Introduction
Osteosarcoma is the most common malignant primary
bone tumor in children and young adults (1). Currently, the 5-year overall survival
rate for patients with non-metastatic osteosarcoma is 60–70%,
whereas the survival rate for patients with metastatic osteosarcoma
is 20–30% (2). At present, the
combination of pre- and postoperative chemotherapy with surgical
resection is the main treatment for osteosarcoma, and the majority
of patients commence similar chemotherapy schedules following
diagnosis. There is currently no active agent for the treatment of
osteosarcoma, therefore, to refine treatment strategies for
osteosarcoma it is necessary to improve our understanding of
osteosarcoma etiology and to identify novel targetable agents as
adjuvant to chemotherapeutics to improve patient outcomes.
Osteosarcoma is characterized by numerical
chromosomal instability (3). A
recent study demonstrated that histone methylation, a mechanism for
modifying chromatin structure, contributes to aberrant
transcriptional regulation and oncogenic signaling pathways in
osteosarcoma (4). Furthermore,
increased expression of histone methyltransferases (HMTs),
including G9a, enhancer of zeste homolog 2 (EZH2), nuclear SET
domain-containing 3 (NSD3), protein arginine N-methyltransferase 1,
and coactivator-associated arginine methyltransferase 1, contribute
to osteosarcoma development (5–10).
These data suggested that genes encoding histone methylation
modifiers, HMTs and histone demethylases (HDMTs), may serve key
roles in the development and progression of OS.
Suppressor of variegation 3–9 homologue 2 (SUV39H2;
also known as lysine N-methyltransferase 2) is a member of the SET
domain-containing HMT family that specifically trimethylates lysine
9 of histone H3 (H3K9me3) (11),
which is a type of post-translational modification that is
associated with transcriptional repression and heterochromatin
formation (12,13). SUV39H2 shares 58% identity with
SUV39H1, another H3K9 methyltransferase belonging to the same
protein family. In adult tissues, SUV39H2 expression is restricted
to the testis, whereas its expression is largely present in
embryonic stem cells (14,15). Notably, SUV39H1- or
SUV39H2-deficient mice exhibit normal viability, whereas
SUV39H−/− null mice exhibit chromosomal instabilities
with abnormally long telomeres and display severely reduced
viability (16,17). SUV39H2 appears to be highly
upregulated in cervical, lung, prostate, bladder and esophageal
cancer, as well as acute lymphoblastic leukemia (18–20).
The function of SUV39H2 in cancer remains to be fully elucidated;
however, it is partially dependent on the regulation of H3K9
methylation status. A limited number of studies have indicated that
SUV39H2 may be associated with cellular senescence through the
methylation of H3K9 (21) and may
be involved in the DNA repair pathway through methylation of
histone H2AX at K134 (18).
Therefore, cells overexpressing SUV39H2 are more resistant to
chemotherapy (19). SUV39H2 has
also been reported to interact with androgen receptor and
melanoma-associated antigen 11, and to increase androgen-dependent
transcriptional activity (22). Our
previous studies demonstrated that the SUV39H2-dependent
methylation of lysine-specific histone demethylase (LSD)-1 at K322
leads to stabilization of LSD1 protein through the inhibition of
its ubiquitination (23) and that
automethylation of SUV39H2 impairs binding affinities to substrate
proteins such as histone H3 and LSD1 (24).
In the present study, SUV39H2 expression was
demonstrated to be upregulated in human osteosarcoma cell lines and
may promote osteosarcoma cancer growth. In addition, reduced
SUV39H2 expression led to the suppression of cancer cell growth
through the induction of G1 phase arrest and increased rates of
apoptosis, which suggested that SUV39H2 may contribute to the
development and progression of osteosarcoma, and may be a novel
targetable gene for the treatment of patients with
osteosarcoma.
Materials and methods
Cell line and cell culture
The 293T cell line, the human osteoblast cell line
hFOB1.19, and the human osteosarcoma cell lines HOS, U2OS and MG-63
were purchased from The Shanghai Institute for Biological Sciences,
Chinese Academy of Cell Resource Center (Shanghai, China). HOS,
U2OS, and MG-63 were grown in minimum essential medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA); 293T and
hFOB1.19 cells were grown in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10%
fetal bovine serum (ScienCell Research Laboratories, Inc., San
Diego, CA, USA) and 1% antibiotic/antimycotic solution (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator
with 5% CO2.
Stable cell line generation
Approximately 40–50% confluent cells were
transfected with pCAGGS-SUV39H2-3×Flag (SUV39H2)-overexpression or
pCAGGS-3×Flag (Mock) vectors using FuGENE® 6
Transfection Reagent (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. Briefly, 6 µl
FuGENE® 6 Transfection Reagent was added to 100 µl
Opti-MEM I reduced-serum medium (Gibco; Thermo Fisher Scientific,
Inc.) and incubated for 5 min at room temperature. Plasmid DNA (1
µg/µl; 2 µg) was added to FuGENE6 Transfection Reagent/Opti-MEM,
and the mixture was incubated for 15 min at room temperature
followed by adding to each well of a 6-well plate. To obtain stably
expressing cells, 293T cells transfected with
SUV39H2-overexpression or Mock vectors were subsequently selected
with geneticin (0.4 mg/ml).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from 1×106 cells
with EASYspin Plus Tissue/Cell RNA Extraction kit (Aidlab
Biotechnologies Co., Ltd., Beijing, China). Extracted RNA was
reverse transcribed to cDNA with ThermoScript First-Strand cDNA
Synthesis kit (Aidlab Biotechnologies Co., Ltd.), according to the
manufacturer's protocol. qPCR was performed using SYBR Premix Ex
Taq (Takara Biotechnology Co., Ltd., Dalian, China) and the ABI
StepOnePlus Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the following thermocycling conditions:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles of
95°C for 5 sec and 60°C for 34 sec. The primer sequences were:
GAPDH forward, 5′-ATGGAAATCCCATCACCATCTT-3′ and reverse
5′-CGCCCCACTTGATTTTGG-3′; and SUV39H2 forward,
5′-AATGGAAAGGATGGCCAGATT-3′ and reverse,
5′-ACGGGCACTTCAGATTTTGC-3′. Each sample was analyzed in duplicate,
and the relative gene expression levels were normalized to GAPDH
expression levels and quantified using the comparative
2−ΔΔCq method (25).
