Introduction
Cholangiocarcinoma (CCA) is an epithelial cell
malignancy originating from cholangiocytes (1,2). CCA,
together with liver cancer, is reported to be the fifth leading
cause of cancer-associated mortality in 2018 in the USA (3). Currently, surgical resection remains
the mainstay of curative treatment, although it is only available
for selected patients (4). Poor
prognosis, even with ideal surgery, is the key problem in CCA. The
5-year overall survival (OS) rate of CCA is 15–40%, and the
post-surgical recurrence rate is 50–60% (5–8).
A number of other therapies have been used in CCA.
Local advanced and metastatic CCA is treated with systemic
chemotherapy; cisplatin and gemcitabine are the common drugs, and
the drug yttrium 90 is used in local radiotherapy (9). Currently, there are few reports on
drug treatments for CCA. Studies suggest that 14-deoxy-11,
12-didehydroandrographolide and curcumin may be the potential drugs
for the treatment of CCA (10,11).
Even with the large number of tested drugs, the results remain
unsatisfactory. The principal causes may be the lack of enrollment
in trials and the limited number of biological profiles of CCA
(12). Hence, novel and effective
drugs are urgently required. Connectivity Map (CMap) is a database
containing 7,000 gene expression data profiles treated by ~1,300
compounds (13). In the field of
drug development, it is helpful to make rapid use of gene
expression profiles associated with diseases. This approach may
identify the principal chemical structure of the majority of drug
molecules and elucidate the possible mechanism of drug action.
To examine novel therapeutics for CCA, the present
study screened the survival-associated genes of CCA, which were
identified based on gene expression profiles provided by The Cancer
Genome Atlas (TCGA) database. Simultaneously, pathways associated
with poor survival in CCA were ascertained through gene-set
functional enrichment analysis. Using the associations already
obtained, the present study put forward a model of the network
aimed at these prognostic-related targets. Finally, through CMap, a
number of molecules associated with the prognosis of CCA were
screened out. In brief, the aim of the present study was to examine
the prognostic landscape and potential therapeutic targets of
CCA.
Materials and methods
Processing of genomic data from TCGA
dataset
RNA sequencing data and clinical information from 45
patients with CCA were download from TCGA portal (https://cancergenome.nih.gov/). The mRNA expression
profiles were extracted, and the data were normalized in the form
of transcripts per kilobase million (TPM). To obtain a more
moderate result, mRNAs with an average TPM <1 were removed.
Survival analysis and functional
annotation
The association between OS and the gene expression
profiles was evaluated by univariate Cox regression analysis, which
was conducted using the survival package in R software version 3.44
(https://CRAN.R-project.org/package=survival).
Subsequently, the prognostic genes were submitted to DAVID
Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/) for functional enrichment
and pathway analyses, which included the Kyoto Encyclopedia of
Genes and Genomes (KEGG) and gene ontology (GO). The functional
enrichment results were displayed using the ggplot2 package in R
software (https://CRAN.R-project.org/package=ggplot2), and
Cytoscape software version 3.61 (http://www.cytoscape.org/). Furthermore, the
protein-protein interactions (PPIs) among these prognostic genes
were generated. Interaction data were downloaded from the STRING
online database (http://string-db.org/), and further visualization was
performed using Cytoscape software.
Gene-drug interaction network
To study the potential survival-associated gene
therapeutic targets based on the available resources, the present
study used the online Drug-Gene Interaction database (DGIdb), which
stores information about druggability and interactions between
drugs and genes from the published literature, databases and web
resources (14). Based on this
information, the drug-gene interaction network was built to
identify druggable targets.
CMap analysis
CMap provides perspectives on small-molecule drugs,
gene expression, and diseases that are associated with each other
at the genome level. Therefore, it allows researchers to link gene
datasets with drugs that are closely associated with the disease
process. The secondary use of drugs in another disease greatly
reduces the time and expense required for drug development. To find
novel drugs to alter the poor OS of patients with CCA, the
survival-associated genes were uploaded to CMap, compared and
ranked via the Kolmogorov-Smirnov test. A drug with a positive
score indicates that there is a similar effect or mechanism of poor
survival for the imported drug. Small molecules with negative
scores were selected for further analysis, as this indicated that
these molecules may work in the improvement of poor survival in
patients with CCA.
Potential targets of
cholangiocarcinoma
Potential therapeutic targets for improving the
survival of patients were also identified. Hub genes of the PPI
network were selected when the number of connections was >5.
Genes that were significant in the development of drugs and
possessed druggability were also identified in the drug-gene
network. Genes obtained by the above two methods simultaneously
were considered to be potential targets for CCA.
Molecular docking analysis
To verify the targeting association between the
molecules and targets, the Sybyl X-2.0 Surflex-Dock program
(Certara, Princeton, NJ, USA) was used to calculate the molecular
docking affinity index. The program obtained detailed information
about the combinations of novel drugs and targets, and the docking
scores were calculated to represent the binding affinities.
Results
Prognostic evaluation of mRNAs in
cholangiocarcinoma
Following omission of the genes with low abundance,
14,458 genes were obtained for prognostic evaluation. The
univariate Cox regression method identified a total of 172 genes
that were significantly associated with the OS of patients with
CCA. Among these genes, 94 genes had a tumor suppressor role, and
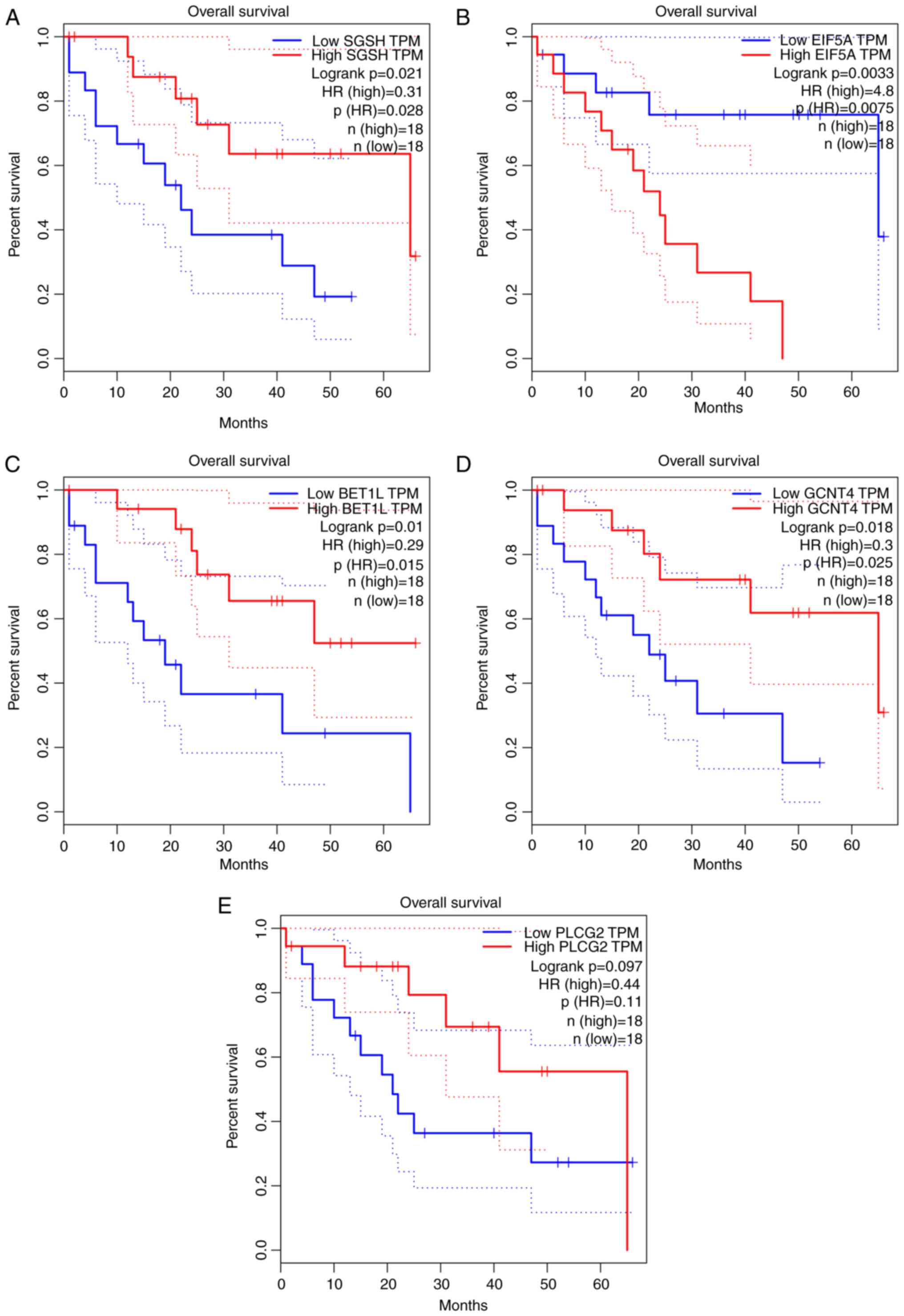
78 were oncogenic genes. The top five most significant
survival-associated genes are displayed in Fig. 1.
