Introduction
Colorectal cancer (CRC) is the third most common
type of cancer worldwide (1). Due
to its rapid progression, invasion and metastasis, CRC is one of
the leading causes of cancer-related deaths globally (2). In China, the newly diagnosed cases
with CRC in 2011 were 310,244, accounting for 9.20% of the overall
new cancer cases (3). Although
treatment for CRC has been developed, the survival rate of CRC
patients remains poor (4). Thus, a
deeper understanding of the molecular and genetic networks in CRC
development and progression and the identification of new molecular
markers or factors are urgent.
Long non-coding RNAs (lncRNAs), generally longer
than 200 nucleotides in length, are widely produced in humans and
they are emerging as important regulators in a wide range of
biological processes, such as proliferation, apoptosis, cell cycle
arrest, cell migration and invasion (5). In recent years, lncRNAs such as ANRIL,
GAS5, UCA1 and HULC were identified as functional factors in tumor
initiation and progression (6–9). In
human CRC, hundreds of novel lncRNAs with dysregulated expression
were found by a genome-wide study and some of them demonstrated
co-regulated expression patterns with their neighboring
protein-coding genes, indicating that they displayed enhancer-like
functions (10). In further
clinical and functional studies, several lncRNAs were found to be
important regulators of tumorigenesis in CRC, including PRNCR1,
MALAT-1, HOTAIR and AK027294, which partly or totally affected
cancer cell proliferation, metastasis, cell cycle progression,
apoptosis and epithelial-mesenchymal transition (EMT) (11–14).
LncRNA ENST00000547547 is a 434-bp transcript on
human chromosome12q15 (RP11-611E13.3–001) that had a distinct
expression pattern in our previous lncRNA microarray assay
(15). However, the potential role
of ENST00000547547 in the development and progression of CRC
remains unknown. In the present study, we have assessed the
expression of ENST00000547547 in CRC tissues and cells. A
functional overexpression model was used to investigate its
function in the proliferation, invasion and migration of CRC cells
in vitro and tumorigenesis abilities in vivo.
Furthermore, we investigated the potential functional mechanism of
ENST00000547547 in regulating CRC cell proliferation, invasion and
migration.
Materials and methods
Tissue collection
CRC tissues and adjacent non-cancerous tissues were
collected from 21 patients who had undergone surgical resection of
CRC at the Cancer Hospital of Hunan from January to September of
2015. The normal tissue samples were 5 cm from the edge of the
tumor and were identified by a pathologist. Prior to the surgical
resections, no preoperative treatment had been administered to
these patients. All tissue samples were immediately frozen in
liquid nitrogen after being surgically resected and then stored at
−80°C until required for the analyses. Prior to sample collection,
written informed consent was obtained from all patients, and all of
the experiments were approved by the Research Ethics Committee of
the Shangyao Hospital of TCM.
Cell lines and culture conditions
Human normal colorectal cell line NCM460 and CRC
cell lines COLO320, HCT116 and LoVo were purchased from Auragene
Bioscience Co. (Changsha, China). NCM460 cells were cultured in
McCoy's 5A medium (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.). COLO320, HCT116 and LoVo cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.). All cells were cultured at 37°C in
humidified air with 5% CO2. All media were supplemented
with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin
(Invitrogen, Shanghai, China).
RNA extraction and qRT-PCR
analyses
Total RNA was extracted from tissues or cultured
cells with TRIzol reagent (Invitrogen, Shanghai, China). For
quantitative RT-PCR (qRT-PCR), cDNA was synthesized using random
primers and a reverse transcription kit (Takara Biotechnology Co.,
Ltd., Dalian, China). qRT-PCR analyses were performed using
SYBR-Green qPCR Mix (Toyobo Life Science, Osaka, Japan). All
protocols were performed according to the manufacturer's
instructions. The qRT-PCR assays and data collection were performed
using an ABI 7300 instrument. Primers for qRT-PCR were synthesized
by Invitrogen (Shanghai, China) and the sequences were as follows:
ENST00000547547 sense, 5′-TTTTCTAAGGCACCAACT-3′ and antisense,
5′-CCAAATGCTCTAAGGGA-3′; ZEB1 sense, 5′-AAATGGAACACCAGATGC-3′ and
antisense, 5′-TTACACCCAGACTGCGTC-3′; Sna1 sense,
5′-GCCTGTCTGCGTGGGTTT-3′ and antisense,
5′-GTGAGTCTGTCAGCCTTTGTCC-3′; vimentin sense,
5′-CCTGAACCTGAGGGAAACTAATC-3′ and antisense,
5′-GAAGTTTCGTTGATAACCTGTCC-3′; E-cadherin sense,
5′-AGGTCTCCTCTTGGCTCTG-3′ and antisense, 5′-AATAGGCTGTCCTTTGTCG-3′;
N-cadherin sense, 5′-GCATCATCATCCTGCTTATCC-3′ and antisense,
5′-GTCCTGGTCTTCTTCTCCTCC-3′; β-actin sense,
5′-AGGGGCCGGACTCGTCATACT-3′ and antisense,
5′-GGCGGCACCACCATGTACCCT-3′. Thermocycling conditions were as
follows: Initial DNA heat denaturation at 95°C for 3 min, followed
by 39 cycles at 95°C for 10 sec and then, annealing and extending
at 60°C for 30 sec in the qRT-qPCR experiment. The expression
levels of the target genes were normalized to the transcription
level of β-actin. Each sample was analyzed in triplicate.
In situ hybridization (ISH)
analysis
Paraffin-embedded sections of CRC and adjacent
non-cancerous tissues were used to detect ENST00000547547
expression using the RNAscope® Assay-Brown (Advanced
Cell Diagnostics, Advanced Cell Diagnostics, Inc., Newark, CA, USA)
according to the manufacturer's instructions with the following
modifications: Antigen retrieval for 15 min, protease digestion for
30 min at 37°C, probe incubation at 42°C overnight. The probe
sequence for ENST00000547547 was 5′-GGTTTACTCTTCCCCTCTGTTC-3′. The
expression level of ENST00000547547 in the nucleus was visualized
with DAB and quantitated using an automatic imaging system (Leica
DMLA; Leica Microsystems, Wetzlar, Germany).