Small interfering (si)RNA
transfection
SUV39H2-specific siRNA oligonucleotide duplexes
(siSUV39H2#1, 5′-CACAGAUUGCUUCUUUCAA-3′; siSUV39H2#2,
5′-CUGGAAUCAGCUUAGUCAA-3′) were synthesized by Biolino Nucleic Acid
Technology Co., Ltd. (Beijing, China). siRNA against enhanced green
fluorescent protein (siEGFP; 5′-GCAGCACGACUUCUUCAAG-3′) and
si-negative control (siNC; 5′-UUCUCCGAACGUGUCACGUTT-3′) were used
as control siRNAs. Osteosarcoma cells with the density of 60–70%
were transfected with siSUV39H2, siEGFP or siNC (100 nM) using
Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.)
and Opti-MEM I reduced-serum medium (Gibco; Thermo Fisher
Scientific, Inc.), according to the manufacturer's, and the
transfected cells were incubated continuously at 37°C for 72–96 h
for further experiments.
RNA-sequencing (RNA-seq) analysis
Indexed libraries from HOS cells incubated with
siEGFP (control) or siSUV39H2 were subjected to RNA-seq on the
Illumina Hiseq 2000 Sequencing platform (Illumina, Inc., San Diego,
CA, USA). Total RNA was extracted as aforementioned, and the
integrity of samples was confirmed with the Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Gene expression levels were quantified using the fragments per
kilobase of transcript per million mapped reads (FPKM)
normalization method (26), and
Cufflinks v 2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/install)
was used for FPKM quantification. Gene expression and differential
transcription between siEGFP- and siSUV39H2-treated cells were
evaluated using Cuffdiff. The lists of significantly differentially
expressed genes (DEGs) were obtained with the thresholds of P≤0.05
and a fold-change ≥2.
Cell viability and colony-formation
assays
Cell proliferation assays were performed using the
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Shanghai, China), according to manufacturer's protocols. Cells
(1×104 cells/well) were plated in 96-well plates the day
before treatment to allow cell attachment and transfected with
siRNAs, as aforementioned. Cell viability was measured 96 h
following transfection by measuring the absorbance at a wavelength
of 450 nm and the relative cell viability was calculated as the
percentage of absorption.
For colony-formation assays, cells were seeded
(2,000 cells/well) in 6-well plates and treated with siRNAs two
times, once every 4 days. After 14 days, the cells were washed with
PBS 3 times, fixed with methanol for 30 min, and stained with 0.1%
crystal violet solution for another 15–20 min at room temperature.
After washing with PBS, the 6-well plates were dried and scanned.
Mock and SUV39H2 stably overexpressing 293T cells (5,000
cells/well) were seeded in 60-mm cell culture dish and cultured for
7 days continuously at 37°C, and subsequently stained with 0.1%
crystal violet as aforementioned. U2OS cells (500 cells/well) in
6-well plates were transfected with pCAGGS-SUV39H2-3×Flag
(SUV39H2)-overexpression or pCAGGS-3×Flag (Mock) vectors using
FuGENE6 Transfection Reagent two times, once every 3 days. Cells
were stained with 0.1% crystal violet at day 10. To observe growth
of Mock and SUV39H2 stably overexpressing 293T cells, equal amounts
of cells (500 cells/well) were seeded into 96-well plate and
incubated at 37°C. Relative viable cell numbers were evaluated
every day for 7 days incubating with CCK-8 for 2 h at 37°C, and the
absorbance was measured at a wavelength of 450 nm everyday.
Western blot analysis
Cells at 80–90% confluence were lysed in
radioimmunoprecipitation assay buffer [50 mM Tris-Cl (pH 7.4), 150
mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonidet-P40 and 0.1
mM PMSF] with complete protease inhibitor cocktail (Roche Applied
Science, Penzberg, Germany). Protein concentrations were determined
by Bicinchoninic Acid Assay, and 20 µg protein was loaded into each
well and separated by 12% SDS-PAGE. The proteins were transferred
onto polyvinylidene fluoride membranes and blocked with 5% milk in
TBS + 0.1% Tween-20 buffer for 1 h at room temperature followed by
the overnight incubation with primary antibodies at 4°C, and
subsequent incubation with secondary antibodies for 1 h at room
temperature. Protein bands were visualized using Tanon High-sig ECL
Western Blotting Substrate (Tanon Science and Technology Co., Ltd.,
Shanghai, China), and the relative density of the protein band of
interest was normalized to β-actin and quantified using Tanon Image
Software v1.0 (Tanon Science and Technology Co., Ltd.). Anti-Flag
(1:8,000; cat. no. F-7425; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), SUV39H2 polyclonal antibody (1:250: cat. no. PA5-11366;
Thermo Fisher Scientific, Inc.) and anti-β-actin (1:10,000; cat.
no. A5441, Sigma-Aldrich; Merck KGaA) were used.
Cell cycle and apoptosis analysis
Osteosarcoma cells at a density of 40–60% were
transfected with siSUV39H2#1 or siEGFP, as aforementioned, and
incubated for 72 h at 37°C. The transfected cells were fixed with
70% ethanol in PBS at 4°C, followed by incubation with 500 µl of
PBS containing 0.5 mg of boiled RNase at 37°C for 30 min.
Subsequently, cells were stained with 50 µg/ml propidium iodide
(PI) and analyzed using a BD FACSAria flow cytometer (BD
Pharmingen; BD Biosciences, Franklin Lakes, NJ, USA) to investigate
cell cycle. The percentage of cells in each group was calculated
using ModFit LT software (Verity Software House, v3.3.11).
Osteosarcoma cells incubated with specific siRNA for 96 h at 37°C
were collected for apoptosis analysis. Apoptosis was analyzed using
fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection kit
(cat. no. BD#556547; BD Biosciences), following the manufacturer's
protocol.