Biological processes of
survival-associated genes
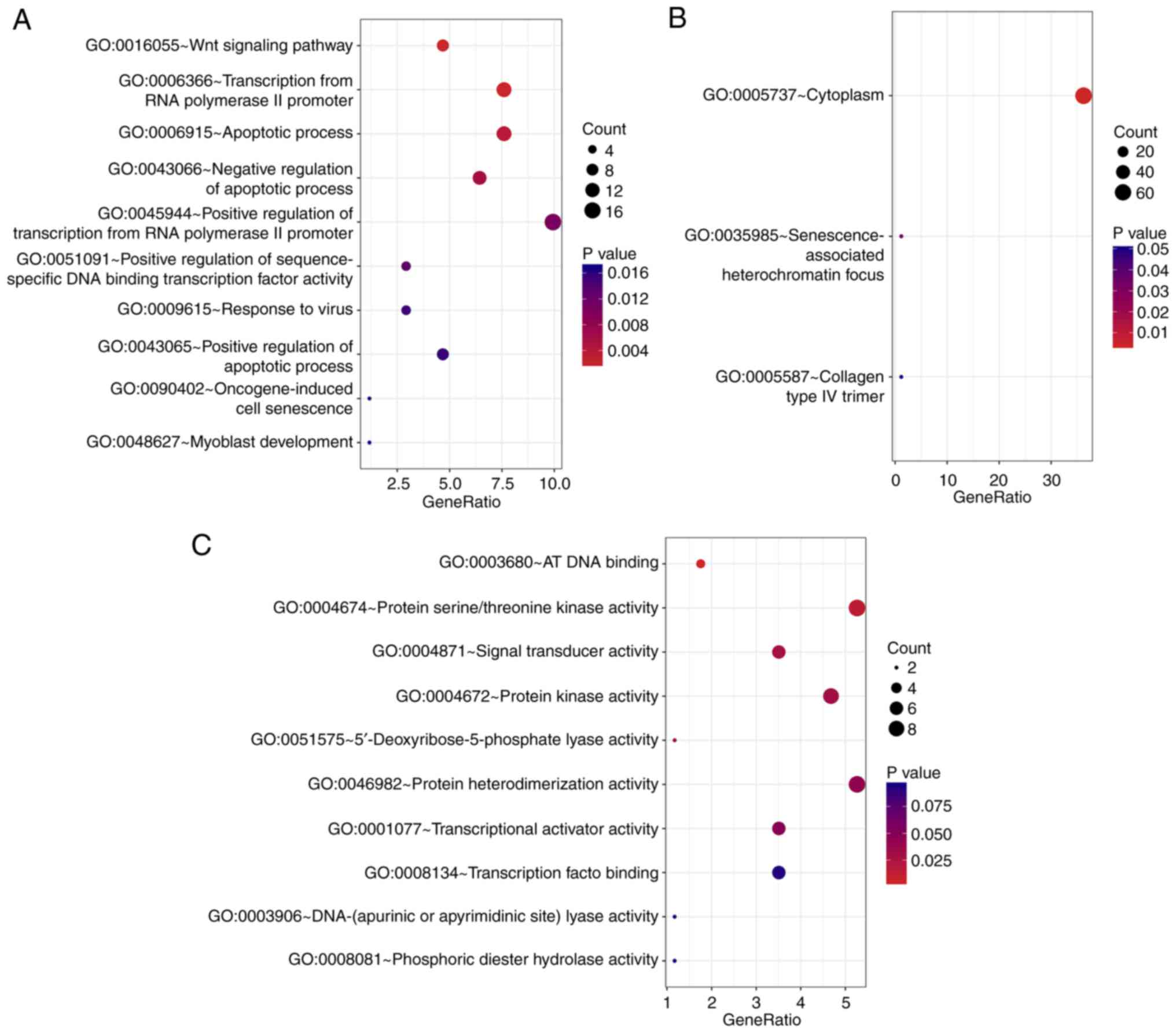
In the biological processes-associated subgroup of
GO terms, the survival-associated genes were significantly enriched
in ‘Wnt signaling pathway’, ‘apoptotic process’ and ‘transcription
from RNA polymerase I promoter’ (Fig.
2A). In addition, the cell component analysis revealed that
these genes were most enriched in ‘cytoplasm’,
‘senescence-associated heterochromatin focus’ and ‘collagen type IV
trimer’ (Fig. 2B). ‘AT DNA
binding’, ‘signal transducer activity’ and ‘protein
serine/threonine kinase activity’ were the three most significant
categories for molecular function (Fig.