Cell transfection
For the overexpression of ENST00000547547, a
pCDNA3.1-ENST00000547547 plasmid was constructed by Auragene
Bioscience Co. The plasmid carrying pCDNA3.1-ENST00000547547 (1
µg/µl) was transfected into the CRC cell lines HCT116 and LoVo
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) at a Lipofectamine 2000:pCDNA3.1-ENST00000547547 ratio of
1:2.
Cell proliferation assay
After pCDNA3.1-ENST00000547547 transfection, the
transfected and control cells were seeded into 96-well plates at an
initial density of 5,000 cells/well. After culture for 24, 48 and
72 h, the cells were treated with 10 µl
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (Sangon Biotech, Co., Ltd., Shanghai, China) by adding it
into each well. The cells were incubated at 37°C for another 4 h,
and then the medium was carefully removed and 150 µl dimethyl
sulfoxide (DMSO) solution (MP Biomedicals, Santa Ana, CA, USA) was
added to lyse the cells for 10 min. Finally, the absorbance was
assessed at 570 nm using a Multiskan MK microplate reader (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). All experiments were
performed in triplicate.
Tumor formation assay in a nude mouse
model
Six 4-week-old male BALB/c nude mice (original
weight, 25 g) were obtained from the Hunan Branch of the Shanghai
Laboratory Animal Center of the Chinese Academy of Sciences
(Changsha, China). The mice were housed on a 12 h light/dark cycle
under pathogen-free conditions and were fed an autoclaved diet
ad libitum. To assess tumor formation, the mice were
subcutaneously injected with 5×106/ml HCT116 cells
stably expressing Lv-NC or Lv-ENST00000547547. For the tumor
formation assay, the tumor volume was calculated as follows: Tumor
volume = (width2 × length)/2. At 28 days after the Lv-NC
and Lv-ENST00000547547 inoculation, the mice were sacrificed and
the tumors derived from each group were used for comparisons. The
protocol was approved by the Ethics Committee on the Animal
Experiments of the Third Xiangya Hospital of Central South
University.
Cell migration and invasion
assays
Cell migration and invasion assays were performed
using Transwell chambers (micropore size, 8 µm, 24-well;
Corning®; Thermo Fisher Scientific, Inc.) without
Matrigel (for migration assays) or with Matrigel (for invasion
assays). Both assays were performed according to the manufacturer's
instructions. Briefly, the treated cells were plated in the upper
chamber at a concentration of 5×104 in 500 µl FBS-free
media. The bottom chamber contained 500 µl media with 10% FBS. The
plates were incubated for 24 h, and then the chamber was fixed and
stained with crystal violet staining solution (Beyotime Institute
of Biotechnology, Haimen, China). The stained cells were imaged
under a microscope at ×100 magnification and the optical density
(OD) values were detected using a microplate reader (Thermo
Multiskan MK3; Thermo Fisher Scientific, Inc.).
Flow cytometric analysis
The CRC cell lines HCT116 and LoVo were transfected
with pCDNA3.1-ENST00000547547 or pCDNA3.1-NC for 48 h and
harvested. Then, the cells were washed three times with cold
phosphate-buffered saline (PBS) and fixed with cold 70% ethanol
overnight. For the cell cycle analysis, the cells were stained with
propidium iodide (PI) (Keygentec, Inc., Nanjing, China) for 30 min
at 4°C in the dark. The cell cycle profiles were assayed using flow
cytometry (BD Biosciences, Franklin Lakes, NJ, USA). For the
apoptosis analysis, the cells were harvested 48 h after
transfection and were stained with Annexin V-fluorescein
isothiocyanate (FITC) and PI (Keygentec, Inc.) for 10 min in the
dark at room temperature. The cells were then examined by flow
cytometry (BD Biosciences). All of the samples were assayed in
triplicate.
Western blot analysis
Cells were lysed using RIPA lysis buffer (Auragene
Bioscience Co.), supplemented with a protease inhibitor cocktail
(Roche Diagnostics, Indianapolis, IN, USA) and phenylmethylsulfonyl
fluoride (PMSF) (Auragene Bioscience Co.). Equal amounts (25 µg) of
protein were loaded on a 10–12% SDS-polyacrylamide separating gel.
The proteins were then transferred to a Immobilon-P polyvinylidene
difluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA). The
membrane was blocked with 3% bovine serum albumin-Tris-buffered
saline with Tween-20 (BSA-TBST) with gentle shaking at room
temperature for 90 min. Then, the membrane was probed with the
indicated primary antibodies with gentle shaking at 4°C overnight.
The membranes were washed with TBST (5×3 min) and incubated with
specific secondary antibodies at room temperature for 1 h. A
β-actin antibody (cat. no. LCA01; Auragene, Changsha, China) was
used as a control and Bcl-2 (1:1,000; cat. no. AM2211; Abzoom,
Changsha, China), Bax (1:500; cat. no. AM2198; Abzoom), PCNA
(1:200; cat. no. AM3547; Abzoom), P21 (1:200; cat. no. B7175; Assay
Biotechnology, California's San Francisco Bay Area, CA, USA),
cyclin D1 (1:1,000; cat. no. ab134175; Abcam, Dallas, TX, USA),
cyclin E1 (1:1,000; cat. no. ab33911; Abcam), P27 (1:200; cat. no.
YM0496; ImmunoWay, Changsha, China), ZEB1 (1:500; cat. no.
21544-1-AP; ProteinTech, Wuhan, China), Sna1 (1:500; cat. no.
ab82846; Abcam), N-cadherin (1:1,000; cat. no. YM0465; ImmunoWay),
vimentin (1:800; cat. no. BM0147; Abzoom) and E-cadherin (1:800;
cat. no. BM0530; Abzoom) antibodies were used for each group. The
second antibody was goat anti-rabbit IgG (H+L)-HRP (1:18,000; cat.
no. SA009; Auragene Bioscience Co.) or goat anti-mouse IgG
(H+L)-HRP(1:18,000; cat. no. SA001; Auragene Bioscience Co.)
incubation for 1 h in room temperature.