Statistical analysis
Statistical analyses were performed using SPSS
version 20.0 (IBM Corp., Armonk, NY, USA). Comparisons between two
groups were analyzed using an independent two-sample t-test
(two-tailed); comparisons among multiple groups were analyzed by
one-way analysis of variance followed by the least significant
difference post hoc test. Experiments were performed in duplicate
or triplicate, and results were presented as mean ± standard
deviation, except for the RT-qPCR experiments, which are presented
as the mean ± standard error of the mean. P<0.05 was considered
to indicate a statistically significant difference.
Results
SUV39H2 expression is increased in
human osteosarcoma cell lines
A number of HMTs and HDMTs are key drivers of cancer
development and progression (27,28).
Therefore, the present study aimed to understand the contribution
of HMTs and HDMTs to osteosarcoma. Among the dozens of examined
HMTs and HDMTs identified, SUV39H2, previously reported to be
upregulated in a set of human cancers (18–20),
was confirmed to be upregulated in osteosarcoma in the present
study. Total RNA was isolated from osteosarcoma cell lines and the
relative expression levels of SUV39H2 mRNA and protein were
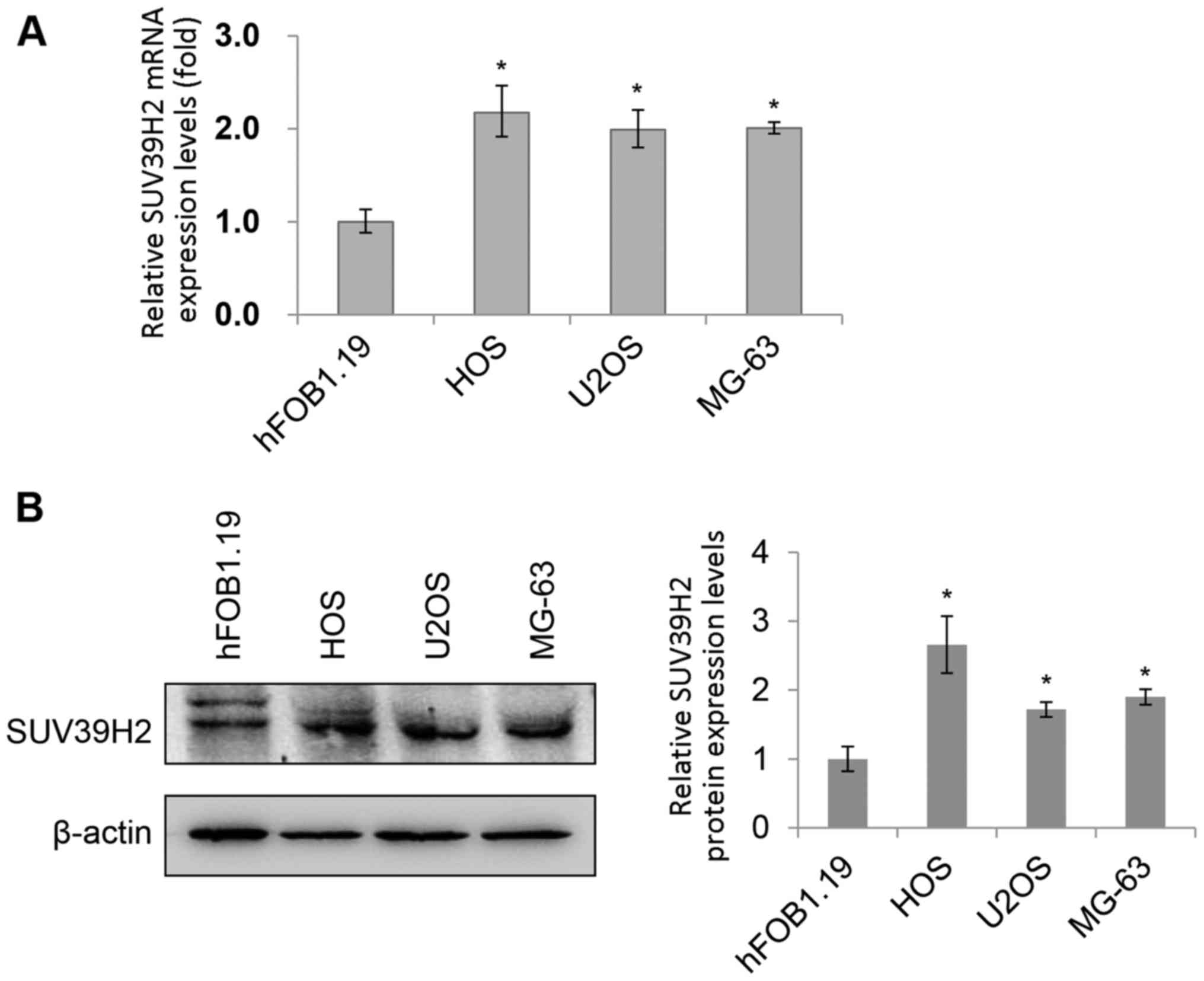
detected by RT-qPCR and western blotting, respectively (Fig. 1A and B, respectively). Notably, the
SUV39H2 mRNA and protein expression levels in all three
osteosarcoma cell lines (HOS, U2OS and MG-63) were significantly
higher compared with those in the normal osteoblast cell line
hFOB1.19.