2C). Furthermore, the KEGG pathway analysis identified a number
of oncogenic pathways, including ‘transcriptional misregulation in
cancer’, which may be the altered pathways in patients with poorer
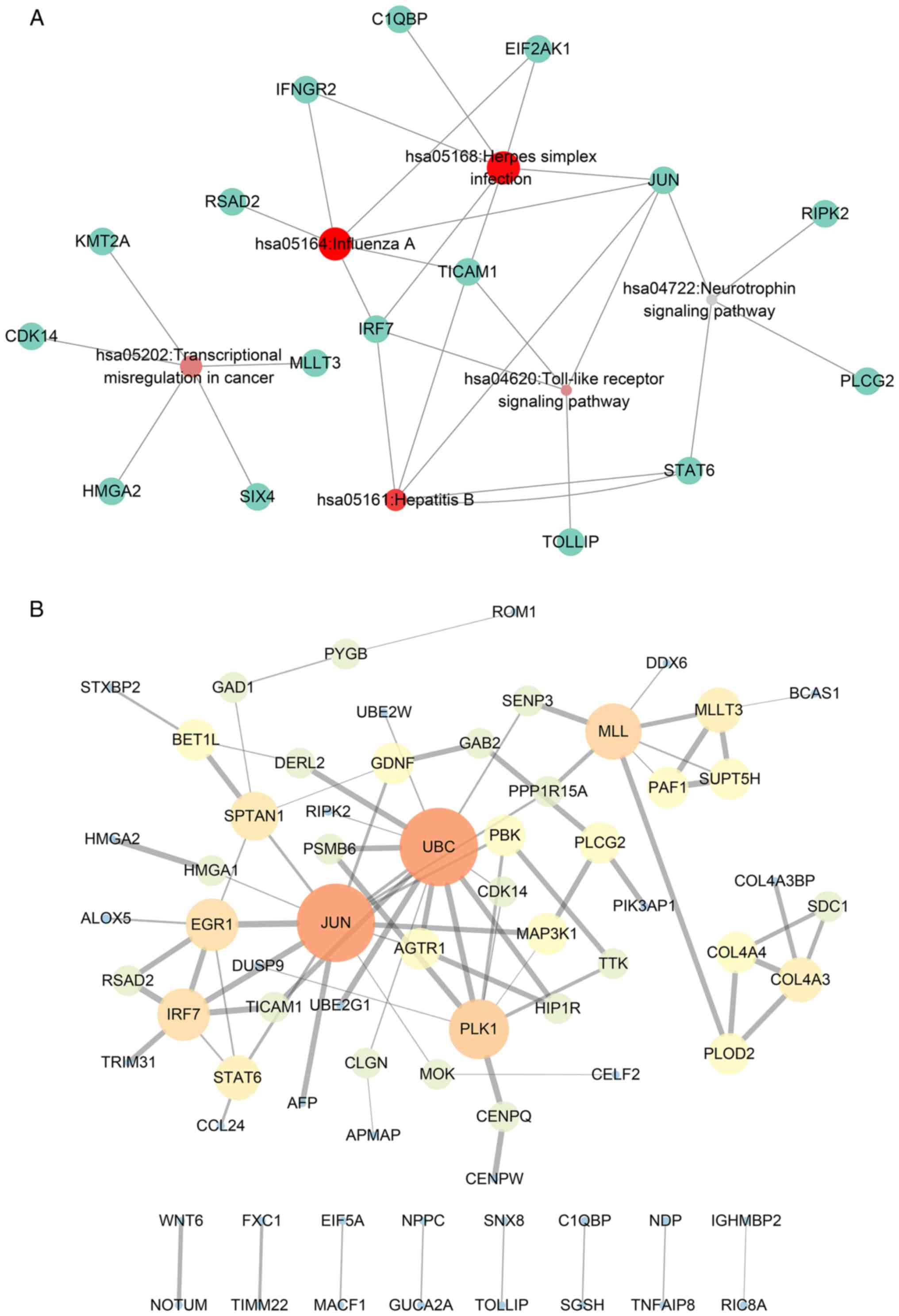
survival (Fig. 3A). The PPI network
revealed that ubiquitin C (UBC), Jun proto-oncogene, AP-1
transcription factor subunit (JUN) and polo-like kinase 1 (PLK1)
ranked as core genes within the group of genes (Fig. 3B), which indicated that these genes
may be the most important in the network.
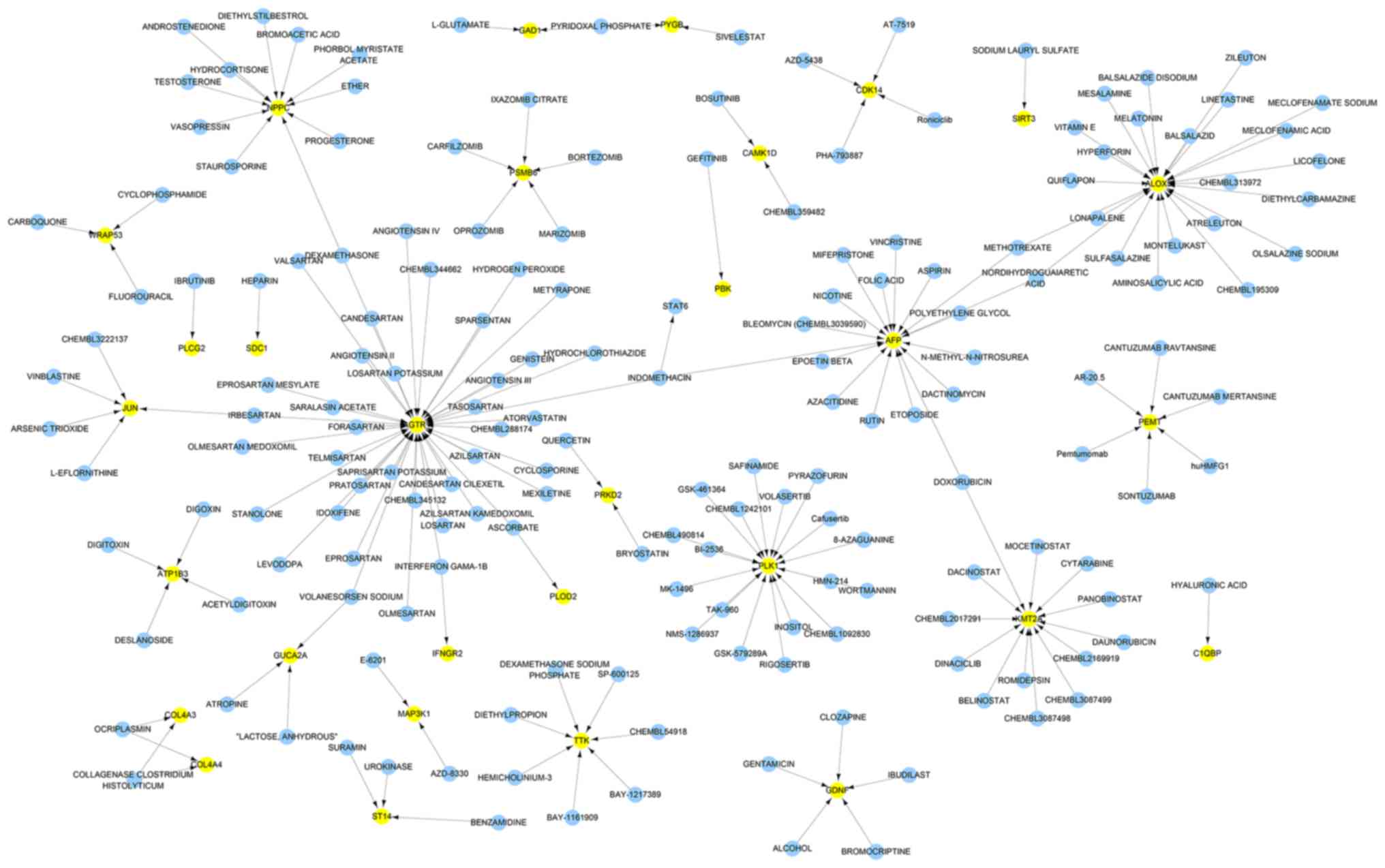
Drug-gene network analysis
Based on the interactions between the drugs and the
effects of unknown molecule drugs, the drug-gene interaction
network was constructed. A total of 31 genes identified to be
putative molecular targets for anti-tumor therapy were involved in
the drug-gene network (Fig. 4).