Statistical analysis
All statistical analyses were performed using SPSS
20.0 (IBM Corp., Armonk, NY, USA). Data are expressed as the mean ±
standard deviation (SD) from at least three independent
experiments. Statistical analyses were determined with Student's
t-test and AVONA as appropriate. All P-values were two-sided and
P<0.05 was considered to indicate a statistically significant
result.
Results
ENST00000547547 is downregulated in
CRC
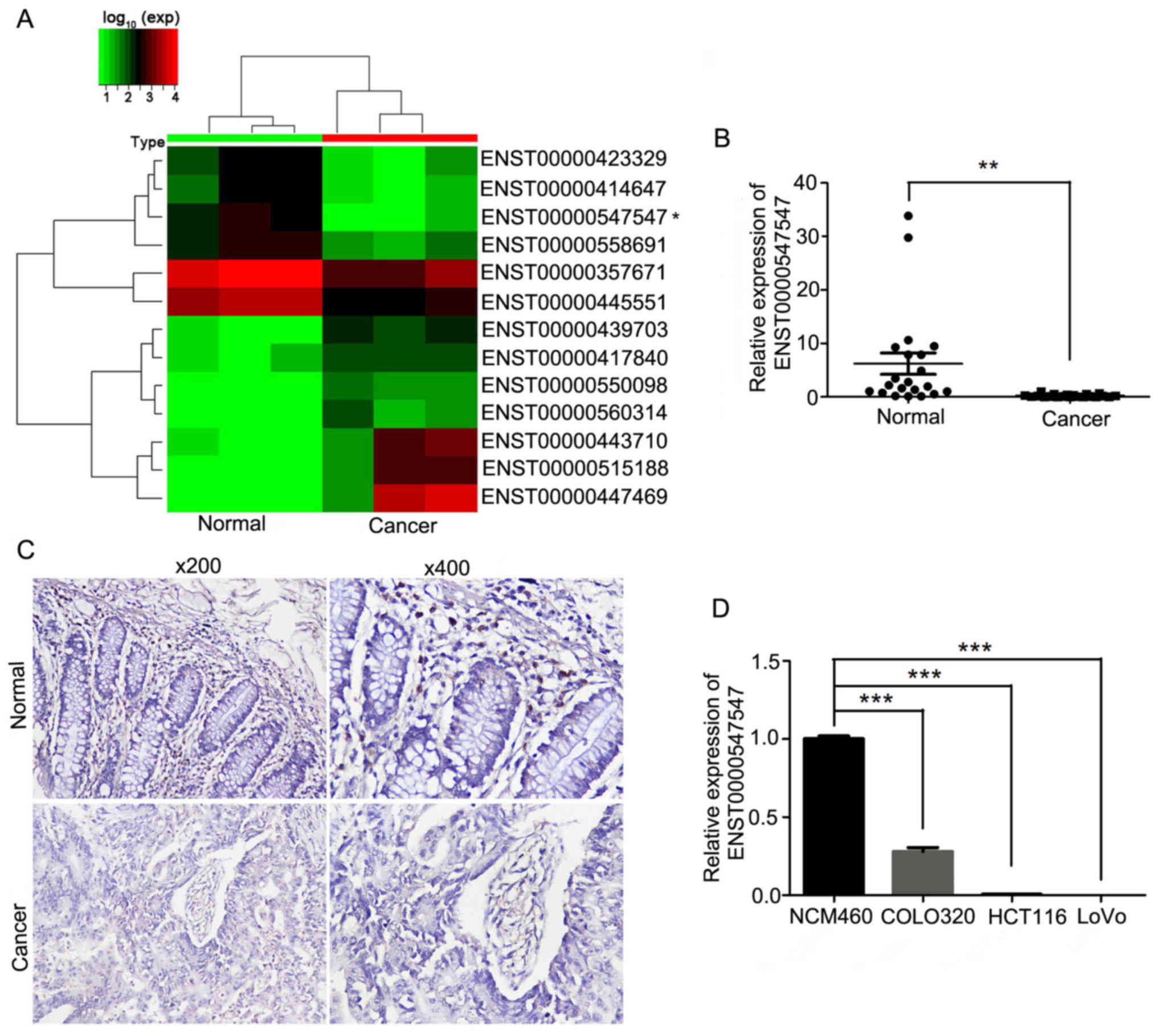
The lncRNA microarray data revealed that lncRNA 3837
was downregulated and lncRNA 1181 was upregulated in CRC tissues.
In addition, the ENST00000547547 level was decreased in the
microarray data in CRC tissues compared to normal tissues (Fig. 1A). Subsequently, its expression was
analyzed by qRT-PCR and in situ hybridization assays in CRC
and adjacent non-cancerous tissues. The qRT-PCR results revealed
that the expression of ENST00000547547 in CRC tissues was
significantly decreased compared to adjacent non-tumor tissues
(Fig. 1B). The in situ
hybridization assay results revealed that the CRC tissues displayed
lower ENST00000547547 staining than the adjacent non-cancerous
tissues (Fig. 1C). The
ENST00000547547 level in the human CRC cell lines COLO320, HCT116
and LoVo, and the normal human colonic epithelial cell line NCM460
was examined by qRT-PCR. As displayed in Fig. 1D, the expression of ENST00000547547
was significantly decreased in CRC cell lines compared to the
expression in normal human colorectal cell lines. These results
indicated that ENST00000547547 was significantly downregulated in
CRC.