siSUV39H2 transfection reduces
osteosarcoma cell viability
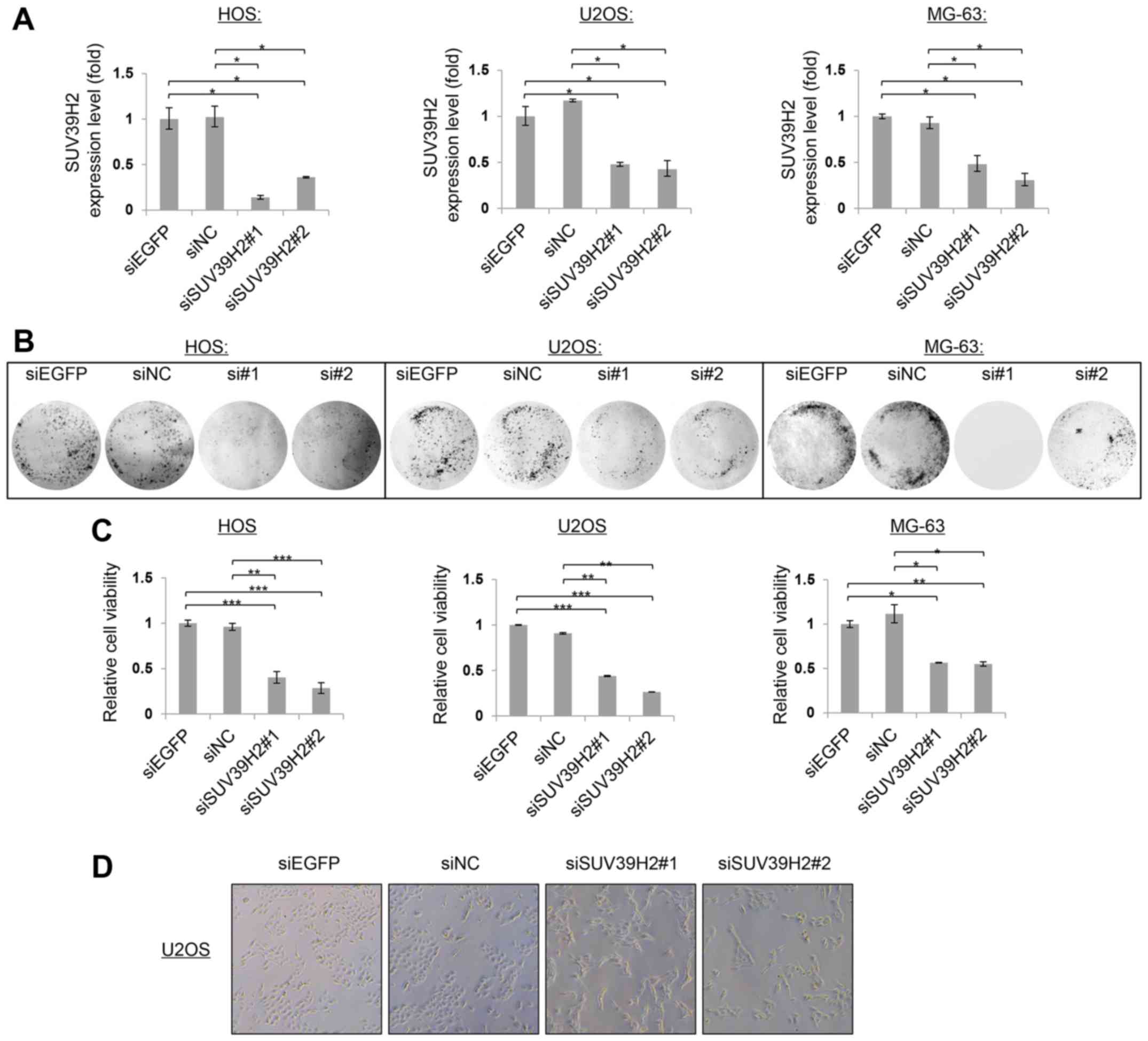
To examine the biological functions of SUV39H2 in
human osteosarcoma, the expression of SUV39H2 mRNA was knocked down
using two independent SUV39H2-specific siRNAs (siSUV39H2#1 and
siSUV39H2#2) in HOS, U2OS and MG63 cells, which was confirmed by
RT-qPCR at 72 h post-transfection (Fig.
2A). SUV39H2 mRNA expression levels were reduced by ~75%
following transfection with siSUV39H2#1 or siSUV39H2#2 compared
with expression levels in the respective siEGFP-transfected or
siNC-transfected control cells.
Colony formation assays were performed to assess the
roles of SUV39H2 on the proliferation of osteosarcoma cells. At 14
days post-transfection, siSUV39H2#1- and siSUV39H2#2-transfected
cells formed notably fewer and smaller colonies compared with the
respective siEGFP- or siNC-treated cells (Fig. 2B). The viability of
siSUV39H2-transfected osteosarcoma cells was measured by CCK-8 at
96 h post-transfection; cells transfected with SUV39H2-specific
siRNAs exhibited a significant decrease in cell viability (~75%)
compared with cells transfected with siEGFP or siNC (Fig. 2C; P<0.05). In addition,
morphological changes were observed in siSUV39H2#1 or siSUV39H2#2