These drugs may have the potential to be effective anti-CCA drugs
in the future.
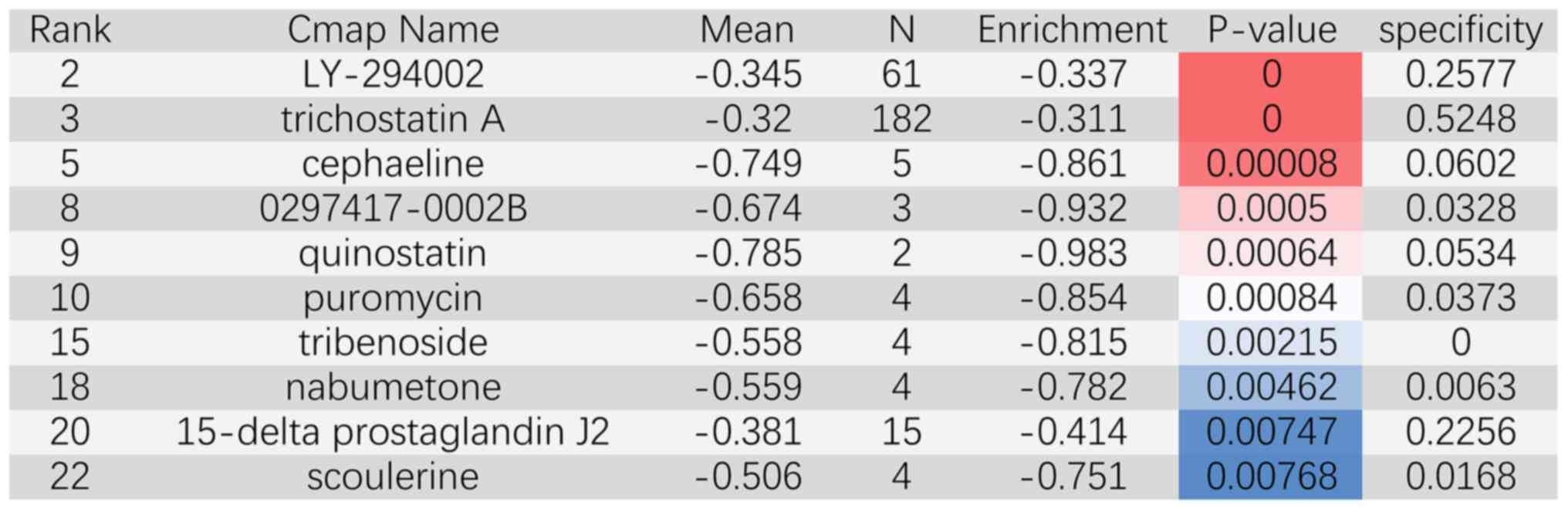
Candidate drugs for CCA therapy
From the small molecules with a negative score that
had potential against CCA, a total of 49 small molecule drugs were
collected by using the CMAP database. The top ten significant novel
drugs were LY-294002, trichostatin A, cephaeline, 0297417-0002B,
quinostatin, puromycin, tribenoside, nabumetone, 15-δ prostaglandin
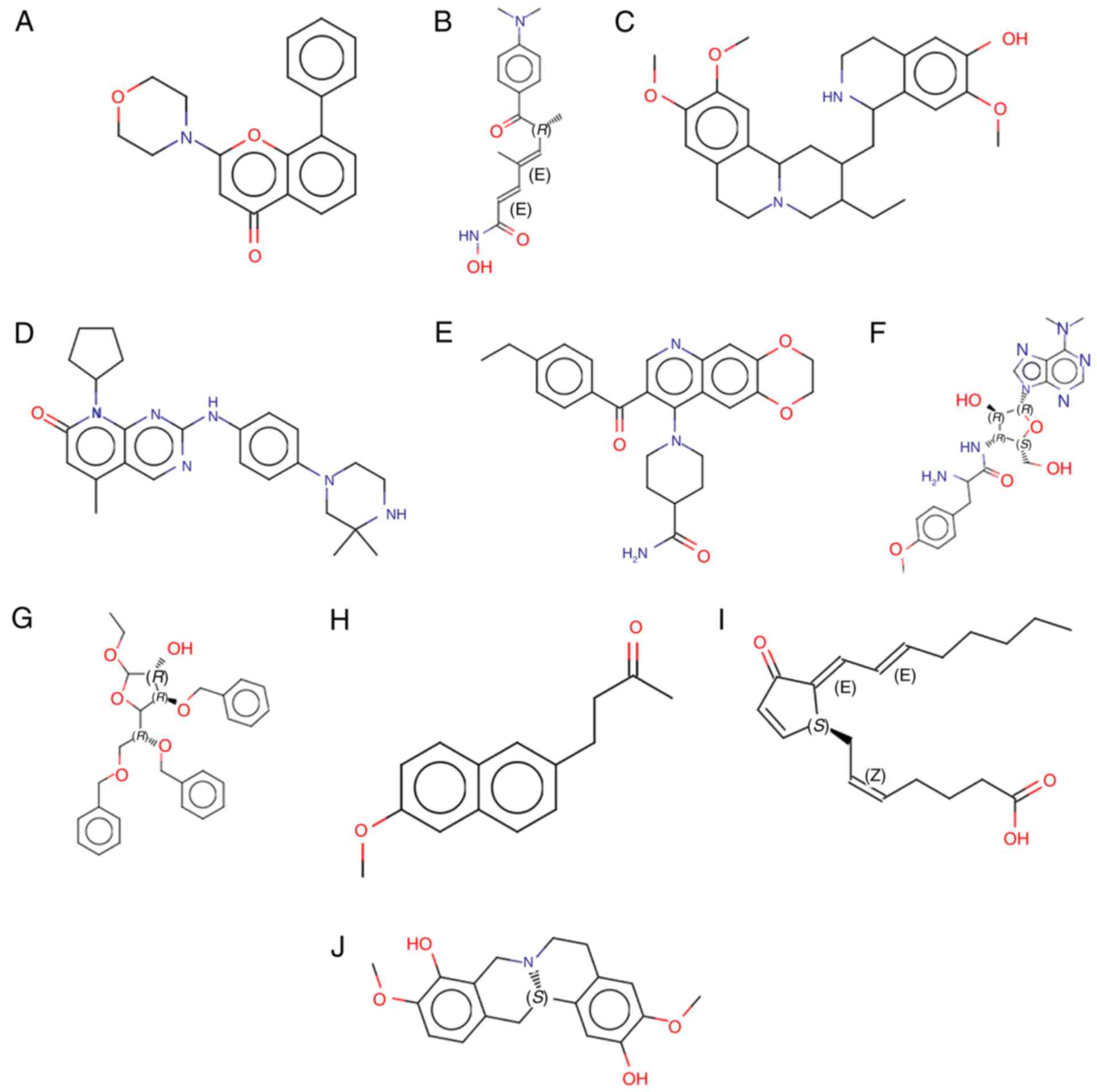
J2 and scoulerine (Fig. 5). The
chemical structures of these drugs are displayed in Fig. 6.