Overexpression of ENST00000547547
inhibits CRC cell proliferation and tumor growth
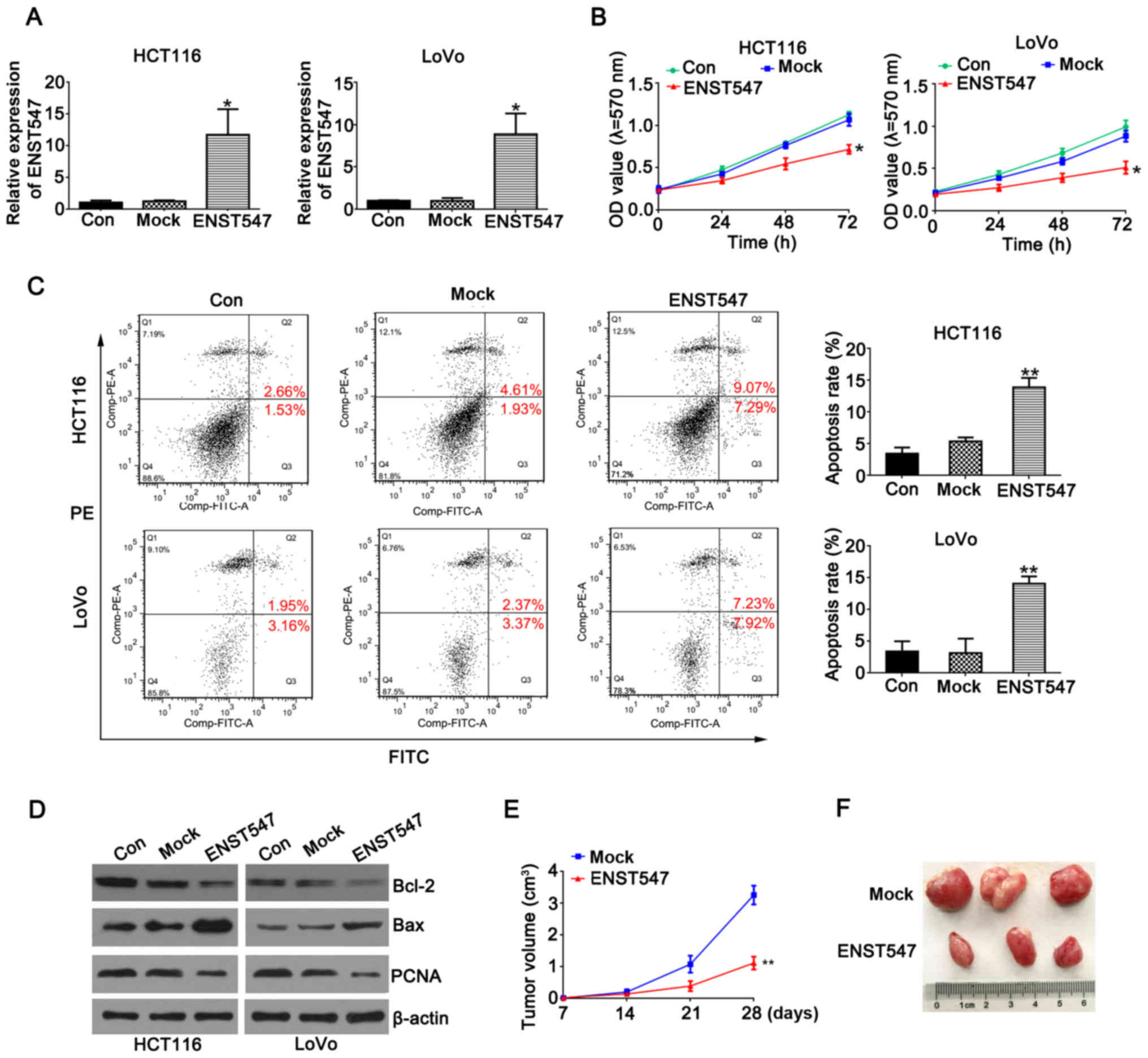
To investigate the potential biological function of
ENST00000547547 in CRC cells, an ENST00000547547-overexpressing
plasmid was constructed and used to transfect HCT116 and LoVo cells
(16). The overexpression
efficiency was verified by qRT-PCR analysis (Fig. 2A).
To determine the effect of ENST00000547547 on CRC
cell proliferation, an MTT assay and flow cytometric analysis were
performed. The MTT results revealed that the overexpression of
ENST00000547547 significantly inhibited cell proliferation in the
HCT116 and LoVo cells compared to the control group (Fig. 2B). As displayed in Fig. 2C, the overexpression of
ENST00000547547 significantly increased cell apoptosis in the
HCT116 and LoVo cells. Various mechanisms have been studied to
explain lncRNA-mediated cell apoptosis and the most important
factor identified is the activation of anti- or pro-apoptotic
regulators. Bcl-2 and Bax proteins are one of the many ways to
induce apoptosis (17), and
proliferating cell nuclear antigen (PCNA) is known as a molecular
marker for proliferation (18,19).
To confirm the role of ENST00000547547-induced apoptosis, the
expression levels of Bcl-2, PCNA and Bax were examined. Western
blot analysis revealed that the protein levels of Bax were
significantly increased in ENST00000547547-overexpressing cells,
whereas the protein levels of Bcl-2 and PCNA were decreased
(Fig. 2D). Collectively, these data
indicated that the overexpression of ENST00000547547 could inhibit
cell growth and promote apoptosis in CRC cells.
To further determine the effect of ENST00000547547
on tumorigenesis, cells stably expressing Lv-NC and
Lv-ENST00000547547 were subcutaneously injected into nude mice.
Nine days after the injection, a palpable tumor could be observed
in both groups. There was a dramatic decrease in tumor volume in
the Lv-ENST00000547547 group compared with the Lv-NC group
(Fig. 2E). In addition, at 28 days,
tumors derived from the Lv-ENST00000547547 group were significantly
smaller than those in the control group (Fig. 2F). These results indicated that the
overexpression of ENST00000547547 significantly inhibited CRC
growth in vitro and in vivo.