under normal microscope (Fig. 2D);
SUV39H2 depleted cells became large and angled. These results
supported the potential oncogenic functions of SUV39H2 in the
development and progression of osteosarcoma. To further determine
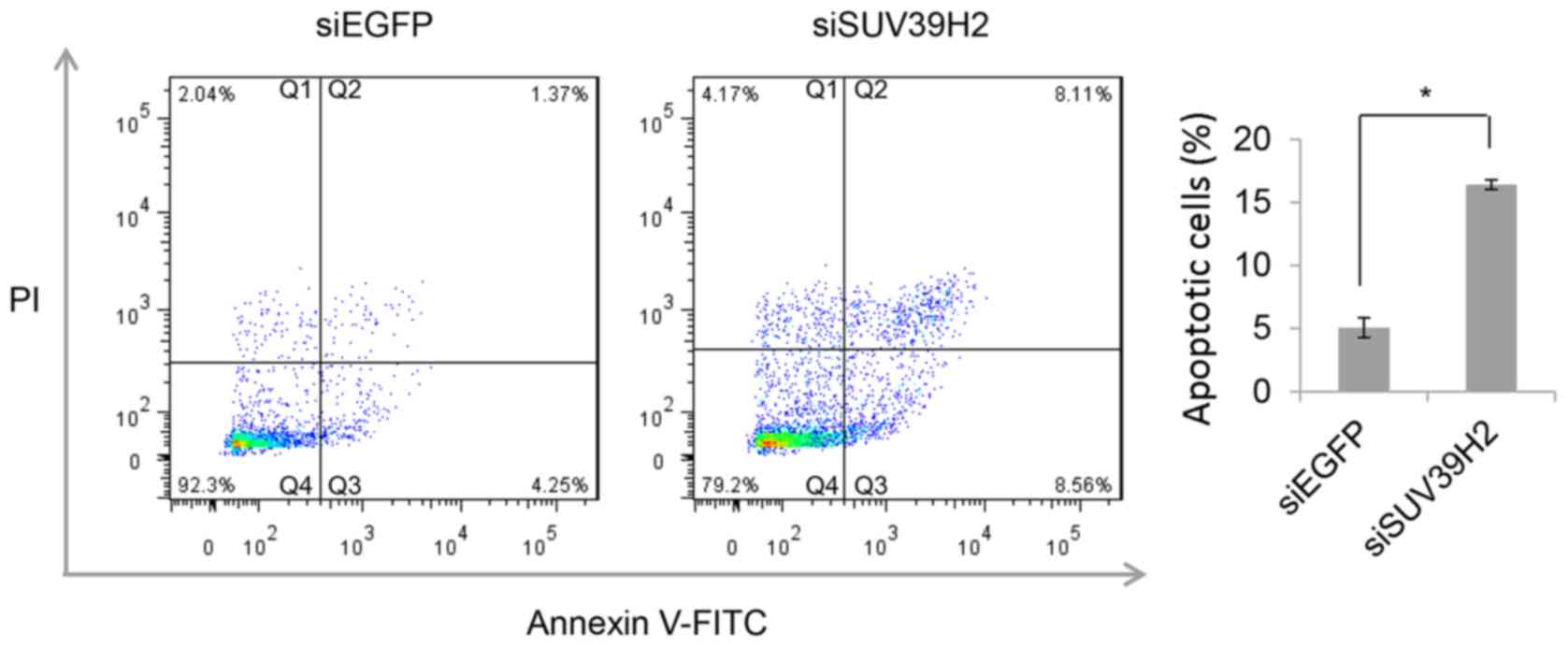
whether the reduction in cell viability was due to induction of
apoptosis, flow cytometric analysis of Annexin V-FITC/PI stained
cells was performed on U2OS cells transfected with either siEGFP or
with siSUV39H2#1 96 h. A higher proportion of Annexin V-FITC
positive cells were detected in siSUV39H2-transfected cells
compared with control siEGFP-treated cells (P<0.05; Fig. 3).
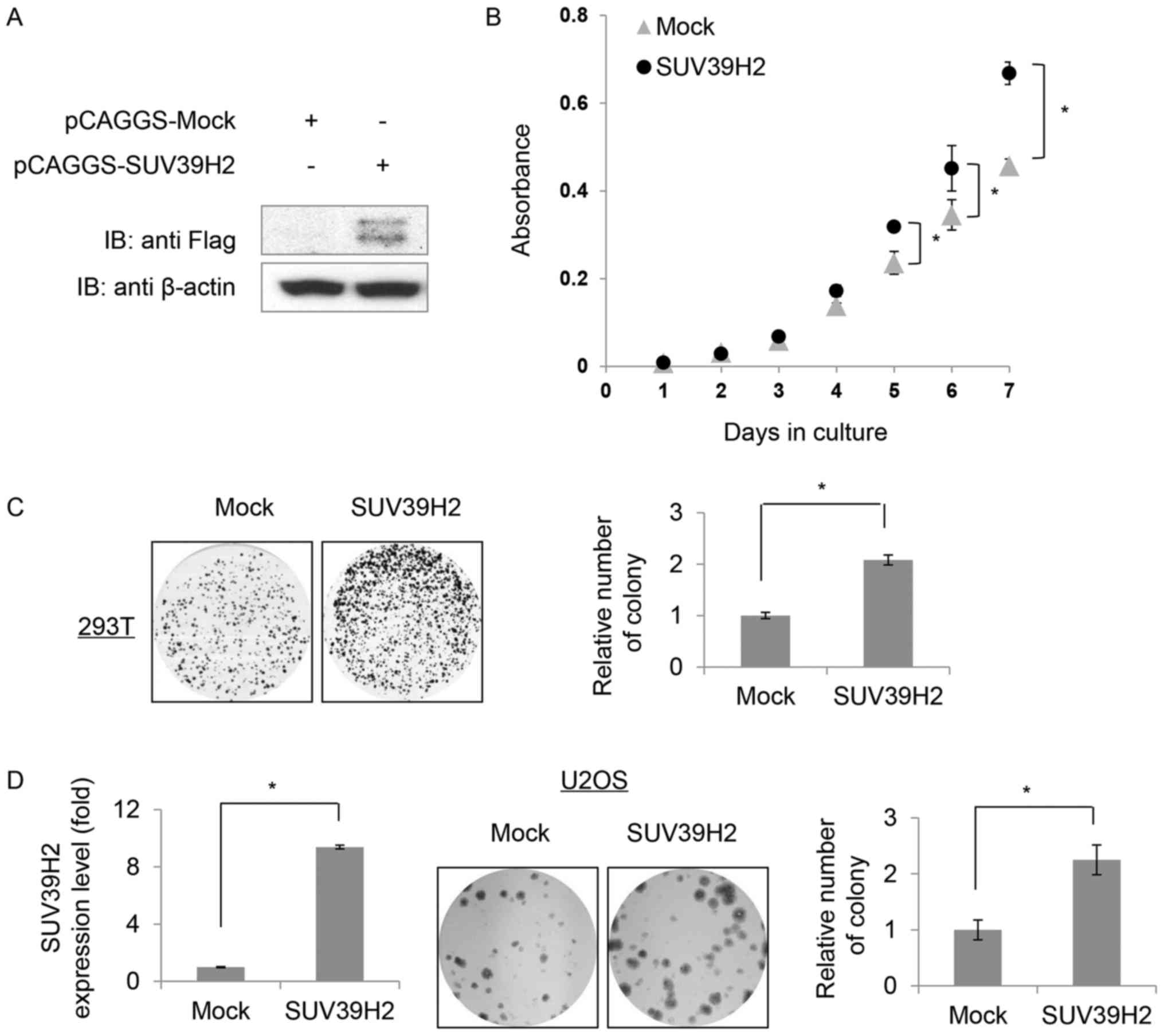
Overexpression of SUV39H2 promotes
cell growth
To further investigate the roles of SUV39H2 in
cells, attempts were made to produce hFOB1.19 cells that stably
expressed SUV39H2. However, following selection with G418
(geneticin) for one month, the expression of SUV39H2 could not be
detected by western blotting (data not shown). Subsequently,
considering the issue of transfection efficiency (29), a 293T cell line was established that
did stably express Flag-tagged SUV39H2, which was confirmed by
western blot analysis (Fig. 4A).