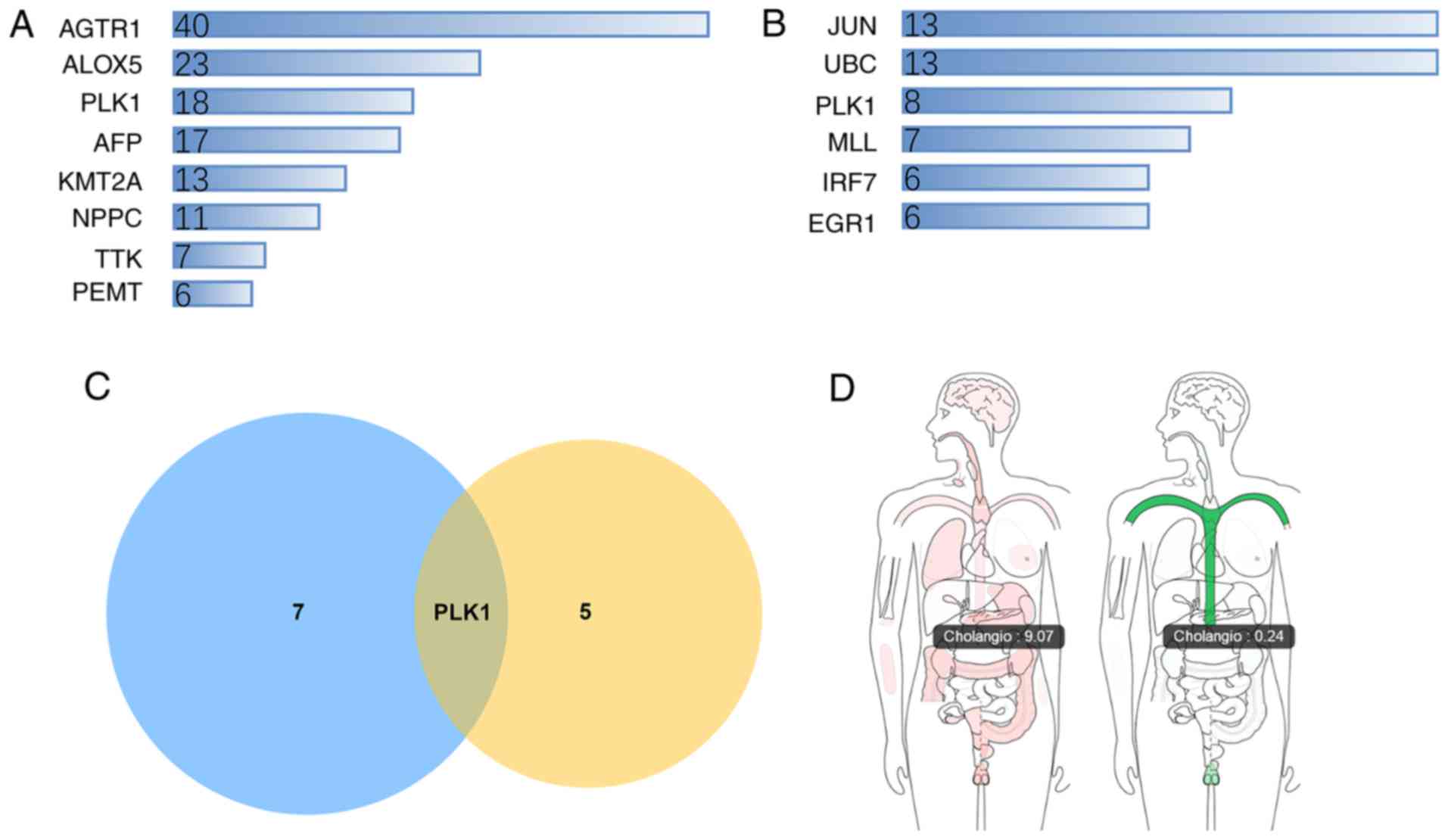
Potential target selection
To select the most significant targets among the
group of survival-associated genes, the PPI network was analyzed,
and it was demonstrated that angiotensin II receptor type 1,
arachidonate 5-lipoxygenase, PLK1, α-fetoprotein, lysine
methyltransferase 2A, natriuretic peptide C, TTK protein kinase and
phosphatidylethanolamine N-methyltransferase were the hub genes
(Fig. 7A). The drug-gene network
analysis demonstrated that JUN, UBC, PLK1, KMT2A, interferon
regulatory factor 7 and early growth response 1 had potential as
drug targets (Fig. 7B). Molecules
targeting these genes may have improved druggability. Notably, PLK1
was simultaneously ranked within the core genes of the two networks
(Fig. 7C). PLK1 was significantly
upregulated in CCA tumor samples compared with non-tumor samples
(Fig. 7D).
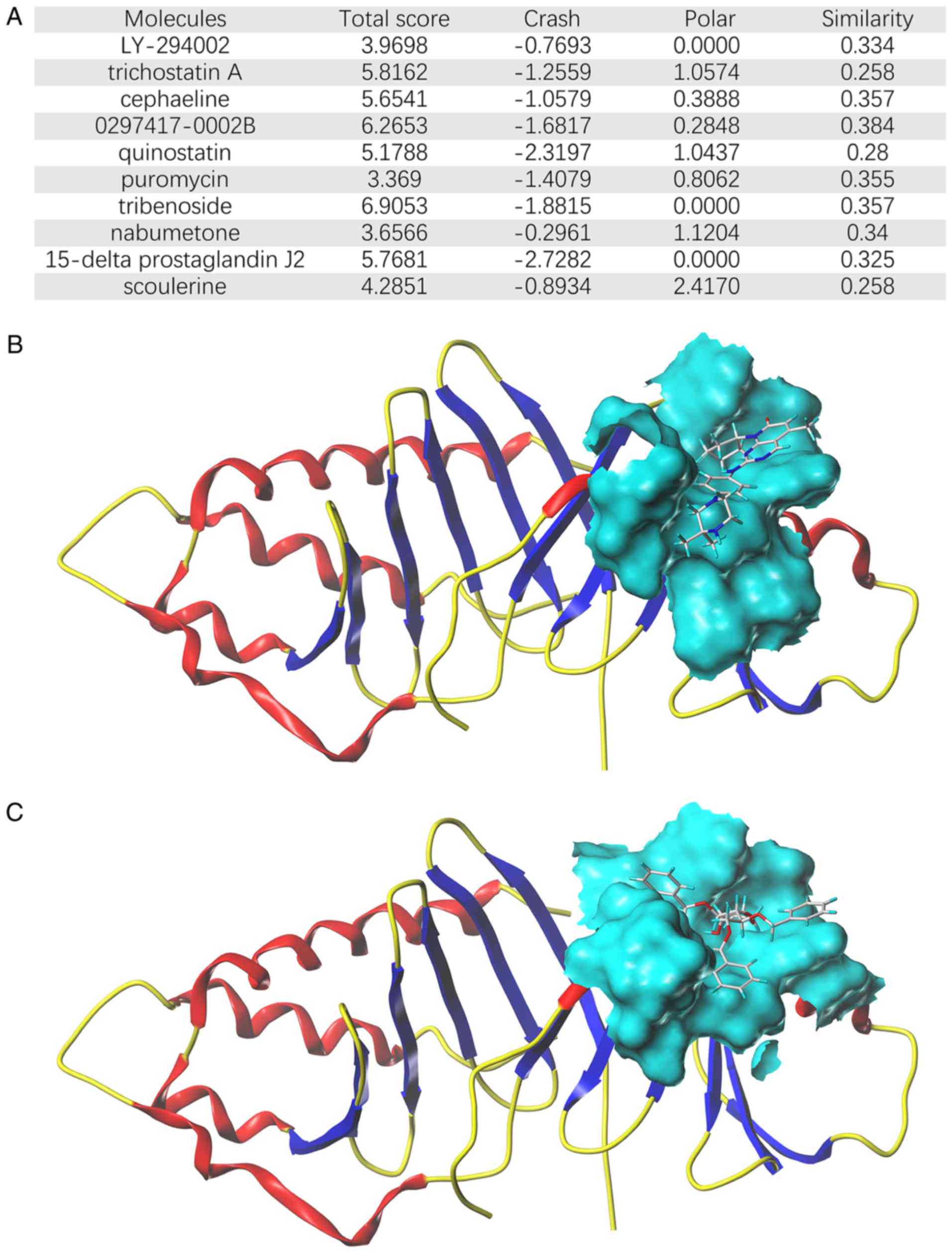
Correlation between PLK1 and compounds
via molecular docking
To examine whether the potential anti-CCA effects of
these molecules were due to direct targeting between the compounds
and genes, molecular docking analysis was performed. PLK1, one of
the hub survival-associated genes, was selected. Notably, the top
ten molecules (Fig. 8A) all exerted
moderate binding capacity with PLK1. 0297417-0002B and tribenoside
exhibited the highest connection scores (Fig. 8B and C). These findings may provide
novel anti-CCA insights for the future.