Overexpression of ENST00000547547
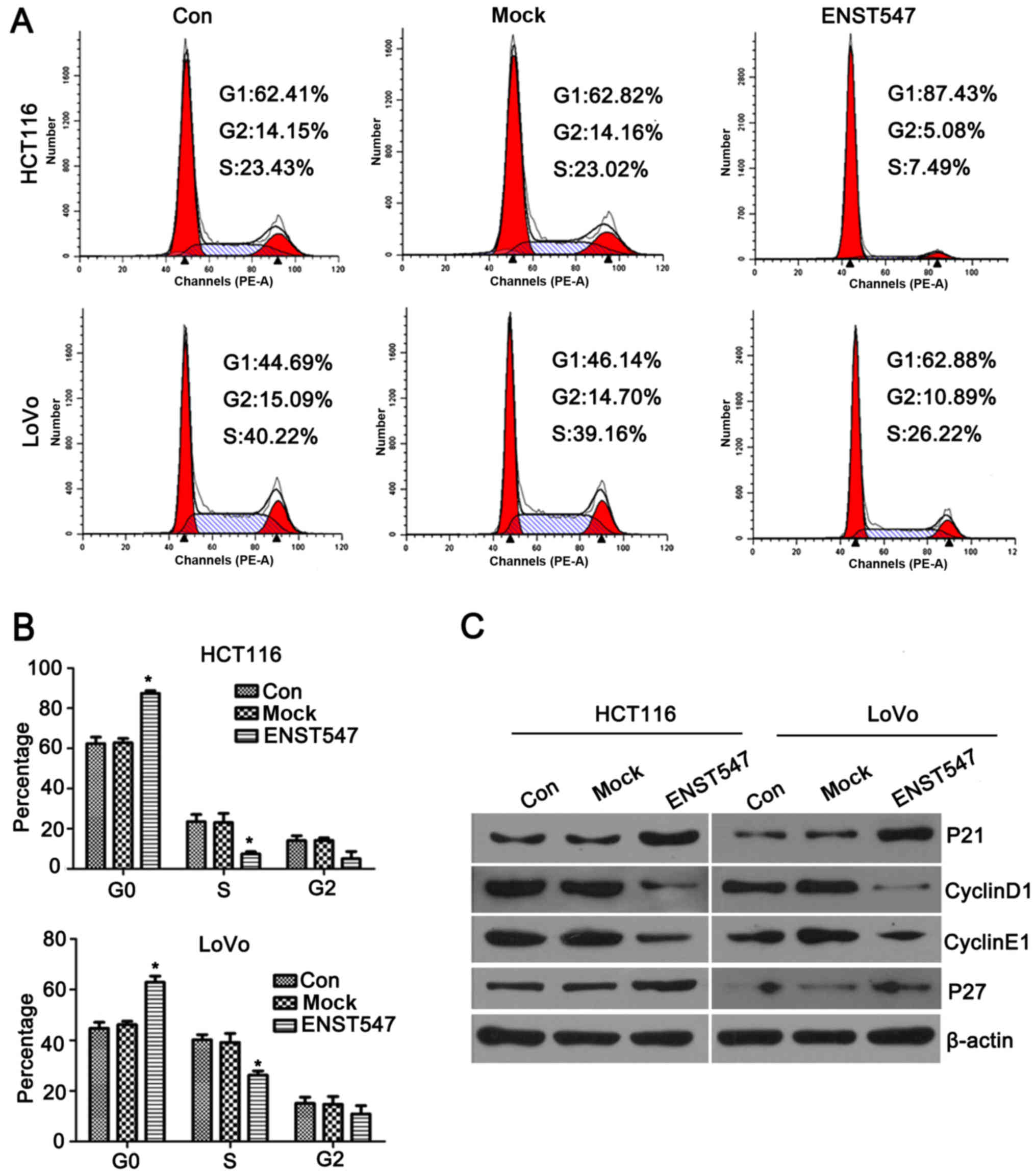
induces G0/G1 phase arrest in vitro
To further assess whether the antiproliferative
effects of ENST00000547547 on CRC cells were mediated by inhibiting
cell cycle progression, the cell cycle was evaluated by flow
cytometry and western blotting. The flow cytometric results
demonstrated that overexpression of ENST00000547547 induced a
significant G1-phase increase and caused an obvious reduction in
the number of cells in the S phase in the HCT116 and LoVo cells
(Fig. 3A and B). Furthermore,
western blotting results revealed that the cell cycle-related
proteins P21 and P27 were significantly increased in the
ENST00000547547-overexpressing cells, whereas the protein levels of
cyclin D1 and E1 were decreased in the HCT116 and LoVo cells
(Fig. 3C). These observations
indicated that ENST00000547547 may induce cell cycle arrest by
affecting the expression levels of cyclins. Collectively, these
data demonstrated that ENST00000547547 induced G0/G1 phase arrest
in CRC cells.
Overexpression of ENST00000547547
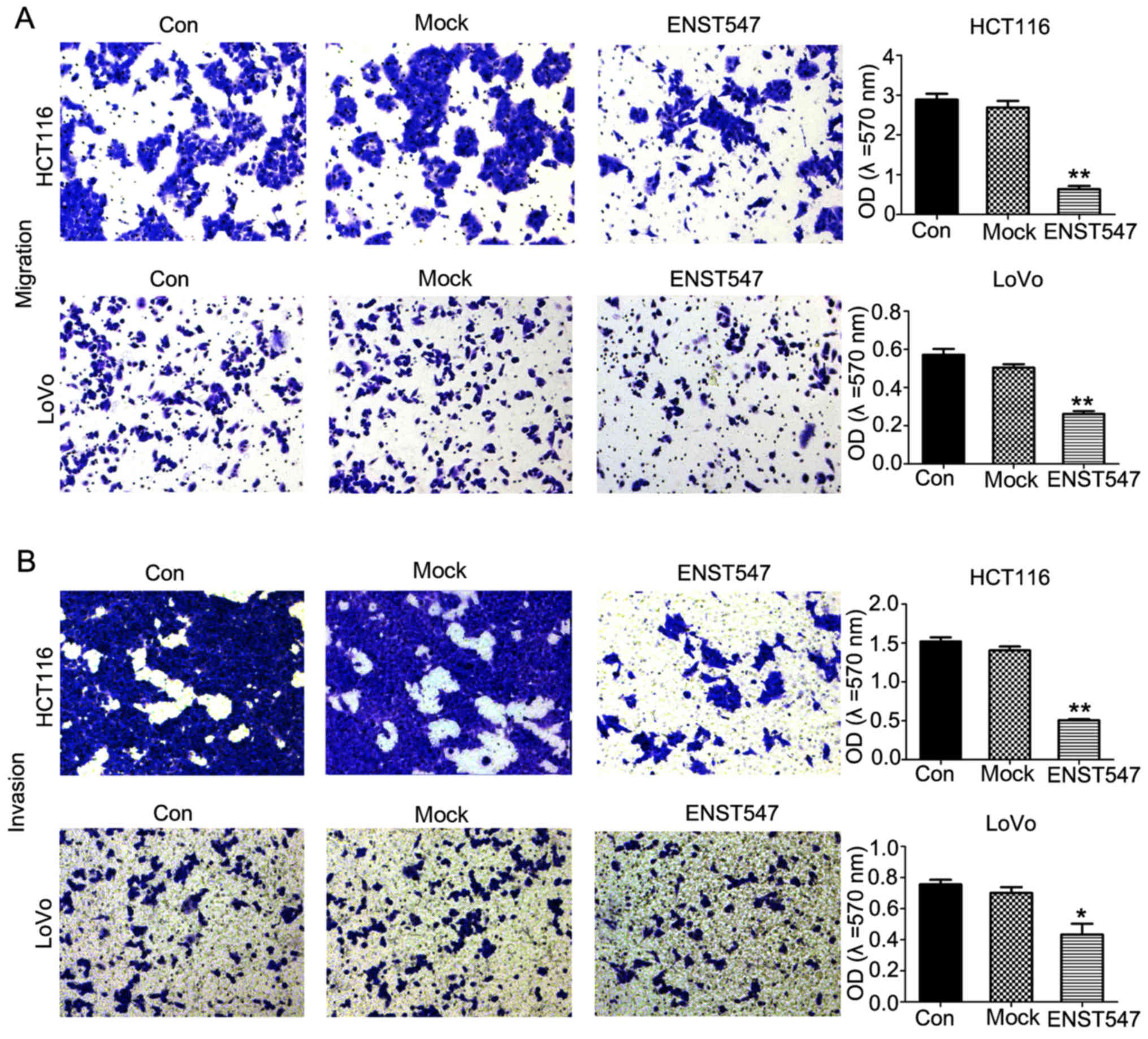
inhibits CRC cell invasion and migration in vitro
To analyze the role of ENST00000547547 in cell
invasion and migration, Transwell assays were performed in HCT116