The SUV39H2-overexpression 293T cells exhibited increased growth
rate compared with control Mock cells (Fig. 4B). Mock or SUV39H2-overexpressing
cells were seeded at equal density (5,000 cells/60 mm dish) and
allowed to proliferate for 7 days. SUV39H2 stably expressing cells
exhibited larger and more numerous colonies compared with Mock
cells (Fig. 4C). To assess the
effects of SUV39H2 overexpression on osteosarcoma cell growth,
SUV39H2 was transiently overexpressed in U2OS cells; increased
SUV39H2 expression promoted cell proliferation compared with Mock
cells (Fig. 4D).
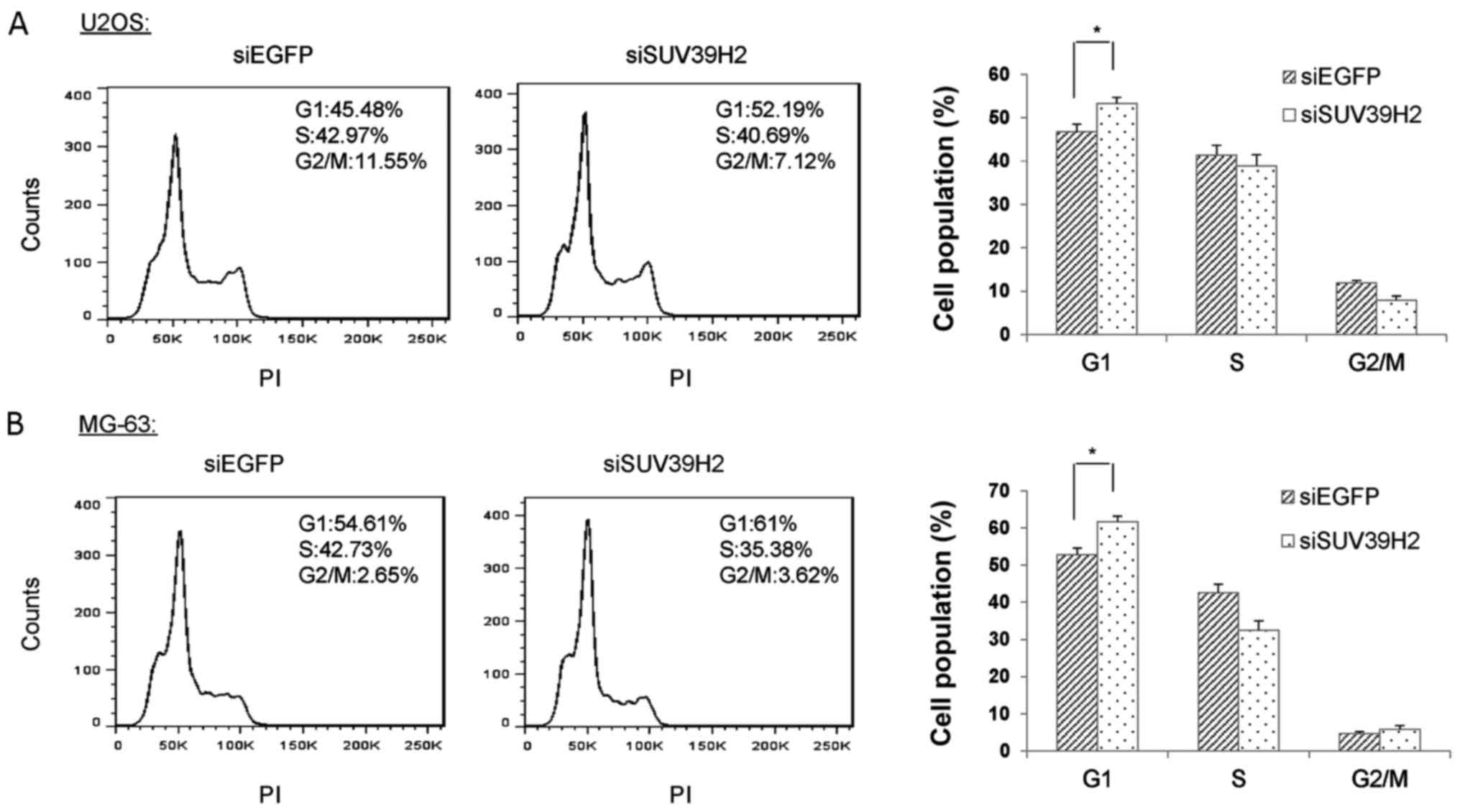
SUV39H2 knockdown leads to G1 phase
arrest in osteosarcoma cells
As knockdown of SUV39H2 led to a reduction of cell
viability and overexpression of SUV39H2 promoted cell
proliferation, it was hypothesized that SUV39H2 may be involved in
the process of cell cycle regulation. To assess the function of
SUV39H2 in the progression of cell cycle, siSUV39H2#1- and
siEGFP-transfected U2OS and MG-63 cells were collected, fixed with
ethanol, stained with propidium iodide (PI) and analyzed using flow
cytometry. The population of cells at the G1 phase in
SUV39H2-depleted U2OS cells was significantly increased compared
with that in siEGFP-treated cells (Fig.
5A). A similar increase in G1 population was observed in MG-63
osteosarcoma cells transfected with siSUV39H2#1 (Fig. 5B). These results indicated that
depletion of SUV39H2 led to G1 cell cycle arrest and suggested that
SUV39H2 may be involved in cell cycle regulation.