Discussion
The present study indicated that certain genes may
be identified as candidate molecular targets that may serve an
essential role in the prognosis and treatment of CCA. The GO
enrichment and KEGG pathway analysis results indicated that these
target genes were closely associated with the relevant signaling
pathways and processes of tumorigenesis, including ‘transcription
from RNA polymerase I promoter’ and ‘Wnt signaling pathway’.
Networks associated with the prognosis of these target genes were
constructed, demonstrating that the genes were potential targets of
a number of drugs. Furthermore, the CMap search identified certain
novel compounds with potential therapeutic value for CCA.
CCA is a bile duct malignancy, which is difficult to
accurately diagnose and has a poor prognosis (15). Patients diagnosed with CCA are
likely to progress into the advanced stages. In the absence of
efficient therapeutic methods in the clinic, the morbidity and
mortality rates of CCA have been increasing gradually in recent
years (1). Therefore, the present
study comprehensively analyzed CCA survival-associated genes, and
key findings emerged regarding target genes [N-sulfoglucosamine
sulfohydrolase (SGSH), eukaryotic translation initiation factor 5A
(EIF5A), Bet1 golgi vesicular membrane trafficking protein-like
(BET1L), glucosaminyl (N-acetyl) transferase 4, core 2 (GCNT4) and
phospholipase C γ2 (PLCG2)] that may be associated with the
prognosis of CCA. SGSH is one of a number of enzymes that are
associated with the lysosomal degradation of heparan sulfate
(16). EIF5A is associated with
protein synthesis and cell proliferation. High eIF5A expression may
be observed in various types of tumors and is responsible for
cancer metastasis (17). Altered
glycosylation is considered to be one of the most common cancer
biomarkers (18). PLCG2 is able to
provide survival signals for cells, inhibit cellular apoptosis, and
indirectly activate protein kinase C, which is frequently highly
expressed in cancer tissues (19).
GO classification and KEGG pathway enrichment
analysis was used in the present investigation to identify
survival-associated genes. Certain notable GO biological processes
were identified, including ‘Wnt signaling pathway’ and
‘transcription from RNA polymerase II promoter’. The Wnt signaling
pathway is essential for the survival of stem cells and the
maintenance of the regenerative ability of tissues (20). The basic role of the Wnt/β-catenin
pathway in development requires regulatory control at multiple
levels, and a deficiency at any level may lead to tumor formation
(21). Previous studies have
demonstrated that the proliferation, invasion and
epithelial-mesenchymal transition of CCA cells are associated with
Wnt/β-catenin signaling (22,23).
Wnt/β-catenin signaling is progressively activated during
cholangiocarcinogenesis, which indicates that it may serve an
essential role in the occurrence and prognosis of CCA.
Transcription via RNA polymerase I is a highly dynamic process that
is tightly regulated at each step of the transcription cycle
(24). RNA polymerase I speeds up
the cell cycle by promoting the transcription of genes, thereby
promoting cell growth and tumor formation.
KEGG pathway analysis demonstrated that the
survival-associated genes were principally involved in the
influenza A, herpes simplex infection and hepatitis B signaling
pathways. When the host is infected with a virus, p53 performs an
inhibitor role in tumorigenesis by promoting the host immune
response and protecting the host from cancer (25). The excessive activation of Ras
signals also increases the degree of clearance of tumor cells
following infection with herpes simplex virus (e.g. R3616)
(26). Bioinformatics analysis was
also performed via PPI network analyses. Based on node
connectivity, genes were classified as hub genes in CCA. UBC, JUN,
and PLK1 were the three hub genes that were essential nodes in the
gene co-expression network.
UBC is also known as HMG20 and code for the
ubiquitin gene. Ubiquitination has a crucial role in protein
degradation, DNA repair, cell cycle regulation, kinase
modification, endocytosis and the regulation of other cell
signaling pathways (27). PLK1 is
also termed STPK13 and encodes a serine/threonine protein kinase
that is part of the CDC5/Polo subfamily. Studies have demonstrated
that PLK1 is overexpressed in various types of tumors, and it is
during the period of mitosis that PLK1 is observed to be highly
expressed (28–30). With the decrease in this protein in
carcinoma cells, PLK1 markedly suppresses cell proliferation and
triggers cellular apoptosis; thus, PLK1 is considered to be a
target for the treatment of carcinoma (31). JUN is regarded as the transforming
gene associated with avian sarcoma virus 17. The protein encoded by
JUN has a high similarity with the viral protein, and the direct
interaction between the protein encoded by JUN and the particular
target DNA sequences may regulate gene expression. In addition to
intron-less genes, JUN is mapped to 1p32-p31, a chromosomal region
involved in translocations and deletions in human malignancies
(32). The antitumor medicine
volasertib is an effective selective PLK1 suppressor, which may
cause selective G2/M blockade and apoptosis in various types of
cancer cells (33,34). Simultaneously, it induces reversible
cell arrest at the G1 and G2 phase while protecting healthy cells
from apoptosis. Volasertib has been used in trials studying the
treatment of acute myeloid leukemia, neoplasms and myelodysplastic
syndrome and may also serve a role in the treatment of CCA
(35–37).