and LoVo cells and the results revealed that ENST00000547547
overexpression greatly decreased cell invasion and migration
compared with the control groups (Fig.
4). These data demonstrated that the overexpression of
ENST00000547547 inhibited the invasion and migration of CRC cells
in vitro.
Overexpression of ENST00000547547
suppresses EMT-inducing gene expression
To investigate whether the expression of
ENST00000547547 regulated the progress of the
epithelial-mesenchymal transition (EMT), we examined the expression
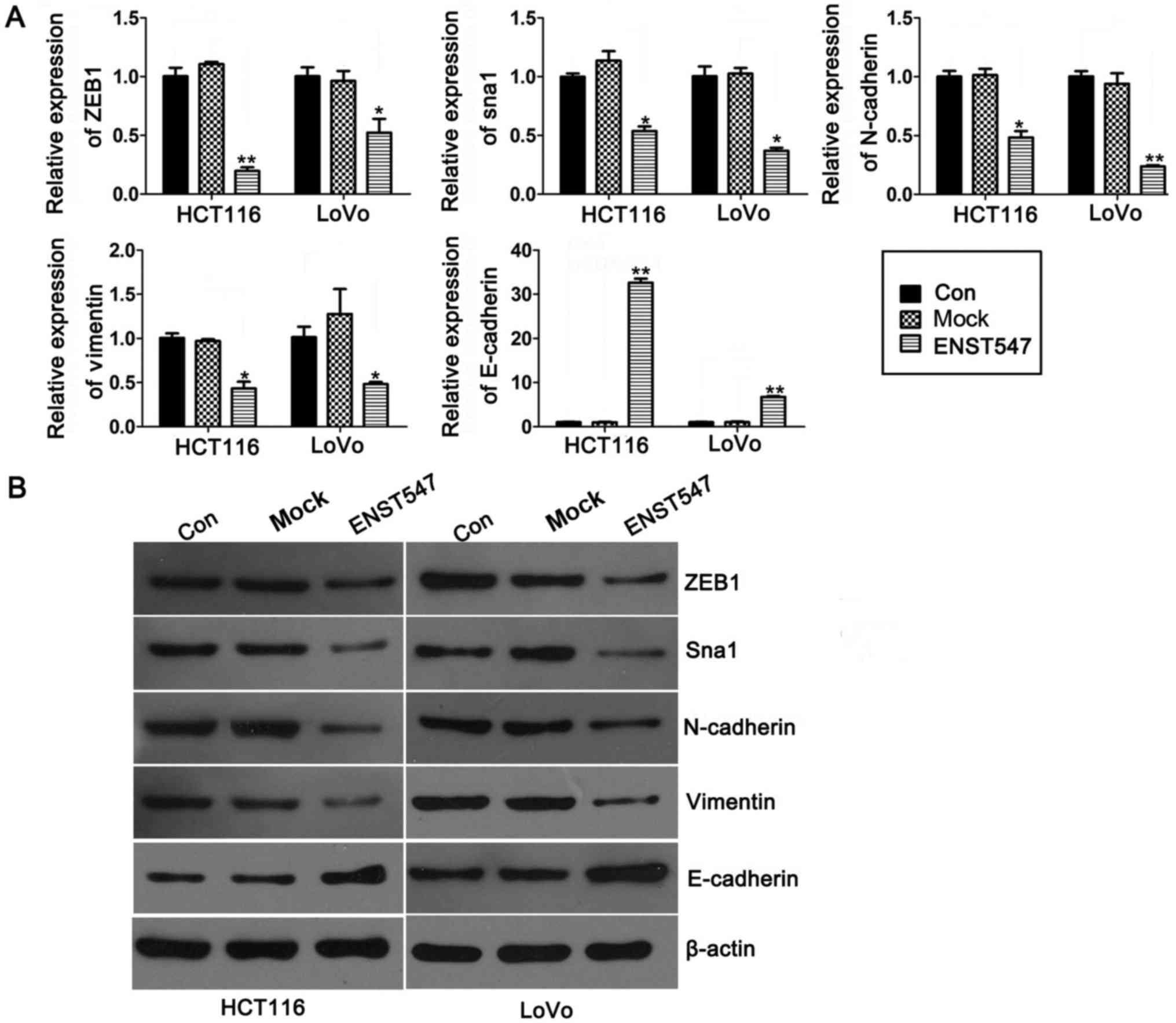
levels of ZEB1, Sna1, N-cadherin, vimentin and E-cadherin in
ENST00000547547-overexpressing HCT116 and LoVo cells. qRT-PCR
analysis indicated that overexpression of ENST00000547547 resulted
in an obvious downregulation in the transcription levels of
EMT-inducing genes (ZEB1, Sna1) and the biomarkers of mesenchymal
cells (N-cadherin and vimentin), as well as a clear upregulation in
the transcription level of E-cadherin, which is a hallmark of
epithelial cells (Fig. 5A). These
results were reconfirmed by western blot analysis (Fig. 5B). These data indicated that
ENST00000547547 inhibited the metastasis of CRC cells by
suppressing the expression of EMT-inducing genes.
