance by activation of
JNK-p53 pathway in non-small cell lung cancer cells. J Exp Clin
Cancer Res. 36:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rogers MS, Foley MA, Crotty TB, Hartmann
LC, Ingle JN, Roche PC and Strehler EE: Loss of immunoreactivity
for human calmodulin-like protein is an early event in breast
cancer development. Neoplasia. 1:220–225. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brooks MD, Bennett RD, Strehler EE, Sebo
TJ, Eckert SE and Carr AB: Human calmodulin-like protein (CLP)
expression in oral squamous mucosa and in malignant transformation.
J Prosthodont. 18:11–16. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang L, Cui M, Zhang L and Song L: FOXM1
facilitates gastric cancer cell migration and invasion by inducing
Cathepsin D. Oncotarget. 8:68180–68190. 2017.PubMed/NCBI
|
|
24
|
Zhu J, Wu G, Li Q, Gong H, Song J, Cao L,
Wu S, Song L and Jiang L: Overexpression of suprabasin is
associated with proliferation and tumorigenicity of esophageal
squamous cell carcinoma. Sci Rep. 6:215492016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmitt AM and Chang HY: Long Noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arab K, Park YJ, Lindroth AM, Schäfer A,
Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et
al: Long noncoding RNA TARID directs demethylation and
activation of the tumor suppressor TCF21 via GADD45A. Mol
Cell. 55:604–614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gooding AJ, Zhang B, Jahanbani FK, Gilmore
HL, Chang JC, Valadkhan S and Schiemann WP: The lncRNA BORG drives
breast cancer metastasis and disease recurrence. Sci Rep.
7:126982017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu X, Huang C, He X, Liu X, Ji J, Zhang E,
Wang W and Guo R: A novel long non-coding RNA, SOX21-AS1, indicates
a poor prognosis and promotes lung adenocarcinoma proliferation.
Cell Physiol Biochem. 42:1857–1869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu X and Li Z: Long non-coding RNA HOTAIR:
A novel oncogene (Review). Mol Med Rep. 12:5611–5618. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen WM, Chen WD, Jiang XM, Jia XF, Wang
HM, Zhang QJ, Shu YQ and Zhao HB: HOX transcript antisense
intergenic RNA represses E-cadherin expression by binding to EZH2
in gastric cancer. World J Gastroenterol. 23:6100–6110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi
K, Gu Y and Fang G: Long non-coding RNA GHET1 promotes gastric
carcinoma cell proliferation by increasing c-Myc mRNA stability.
FEBS J. 281:802–813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Bo P, Liu L, Zhang X and Li J:
Overexpression of long non-coding RNA GHET1 promotes the
development of multidrug resistance in gastric cancer cells. Biomed
Pharmacother. 92:580–585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu
QL, Su LP, Liu BY, Li C and Zhu Z: Long noncoding RNA UCA1 promotes
tumour metastasis by inducing GRK2 degradation in gastric cancer.
Cancer Lett. 408:10–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
López-Sánchez LM, Jurado-Gámez B,
Feu-Collado N, Valverde A, Cañas A, Fernández-Rueda JL, Aranda E
and Rodríguez-Ariza A: Exhaled breath condensate biomarkers for the
early diagnosis of lung cancer using proteomics. Am J Physiol Lung
Cell Mol Physiol. 313:L664–L676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Trzoss L, Fukuda T, Costa-Lotufo LV,
Jimenez P, La Clair JJ and Fenical W: Seriniquinone, a selective
anticancer agent, induces cell death by autophagocytosis, targeting
the cancer-protective protein dermcidin. Proc Natl Acad Sci USA.
111:14687–14692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Moreira DF, Strauss BE, Vannier E and
Belizário JE: Genes up- and down-regulated by dermcidin in breast
cancer: A microarray analysis. Genet Mol Res. 7:925–932. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brauer HA, D'Arcy M, Libby TE, Thompson
HJ, Yasui YY, Hamajima N, Li CI, Troester MA and Lampe PD:
Dermcidin expression is associated with disease progression and
survival among breast cancer patients. Breast Cancer Res Treat.
144:299–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bancovik J, Moreira DF, Carrasco D, Yao J,
Porter D, Moura R, Camargo A, Fontes-Oliveira CC, Malpartida MG,
Carambula S, et al: Dermcidin exerts its oncogenic effects in
breast cancer via modulation of ERBB signaling. BMC Cancer.
15:702015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lowrie AG, Dickinson P, Wheelhouse N,
Stewart GD, Ross AJ, Forster T and Ross JA: Proteolysis-inducing
factor core peptide mediates dermcidin-induced proliferation of
hepatic cells through multiple signalling networks. Int J Oncol.
39:709–718. 2011.PubMed/NCBI
|
|
43
|
Shen SL, Qiu FH, Dayarathna TK, Wu J,
Kuang M, Li SS, Peng BG and Nie J: Identification of Dermcidin as a
novel binding protein of Nck1 and characterization of its role in
promoting cell migration. Biochim Biophys Acta. 1812:703–710. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stewart GD, Lowrie AG, Riddick AC, Fearon
KC, Habib FK and Ross JA: Dermcidin expression confers a survival
advantage in prostate cancer cells subjected to oxidative stress or
hypoxia. Prostate. 67:1308–1317. 2007. View Article : Google Scholar : PubMed/NCBI
|