Through CMap analysis, 10 drugs (LY-294002,
trichostatin A, Cephaeline, 0297417-0002B, quinostatin, puromycin,
tribenoside, nabumetone, 15-δ prostaglandin J2, and scoulerine)
were identified that may have potential novel efficacy against the
poor prognosis of CCA. Trichostatin A induces hyperacetylation of
the core histone followed by structural modulation of chromatin,
and finally causes the promotion of selective gene transcription
and the suppression of tumor growth by reversibly inhibiting
deacetylases. Currently, trichostatin A has been used in the
treatment of laryngeal cancer and as a novel biomarker for
nasopharyngeal carcinoma, and is also used to prevent the side
effects of cisplatin in the treatment of cancer (38). It may have positive effects on CCA
treatment. LY-294002 is a specific inhibitor of
phosphatidylinositol 3-kinase (PI3K), which serves a role in the
treatment of cancer by affecting the PI3K/RAC-α
serine/threonine-protein kinase signaling pathway (39). Tribenoside and nabumetone are
anti-inflammatory drugs, which may be used to inhibit the synthesis
of prostaglandins and thrombin precursors to anti-inflammatory,
antipyretic and analgesic effect. Among them, nabumetone has been
used to treat acute and chronic osteoarthritis and rheumatoid
arthritis. Tribenoside has been used in the clinical treatment of
hemorrhoidal disease associated with inflammation, coagulation,
varicose veins and wounds (40,41).
Puromycin is a conversion inhibitor in biosynthesis and inhibits
protein synthesis, and has anti-tumor, anti-parasitic and
antibiotic effects (42).
Cephaeline and scoulerine are associated with alkaloid synthesis
(43). L-scoulerine antagonizes the
dopamine D2 receptor to inhibit decreases in prepulse inhibition,
which is caused by MK-801. It may be a potential cure for
schizophrenia (44). 15-δ
prostaglandin J2, 0297417-0002B and quinostatin have no clinical
applications at present. 15-δ prostaglandin J2 is a nuclear
factor-κb inhibitor (45). The
above drugs may affect CCA in a variety of small molecular
pathways.
It was further proposed that PLK1 may be a potential
therapeutic target at the genome level. Previously, Thrum et
al (46) reported that the PLK1
inhibitor BI 2536 may be active against CCA cells in vitro.
The present study further demonstrated that PLK1 was significantly
upregulated in CCA tissues and its overexpression was indicative of
poor survival. Furthermore, novel drug selection based on
prognosis-associated genes may provide a comprehensive insight into
anti-CCA therapy. The molecules identified in the present study all
displayed high binding affinity with PLK1.
However, there remain a number of limitations to the
present study. First, various novel potential drugs for CCA were
identified based on alterations in the genome expression landscape
of patients with CCA; however, examination of the functional
characterization and underlying molecular mechanisms is required in
the future. Second, despite the fact that the molecular docking
analysis provided binding forces between molecules and targets, the
complex mechanisms and models of the specific interactions ought to
be confirmed by future experiments.
In conclusion, the present study first screened the
differentially expressed genes involved in the prognosis of CCA.
Second, GO and KEGG gene enrichment analysis was used to determine
the pathway of prognosis in CCA. A network targeted at prognostic
targets was built. Finally, certain molecules were screened using
the CMap database for the prognosis of CCA as a whole. These
molecules may offer novel insights into the prognosis of CCA and
may offer perspectives on the future innovative treatment of
CCA.
Acknowledgements
The authors would like to thank the CMap, DGIdb and
TCGA databases for the availability of the data.
Funding
The present study was supported by grants from the
Guangxi Science and Technology Program (grant no. GuiKeAB17195020),
the Fund of National Natural Science Foundation of China (grant
nos. NSFC81060202, NSFC81860319 and NSFC81260222) and Innovation
Project of Guangxi Graduate Education (grant nos. YCSW2018104 and
YCSW2017105).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The study was designed by GC, HY, YunH and PL. PL,
XZZ, XDW, JJL, RQZ, YuH, YQJ and XWH were involved in the
statistical analysis. PL and HY wrote the draft and GC, HY and YunH
corrected the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kaneko R, Sato Y and Kobayashi Y:
Cholangiocarcinoma prognosis varies over time depending on tumor
site and pathology. J Gastrointestin Liver Dis. 27:59–66.
2018.PubMed/NCBI
|
|
2
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ikeno Y, Seo S, Iwaisako K, Yoh T,
Nakamoto Y, Fuji H, Taura K, Okajima H, Kaido T, Sakaguchi S and
Uemoto S: Preoperative metabolic tumor volume of intrahepatic
cholangiocarcinoma measured by 18F-FDG-PET is associated
with the KRAS mutation status and prognosis. J Transl Med.
16:952018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
DeOliveira ML, Cunningham SC, Cameron JL,
Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ and Schulick
RD: Cholangiocarcinoma: Thirty-one-year experience with 564
patients at a single institution. Ann Surg. 245:755–762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Li J, Xia Y, Gong R, Wang K, Yan
Z, Wan X, Liu G, Wu D, Shi L, et al: Prognostic nomogram for
intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin
Oncol. 31:1188–1195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Endo I, Gonen M, Yopp AC, Dalal KM, Zhou
Q, Klimstra D, D'Angelica M, DeMatteo RP, Fong Y, Schwartz L, et
al: Intrahepatic cholangiocarcinoma: Rising frequency, improved
survival, and determinants of outcome after resection. Ann Surg.
248:84–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alvaro D, Crocetti E, Ferretti S, Bragazzi
MC and Capocaccia R: AISF Cholangiocarcinoma committee: Descriptive
epidemiology of cholangiocarcinoma in Italy. Dig Liver Dis.
42:490–495. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sombut S, Bunthawong R, Sirion U, Kasemsuk
T, Piyachaturawat P, Suksen K, Suksamrarn A and Saeeng R: Synthesis
of 14-deoxy-11,12-didehydroandrographolide analogues as potential
cytotoxic agents for cholangiocarcinoma. Bioorg Med Chem Lett.
27:5139–5143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu RW, Carey EJ, Lindor KD and Tabibian
JH: Curcumin in hepatobiliary disease: Pharmacotherapeutic
properties and emerging potential clinical applications. Ann
Hepatol. 16:835–841. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tampellini M, La Salvia A and Scagliotti
GV: Novel investigational therapies for treating biliary tract
carcinoma. Expert Opin Investig Drugs. 25:1423–1436. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lamb J: The Connectivity Map: A new tool
for biomedical research. Nat Rev Cancer. 7:54–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wagner AH, Coffman AC, Ainscough BJ, Spies
NC, Skidmore ZL, Campbell KM, Krysiak K, Pan D, McMichael JF,
Eldred JM, et al: DGIdb 2.0: Mining clinically relevant drug-gene
interactions. Nucleic Acids Res. 44:D1036–D1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sangsin A, Saiudom D, Pongmanee S,
Saengsin J, Leerapun T and Murakami H: Natural history and
prognostic factors of cholangiocarcinoma with spinal metastasis: A
10-year single center study. Clin Spine Surg. 31:E160–E165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ouesleti S, Coutinho MF, Ribeiro I, Miled
A, Mosbahi DS and Alves S: Update of the spectrum of
mucopolysaccharidoses type III in Tunisia: Identification of three
novel mutations and in silico structural analysis of the missense
mutations. World J Pediatr. 13:374–380. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pelechano V and Alepuz P: eIF5A
facilitates translation termination globally and promotes the
elongation of many non polyproline-specific tripeptide sequences.
Nucleic Acids Res. 45:7326–7338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jones D, Woyach JA, Zhao W, Caruthers S,
Tu H, Coleman J, Byrd JC, Johnson AJ and Lozanski G: PLCG2 C2
domain mutations co-occur with BTK and PLCG2 resistance mutations
in chronic lymphocytic leukemia undergoing ibrutinib treatment.