Discussion
Colorectal cancer (CRC) is the third most common
cancer and the fourth highest cause of cancer-related deaths
worldwide, with more than 1 million individuals suffering from CRC;
694,000 people died of CRC in 2012 (20). To clarify the mechanism of CRC
pathogenesis, numerous protein-coding genes and non-protein-coding
genes have been shown to regulate CRC initiation and development
(21,22). As non-coding transcripts, lncRNAs
play critical roles in every aspect of cancer progression, such as
initiation, invasion and migration, and could function as oncogenes
or tumor suppressors (11,23). In CRC, lncRNAs have been identified
as regulators that widely function in CRC cell proliferation,
metastasis, cell cycle progression, apoptosis and EMT (12–14).
In our lncRNA microarray data, we observed that lncRNA
ENST00000547547, a 434-bp transcript on human chromosome12q15
(RP11-611E13.3–001), was lower in CRC tissues. In addition, it was
also downregulated in adrenocortical carcinoma (ACC), pancreatic
adenocarcinoma (PAAD), kidney chromophobe (KICH) and thyroid
carcinoma (THCA) than in normal tissues as analyzed by the TCGA
database data using GEPIA software (http://gepia.cancer-pku.cn/) (24). However, the role that
ENST00000547547 plays in CRC development remained unknown.
In the present study, we provided the first evidence
that ENST00000547547 was significantly downregulated in CRC tissues
and human CRC cell lines. Furthermore, (23), the overexpression of ENST00000547547
significantly inhibited CRC cell proliferation in vitro and
tumorigenesis in vivo. Although lncRNAs function widely in
the developmental processes of various tumors, their precise
regulatory mechanisms remain largely unknown. A previous study
(25) revealed that the lncRNA
Loc554202 decreased CRC cell proliferation by promoting significant
arrest in the G0/G1 phase and CRC cell apoptosis. However, a study
of the lncRNA PRNCR1 indicated that the regulation of CRC cell
proliferation was necessarily associated with affecting cell
apoptosis (11). Our results
revealed that the overexpression of ENST00000547547 significantly
induced both cell cycle arrest in the G0/G1 phase and apoptosis in
CRC cell lines. In the cell cycle progression, P21 and P27, Cip/Kip
proteins, are well known for their negative role in the cell cycle
(26,27). In contrast, PCNA and cyclins (cyclin
D1 and E1) positively regulate the cell cycle by promoting DNA
replication and pushing cells from the G1 to the S phase,
respectively (28–30). Our results indicated that
ENST00000547547 induced cell cycle arrest by upregulating the
expression of P21 and P27 and downregulating the expression of
cyclin D1 and E1. During apoptosis in tumor cells, the Bcl-2 family
is critical to the regulation of apoptosis, including both death
antagonists such as Bcl-2 and death agonists such as Bax (31–33).
Our results demonstrated that the overexpression of ENST00000547547
decreased the expression of Bcl-2 while concomitantly, increased
the expression of Bax. Along with the above-mentioned study on its
effects on cell cycle, these data indicated that ENST00000547547
may be another critical effector in the regulation of the cell
cycle and apoptosis in CRC cells, similar to the lncRNA
Loc554202.
EMT is proposed to play a key role in the
acquisition of migratory or invasive capacities by cancer cells
(34,35). Previous studies indicated that
decreased expression of E-cadherin and increased expression of
N-cadherin and vimentin are considered to be fundamental events in
EMT (36,37). Furthermore, ZEB1 and Sna1 can
supress E-cadherin directly or indirectly, and they can be
considered EMT transcription factors (38). Our results indicated that the
inhibition of invasion and migration in the
ENST00000547547-overexpressing CRC cells was associated with the
suppression of EMT-inducing genes, indicating that ENST00000547547
may be a pleiotropic suppressor participating in EMT. And the high
levels of expression of any lncRNA cause stress that may serve to
induce apoptosis, such as oxidative stress-induced apoptosis
(39). However, the functioning
role of silencing the ENST00000547547 expression level in CRC cells
was not performed, and this will be the aim of our future studies.
In conclusion, the present study provided the first evidence that
the lncRNA ENST00000547547 was significantly downregulated in CRC
tissues and cells and that its overexpression inhibited cell
proliferation, migration, invasion and EMT progression, caused cell
cycle arrest at the G1 to the S phase transition in CRC cancer
cells and inhibited tumorigenesis in vivo. Collectively, our
study is the first to indicate that the lncRNA ENST00000547547 acts
as a tumor suppressor in CRC and that it could be a new candidate
biomarker for CRC prognosis and a potential therapeutic target for
molecular cancer therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the grant from
the Natural Science Foundation of Hunan Province, China (grant no.
2018JJ6073), the Hunan Provincial Health and Family Planning
Commission, China (no. C20180838) and the Shaoyang Science and
Technology Bureau of Hunan Province, China (no. 2017GX35).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XA, XL and CL conceived and designed the study. XA,
QL, and XS performed the experiments. JL, GH and CC performed the
experiments of the in situ hybridization (ISH)
analysis and wrote the paper. JL, GX, BL, MC, WZ and HW
performed the experiments of the tumor formation assay in a nude
mouse model, reviewed and edited the manuscript. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Shangyang Hospital of TCM(Shaoyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
mock
|
pcDNA3.1 plasmid transfected group or
Lv-NC stable cells
|
|
ENST547
|
pcDNA3.1-ENST00000547547 plasmid
transfected group or Lv-ENST00000547547 stable cells
|
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A and Ward E:
International trends in colorectal cancer incidence rates. Cancer
Epidemiol Biomarkers Prev. 18:1688–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu S, Zheng R, Zhang M, Zhang S, Sun X
and Chen W: Incidence and mortality of colorectal cancer in China,
2011. Chin J Cancer Res. 27:22–28. 2015.PubMed/NCBI
|
|
4
|
Schreuders EH, Ruco A, Rabeneck L, Schoen
RE, Sung JJ, Young GP and Kuipers EJ: Colorectal cancer screening:
A global overview of existing programmes. Gut. 64:1637–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of
p15INK4B tumor suppressor gene. Oncogene. 30:1956–1962.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen
W and Wei Q: Downregulation of LncRNA GAS5 causes trastuzumab
resistance in breast cancer. Oncotarget. 7:27778–27786.