To identify genes associated with cell cycle that
are also mediated by SUV39H2, RNA-seq gene expression analysis was
performed in HOS cells treated with siSUV39H2#1 or siEGFP. A total
of 438 DEGs, including a set of genes related to the G1/S cell
cycle transition (DNA polymerase ε2, cyclin E1, origin recognition
complex subunit 1, cyclin dependent kinase inhibitor 2D, proteasome
inhibitor subunit 1 and RB-binding protein 8) were identified,
supporting the possibility that SUV39H2 serves an essential role in
cell cycle.
Discussion
Histone methylation serves key roles in normal
mammalian development and in regulating gene expression (30,31).
In addition to the importance of histone methylation in normal
physiological functions, previous studies have indicated that its
deregulation is deeply involved in the development of human cancer,
and global levels of some histone methylation events were
correlated with an increase of cancer recurrence and poor survival
(27,28). In particular, recent studies
suggested that aberrant expression of HMTs and HDMTs may serve
crucial roles in human carcinogenesis (32,33).
A limited number of studies have indicated that it
may be plausible that targeting abnormal HMTs and HDMTs in cancer
cells may partly contribute to the treatment of osteosarcoma. For
example, EZH2 methyltransferase, which trimethylates H3K27 serves
oncogenic roles in human osteosarcoma (6), and treatment with EZH2 inhibitor leads
to a marked reduction of cell viability in osteosarcoma (34); increased expression of the HMT NSD3
has been observed in osteosarcoma, and NSD3 is likely to serve key
roles in the development of osteosarcoma (8). Another study reported that LSD1 is
overexpressed in osteosarcoma, and treatment of osteosarcoma cells
with the LSD1 inhibitor tranylcypromine reduced cell growth
(35). Therefore, identification
and clarification of the biological functions of aberrant histone
methylation modifiers in osteosarcoma may attribute to identifying
novel targetable genes and pathways in osteosarcoma.
SUV39H2 trimethylates H3K9, which is an important
feature of cellular senescence and is essential for the viability
of cells (12,13). Suv39h1 and Suv39h2 knockout mice are
lethal, and exhibit abnormally long telomeres with reduced binding
to the chromobox (Cbx) proteins Cbx1, Cbx3 and Cbx5 (17,36).
SUV39H2 was reported to be involved in acute stress-induced
increase of H3K9 methylation in the hippocampus (37), and also serves a crucial role in DNA
repair following double-stranded breakage (18). SUV39H2 is characterized as an
embryonic and testis-specific HMT, which is undetectable in other
tissues (14). It has been
demonstrated that SUV39H2 is upregulated in multiple types of
cancer, and serves oncogenic functions in human cancer (18,19);
however, there are no selective inhibitors reported for SUV39H2, a
potentially important therapeutic target.
The present study examined the expression of various
histone methylation modifiers and demonstrated that the expression
of SUV39H2 was increased in osteosarcoma cancer cell lines, which
is in agreement with the previously reported upregulation of
SUV39H2 in osteosarcoma tissue samples (18). However, the expression of SUV39H2 in
osteosarcoma tissue samples was not examined in the present study.
Reduced expression of SUV39H2 led to a drastic reduction of
osteosarcoma cell viability, and overexpression of SUV39H2 promoted
cell growth, which indicated that SUV39H2 may serve crucial roles
in osteosarcoma development. Furthermore, knockdown of SUV39H2
caused G1 arrest in osteosarcoma cells and induced apoptosis. To
further assess the role of SUV39H2 in apoptosis, RNA-seq analysis
was performed to identify downstream target genes of. Expression
profiling through RNA-seq in HOS cells following SUV39H2 knockdown
resulted in 438 DEGs including a set of genes related to the G1/S
cell cycle transition that supported these observations. These
results indicated the importance of SUV39H2 in osteosarcoma cells
and provided some evidence in support of the hypothesis that
SUV39H2 may be a promising therapeutic approach for the treatment
of osteosarcoma.
Acknowledgements
The authors would like to thank Professor Ryuji
Hamamoto, who provided us the expression plasmids.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81603152), The
Industry-Academia Cooperation Innovation Fund Project of Jiangsu
Province (grant no. BY2016030-11) and The Jiangsu Education
Department (grant no. 16KJD310001).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LP participated in the whole project and performed
most of the work. XY performed most of the additional revised
experiments. MZ and XQ participated in the overall design of this
study and proposed helpful ideas. XX and RK performed flow
cytometry-related experiments. ZL was involved in the conception of
the study and also supervised the quality of all the work
throughout the entire process. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hagleitner MM, Coenen MJ, Gelderblom H,
Makkinje RR, Vos HI, de Bont ES, van der Graaf WT, Schreuder HW,
Flucke U, van Leeuwen FN, et al: A first step toward personalized
medicine in osteosarcoma: Pharmacogenetics as predictive marker of
outcome after chemotherapy-based treatment. Clin Cancer Res.
21:3436–3441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin JW, Squire JA and Zielenska M: The
genetics of osteosarcoma. Sarcoma. 2012:6272542012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: Emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye K, Wang S, Wang J, Han H, Ma B and Yang
Y: Zebularine enhances apoptosis of human osteosarcoma cells by
suppressing methylation of ARHI. Cancer Sci. 107:1851–1857. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lv YF, Yan GN, Meng G, Zhang X and Guo QN:
Enhancer of zeste homolog 2 silencing inhibits tumor growth and
lung metastasis in osteosarcoma. Sci Rep. 5:129992015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu MH, Fan MF and Yu XD: NSD2 promotes
osteosarcoma cell proliferation and metastasis by inhibiting
E-cadherin expression. Eur Rev Med Pharmacol Sci. 21:928–936.
2017.PubMed/NCBI
|
|
8
|
Liu Z, Piao L, Zhuang M, Qiu X, Xu X,
Zhang D, Liu M and Ren D: Silencing of histone methyltransferase
NSD3 reduces cell viability in osteosarcoma with induction of
apoptosis. Oncol Rep. 38:2796–2802. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hsu JH, Hubbell-Engler B, Adelmant G,
Huang J, Joyce CE, Vazquez F, Weir BA, Montgomery P, Tsherniak A,
Giacomelli AO, et al: PRMT1-mediated translation regulation is a
crucial vulnerability of cancer. Cancer Res. 77:4613–4625. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li S, Cheng D, Zhu B and Yang Q: The
overexpression of CARM1 promotes human osteosarcoma cell
proliferation through the pGSK3β/β-catenin/cyclinD1 signaling
pathway. Int J Biol Sci. 13:976–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schuhmacher MK, Kudithipudi S, Kusevic D,
Weirich S and Jeltsch A: Activity and specificity of the human
SUV39H2 protein lysine methyltransferase. Biochim Biophys Acta.