Leukemia. 31:1645–1647. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ely KA, Bischoff LA and Weiss VL: Wnt
signaling in thyroid homeostasis and carcinogenesis. Genes. 9:pii:
E2042018. View Article : Google Scholar
|
|
21
|
Jiang J, Protopopov A, Sun R, Lyle S and
Russell M: Genomic profiling on an unselected solid tumor
population reveals a highly mutated Wnt/β-catenin pathway
associated with oncogenic EGFR mutations. J Pers Med. 8:pii:
E132018. View Article : Google Scholar
|
|
22
|
Mao X, Duan X and Jiang B: Fascin induces
epithelial-mesenchymal transition of cholangiocarcinoma cells by
regulating wnt/β-catenin signaling. Med Sci Monit. 22:3479–3485.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noll AT, Cramer T, Damink Olde SW and
Schaap FG: Cholangiocarcinoma, gone without the Wnt? World J
Hepatol. 8:1093–1096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Steurer B, Janssens RC, Geverts B, Geijer
ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen
WA, et al: Live-cell analysis of endogenous GFP-RPB1 uncovers rapid
turnover of initiating and promoter-paused RNA Polymerase I. Proc
Natl Acad Sci USA. 155:E4368–E4376. 2018. View Article : Google Scholar
|
|
25
|
Yan W, Wei J, Deng X, Shi Z, Zhu Z, Shao
D, Li B, Wang S, Tong G and Ma Z: Transcriptional analysis of
immune-related gene expression in p53-deficient mice with increased
susceptibility to influenza A virus infection. BMC Med Genomics.
8:522015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Esfandyari T, Tefferi A, Szmidt A, Lain T,
Zwolak P, Lasho T, Lee PW and Farassati F: Transcription factors
down-stream of Ras as molecular indicators for targeting
malignancies with oncolytic herpes virus. Mol Oncol. 3:464–468.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Munari F, Bortot A, Zanzoni S, D'Onofrio
M, Fushman D and Assfalg M: Identification of primary and secondary
UBA footprints on the surface of ubiquitin in cell-mimicking
crowded solution. FEBS Lett. 591:979–990. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin P, Wen DY, Dang YW, He Y, Yang H and
Chen G: Comprehensive and integrative analysis reveals the
diagnostic, clinicopathological and prognostic significance of
polo-like kinase 1 in hepatocellular carcinoma. Cell Physiol
Biochem. 47:925–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Wang H, Sun Z, Guo Q, Shi H and Jia
Y: The clinical and prognostic value of polo-like kinase 1 in lung
squamous cell carcinoma patients: Immunohistochemical analysis.
Biosci Rep. Jul 19;pii: BSR20170852. 2017.(Epub ahead of print).
View Article : Google Scholar
|
|
30
|
Lin P, Xiong DD, Dang YW, Yang H, He Y,
Wen DY, Qin XG and Chen G: The anticipating value of PLK1 for
diagnosis, progress and prognosis and its prospective mechanism in
gastric cancer: A comprehensive investigation based on
high-throughput data and immunohistochemical validation.
Oncotarget. 8:92497–92521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gheghiani L, Loew D, Lombard B, Mansfeld J
and Gavet O: PLK1 activation in late G2 sets up commitment to
mitosis. Cell Rep. 19:2060–2073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin S, Hoffmann K, Gao C, Petrulionis M,
Herr I and Schemmer P: Melatonin promotes sorafenib-induced
apoptosis through synergistic activation of JNK/c-jun pathway in
human hepatocellular carcinoma. J Pineal Res. 62:2017. View Article : Google Scholar
|
|
33
|
Dong J, Park SY, Nguyen N, Ezhilarasan R,
Martinez-Ledesma E, Wu S, Henry V, Piao Y, Tiao N, Brunell D, et
al: The polo-like kinase 1 inhibitor volasertib synergistically
increases radiation efficacy in glioma stem cells. Oncotarget.
9:10497–10509. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng DW, Xue YQ, Li Y, Di JM, Qiu JG,
Zhang WJ, Jiang QW, Yang Y, Chen Y, Wei MN, et al: Volasertib
suppresses the growth of human hepatocellular carcinoma in vitro
and in vivo. Am J Cancer Res. 6:2476–2488. 2016.PubMed/NCBI
|
|
35
|
Gopalakrishnan B, Cheney C, Mani R, Mo X,
Bucci D, Walker A, Klisovic R, Bhatnagar B, Walsh K, Rueter B, et
al: Polo-like kinase inhibitor volasertib marginally enhances the
efficacy of the novel Fc-engineered anti-CD33 antibody BI 836858 in
acute myeloid leukemia. Oncotarget. 9:9706–9713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X: Targeting Polo-like kinases: A
promising therapeutic approach for cancer treatment. Transl Oncol.
8:185–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Van den Bossche J, Lardon F,
Deschoolmeester V, De Pauw I, Vermorken JB, Specenier P, Pauwels P,
Peeters M and Wouters A: Spotlight on volasertib: Preclinical and
clinical evaluation of a promising Plk1 inhibitor. Med Res Rev.
36:749–786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sanaei M, Kavoosi F, Roustazadeh A and
Golestan F: Effect of genistein in comparison with trichostatin a
on reactivation of DNMTs genes in hepatocellular carcinoma. J Clin
Transl Hepatol. 6:141–146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shang Y, Zhou Q, Wang T, Jiang Y, Zhong Y,
Qian G, Zhu T, Qiu X and An J: Airborne nitro-PAHs induce Nrf2/ARE
defense system against oxidative stress and promote inflammatory
process by activating PI3K/Akt pathway in A549 cells. Toxicol In
Vitro. 44:66–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kikkawa Y, Takaki S, Matsuda Y, Okabe K,
Taniguchi M, Oomachi K, Samejima T, Katagiri F, Hozumi K and Nomizu
M: The influence of Tribenoside on expression and deposition of
epidermal laminins in HaCaT cells. Biol Pharm Bull. 33:307–310.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kubicsek T, Kazy Z and Czeizel AE:
Teratogenic potential of tribenoside, a drug for the treatment of
haemorrhoids and varicose veins-a population-based case-control
study. Reprod Toxicol. 31:464–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bhukhai K, de Dreuzy E, Giorgi M, Colomb
C, Negre O, Denaro M, Gillet-Legrand B, Cheuzeville J, Paulard A,
Trebeden-Negre H, et al: Ex Vivo selection of transduced
hematopoietic stem cells for gene therapy of β-hemoglobinopathies.
Mol Ther. 26:480–495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chang L, Hagel JM and Facchini PJ:
Isolation and characterization of O-methyltransferases Involved in
the biosynthesis of glaucine in glaucium flavum. Plant Physiol.
169:1127–1140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mi G, Gao Y, Yan H, Jin X, Ye E, Liu S,
Gong Z, Yang H and Yang Z: l-Scoulerine attenuates behavioural
changes induced by methamphetamine in zebrafish and mice. Behav
Brain Res. 298:97–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shi J, Jiang S, Qiu D, Le W, Wang X, Lu Y
and Liu Z: Rapid identification of potential drugs for diabetic
nephropathy using whole-genome expression profiles of glomeruli.
Biomed Res Int. 2016:16347302016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Thrum S, Lorenz J, Mössner J and Wiedmann
M: Polo-like kinase 1 inhibition as a new therapeutic modality in
therapy of cholangiocarcinoma. Anticancer Res. 31:3289–3299.
2011.PubMed/NCBI
|