2016.PubMed/NCBI
|
|
8
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27Kip1. Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Y, Guo Q, Chen J, Hu J, Wang S and
Sun Y: Role of long non-coding RNA HULC in cell proliferation,
apoptosis and tumor metastasis of gastric cancer: A clinical and
in vitro investigation. Oncol Rep. 31:358–364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Liu B, Yang R, Guo Y, Li F, Wang L
and Hu H: Integrated analysis of long non-coding RNAs in human
colorectal cancer. Oncotarget. 7:23897–23908. 2016.PubMed/NCBI
|
|
11
|
Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li
M, Xu L and Yin R: Upregulation of long non-coding RNA PRNCR1 in
colorectal cancer promotes cell proliferation and cell cycle
progression. Oncol Rep. 35:318–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep.
35:479–487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Niu H, Hu Z, Liu H, Hu G, Yang B, Wu S and
Li F: Long non-coding RNA AK027294 involves in the process of
proliferation, migration, and apoptosis of colorectal cancer cells.
Tumor Biol. 37:10097–10105. 2016. View Article : Google Scholar
|
|
15
|
Li J, Li X, Cen C, Ai X, Lin C and Hu G:
The long non-coding RNA ENST00000547547 reduces 5-fluorouracil
resistance of colorectal cancer cells via competitive binding to
microRNA-31. Oncol Rep. 39:217–226. 2018.PubMed/NCBI
|
|
16
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C, et al: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Portt L, Norman G, Clapp C, Greenwood M
and Greenwood MT: Anti-apoptosis and cell survival: A review.
Biochim Biophys Acta. 1813:238–259. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Punchihewa C, Inoue A, Hishiki A, Fujikawa
Y, Connelly M, Evison B, Shao Y, Heath R, Kuraoka I, Rodrigues P,
et al: Identification of small molecule proliferating cell nuclear
antigen (PCNA) inhibitor that disrupts interactions with PIP-box
proteins and inhibits DNA replication. J Biol Chem.
287:14289–14300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SC: PCNA: A silent housekeeper or a
potential therapeutic target? Trends Pharmacol Sci. 35:178–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Punt CJ, Koopman M and Vermeulen L: From
tumour heterogeneity to advances in precision treatment of
colorectal cancer. Nat Rev Clin Oncol. 14:235–246. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu
K, Yu J and Sung JJ: MicroRNA in colorectal cancer: From benchtop
to bedside. Carcinogenesis. 32:247–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding J, Lu B, Wang J, Wang J, Shi Y, Lian
Y, Zhu Y, Wang J, Fan Y, Wang Z, et al: Long non-coding RNA
Loc554202 induces apoptosis in colorectal cancer cells via the
caspase cleavage cascades. J Exp Clin Cancer Res. 34:1002015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Coqueret O: New roles for p21 and p27
cell-cycle inhibitors: A function for each cell compartment? Trends
Cell Biol. 13:65–70. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Jing Z, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS5 enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cayrol C, Knibiehler M and Ducommun B: p21
binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient
cells. Oncogene. 16:311–320. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Zhang H, Shi L, Zhang W, Yuan J,
Chen X, Liu J, Zhang Y and Wang Z: Inhibition of Rac1 activity
induces G1/S phase arrest through the GSK3/cyclin D1 pathway in
human cancer cells. Oncol Rep. 32:1395–1400. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rosenberg E, Demopoulos RI,
Zeleniuch-Jacquotte A, Yee H, Sorich J, Speyer JL and Newcomb EW:
Expression of cell cycle regulators p57KIP2, cyclin D1,
and cyclin E in epithelial ovarian tumors and survival. Hum Pathol.
32:808–813. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang JL, Liu D, Zhang ZJ, Shan S, Han X,
Srinivasula SM, Croce CM, Alnemri ES and Huang Z: Structure-based
discovery of an organic compound that binds Bcl-2 protein and
induces apoptosis of tumor cells. Proc Natl Acad Sci USA.
97:7124–7129. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yip KW and Reed JC: Bcl-2 family proteins
and cancer. Oncogene. 27:6398–6406. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Koda M, Przystupa W, Jarzabek K, Wincewicz
A, Kanczuga-Koda L, Tomaszewski J, Sulkowska M, Wolczynski S and
Sulkowski S: Expression of insulin-like growth factor-I receptor,
estrogen receptor alpha, Bcl-2 and Bax proteins in human breast
cancer. Oncol Rep. 14:93–98. 2005.PubMed/NCBI
|
|
34
|
Kramer N, Walzl A, Unger C, Rosner M,
Krupitza G, Hengstschläger M and Dolznig H: In vitro cell migration
and invasion assays. Mutat Res. 752:10–24. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Singh A and Settleman J: E MT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li L and Li W: Epithelial-mesenchymal
transition in human cancer: Comprehensive reprogramming of
metabolism, epigenetics, and differentiation. Pharmacol Ther.
150:33–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nalla AK, Estes N, Patel J and Rao JS:
N-cadherin mediates angiogenesis by regulating monocyte
chemoattractant protein-1 expression via PI3K/Akt signaling in
prostate cancer cells. Exp Cell Res. 317:2512–2521. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang D, Lee H, Haspel JA and Jin Y: Long
noncoding RNA FOXD3-AS1 regulates oxidative stress-induced
apoptosis via sponging microRNA-150. FASEB J. 31:4472–4481. 2017.
View Article : Google Scholar : PubMed/NCBI
|