1849:55–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang K, Mosch K, Fischle W and Grewal SI:
Roles of the Clr4 methyltransferase complex in nucleation,
spreading and maintenance of heterochromatin. Nat Struct Mol Biol.
15:381–388. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al-Sady B, Madhani HD and Narlikar GJ:
Division of labor between the chromodomains of HP1 and Suv39
methylase enables coordination of heterochromatin spread. Mol Cell.
51:80–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Carroll D, Scherthan H, Peters AH,
Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A,
Jerratsch M, et al: Isolation and characterization of Suv39h2, a
second histone H3 methyltransferase gene that displays
testis-specific expression. Mol Cell Biol. 20:9423–9433. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bulut-Karslioglu A, De La Rosa-Velázquez
IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C,
Winter GE, Engist B, Gerle B, et al: Suv39h-dependent H3K9me3 marks
intact retrotransposons and silences LINE elements in mouse
embryonic stem cells. Mol Cell. 55:277–290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peters AH, O'Carroll D, Scherthan H,
Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner
M, Kohlmaier A, et al: Loss of the Suv39h histone
methyltransferases impairs mammalian heterochromatin and genome
stability. Cell. 107:323–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
García-Cao M, O'Sullivan R, Peters AH,
Jenuwein T and Blasco MA: Epigenetic regulation of telomere length
in mammalian cells by the Suv39h1 and Suv39h2 histone
methyltransferases. Nat Genet. 36:94–99. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sone K, Piao L, Nakakido M, Ueda K,
Jenuwein T, Nakamura Y and Hamamoto R: Critical role of lysine 134
methylation on histone H2AX for γ-H2AX production and DNA repair.
Nat Commun. 5:56912014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mutonga M, Tamura K, Malnassy G, Fulton N,
de Albuquerque A, Hamamoto R, Stock W, Nakamura Y and Alachkar H:
Targeting suppressor of variegation 3–9 homologue 2 (SUV39H2) in
acute lymphoblastic leukemia (ALL). Transl Oncol. 8:368–375. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carvalho Alves-Silva J, do Amaral Rabello
D, Oliveira Bravo M, Lucena-Araujo A, Madureira de Oliveira D,
Morato de Oliveira F, Magalhaes Rego E, Pittella-Silva F and
Saldanha-Araujo F: Aberrant levels of SUV39H1 and SUV39H2
methyltransferase are associated with genomic instability in
chronic lymphocytic leukemia. Environ Mol Mutagen. 58:654–661.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Braig M, Lee S, Loddenkemper C, Rudolph C,
Peters AH, Schlegelberger B, Stein H, Dörken B, Jenuwein T and
Schmitt CA: Oncogene-induced senescence as an initial barrier in
lymphoma development. Nature. 436:660–665. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Askew EB, Bai S, Parris AB, Minges JT and
Wilson EM: Androgen receptor regulation by histone
methyltransferase Suppressor of variegation 3–9 homolog 2 and
Melanoma antigen-A11. Mol Cell Endocrinol. 443:42–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Piao L, Suzuki T, Dohmae N, Nakamura Y and
Hamamoto R: SUV39H2 methylates and stabilizes LSD1 by inhibiting
polyubiquitination in human cancer cells. Oncotarget.
6:16939–16950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piao L, Nakakido M, Suzuki T, Dohmae N,
Nakamura Y and Hamamoto R: Automethylation of SUV39H2, an oncogenic
histone lysine methyltransferase, regulates its binding affinity to
substrate proteins. Oncotarget. 7:22846–22856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chi P, Allis CD and Wang GG: Covalent
histone modifications - miswritten, misinterpreted and mis-erased
in human cancers. Nat Rev Cancer. 10:457–469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jacobsen LB, Calvin SA, Colvin KE and
Wright M: FuGENE 6 Transfection Reagent: The gentle power. Methods.
33:104–112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martin C and Zhang Y: The diverse
functions of histone lysine methylation. Nat Rev Mol Cell Biol.
6:838–849. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Biggar KK and Li SS: Non-histone protein
methylation as a regulator of cellular signalling and function. Nat
Rev Mol Cell Biol. 16:5–17. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hamamoto R, Saloura V and Nakamura Y:
Critical roles of non-histone protein lysine methylation in human
tumorigenesis. Nat Rev Cancer. 15:110–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiong X, Zhang J, Liang W, Cao W, Qin S,
Dai L, Ye D and Liu Z: Fuse-binding protein 1 is a target of the
EZH2 inhibitor GSK343, in osteosarcoma cells. Int J Oncol.
49:623–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bennani-Baiti IM, Machado I,
Llombart-Bosch A and Kovar H: Lysine-specific demethylase 1
(LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug
target in chondrosarcoma, Ewing's sarcoma, osteosarcoma, and
rhabdomyosarcoma. Hum Pathol. 43:1300–1307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dang-Nguyen TQ, Haraguchi S, Furusawa T,
Somfai T, Kaneda M, Watanabe S, Akagi S, Kikuchi K, Tajima A and
Nagai T: Downregulation of histone methyltransferase genes SUV39H1
and SUV39H2 increases telomere length in embryonic stem-like cells
and embryonic fibroblasts in pigs. J Reprod Dev. 59:27–32.
2013.PubMed/NCBI
|
|
37
|
Hunter RG, Murakami G, Dewell S, Seligsohn
M, Baker ME, Datson NA, McEwen BS and Pfaff DW: Acute stress and
hippocampal histone H3 lysine 9 trimethylation, a retrotransposon
silencing response. Proc Natl Acad Sci USA. 109:17657–17662. 2012.
View Article : Google Scholar : PubMed/NCBI
|