Introduction
Ovarian cancer (OC) is the leading cause of
mortality associated with gynecological malignancies worldwide
(1). Frequent metastasis is
responsible for the rapid recurrence and poor survival associated
with OC (2,3). Therefore, identifying molecular
markers involved in the progression of OC metastasis may provide
potential targets for the treatment of OC.
SMAD specific E3 ubiquitin protein ligase 1
(SMURF1), a member of the homologous to the E6-AP carboxyl terminus
(HECT) family of E3 ubiquitin ligases, participates in a variety of
physiological and pathological processes, including cell motility,
cell proliferation and inflammatory responses (4,5).
Recently, aberrant SMURF1 expression has been associated with the
development of numerous cancers (6–8). There
is increasing interest in the prominent role that SMURF1 serves in
promoting recurrent tumor metastasis (9,10).
SMURF1 expression has been reported to be upregulated in breast
cancer (7,11). Inhibition of SMURF1 expression
decreased breast cancer cell invasion and migration (7,11). In
prostate cancer, SMURF1-mediated regulation of the tumor suppressor
ovarian carcinoma 2/disabled homolog 2 interacting protein (DAB2IP)
was reported to control tumor cell proliferation and migration
(12). Elevated expression levels
of ubiquitin ligase E3 genes, including SMURF1, SMURF2 and WW
domain containing E3 ubiquitin protein ligase may underlie the
mechanisms of occurrence, development and metastasis of prostate
cancer (13). In pancreatic cancer,
SMURF1 amplification has been detected in primary human pancreatic
cancer tissues (14). Knockdown of
SMURF1 in a pancreatic cancer cell line (AsPC-1) with focal
amplification did not alter cell growth, but led to reduced cell
invasion (14). The results from
these studies indicated that SMURF1 may serve a role in cancer cell
migration and invasion; however, the underlying pathophysiological
mechanisms contributing to the progression of OC metastasis remains
unknown. Therefore, the present study aimed to determine whether
SMURF1 was involved in the development of OC metastasis.
Materials and methods
Patient samples
A total of 80 ovarian serous cystadenocarcinoma
samples were collected from patients with age from 49 to 71 years
between May 2007 and December 2013 at the Department of Obstetrics
and Gynecology, the People's Hospital of Zhengzhou University
(Zhengzhou, China). None of the patients had received preoperative
treatments, including chemotherapy or radiotherapy. Tumour tissues
were analyzed via histopathological analysis by two experienced
pathologists independently in a blinded manner. The International
Federation of Gynecology and Obstetrics (FIGO) staging system were
used to classify the ovarian serous cystadenocarcinoma samples
(15). Clinical information of the
patients is presented in Table I.
The patients were followed-up for a median duration of 45.9 months
(range, 6–60 months). The present study was approved by the Life
Sciences Ethics Committee of Zhengzhou University and informed
consent was obtained from each patient.
 | Table I.SMURF1 expression and
clinicopathological characteristics in OC. |
Table I.
SMURF1 expression and
clinicopathological characteristics in OC.
|
|
| SMURF1
expression |
|
|---|
|
|
|
|
|
|---|
|
| Total cases | High | Low | P-value |
|---|
| Age (years) | 80 | 42 | 38 |
|
|
≤60 | 39 | 21 | 18 | 0.431 |
|
>60 | 41 | 21 | 20 |
|
| TNM stage |
|
I–II | 26 | 8 | 18 | 0.0043a |
|
III–IV | 54 | 34 | 20 |
|
|
Differentiation |
|
Well | 17 | 7 | 10 | 0.373 |
|
Moderate | 19 | 11 | 8 |
|
|
Poor | 44 | 24 | 20 |
|
| Lymph node
metastasis |
|
Yes | 46 | 30 | 16 | 0.0032a |
| No | 34 | 12 | 22 |
|
| Residual tumor size
size (cm) |
| ≤1 | 42 | 22 | 20 | 0.464 |
|
>1 | 38 | 20 | 18 |
|
Cell lines and cell culture
SKOV3 and OVCAR3 cell lines were purchased from the
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences (Shanghai, China). Cells were cultured at 37°C in an
atmosphere of 5% CO2 in Dulbecco's modified Eagle's
medium (DMEM)/F-12 (Biological Industries, Cromwell, CT, USA)
supplemented with 10% fetal bovine serum (FBS; Biological
Industries).
Lentivirus production and
transfection
The SMURF1-expressing lentivirus vector, LV5-SMURF1,
was constructed by the insertion of a full-length SMURF1 cDNA into
the LV5 vector (Shanghai GenePharma Co., Ltd., Shanghai, China) via
the NotI and BamHI cloning sites. The LV5-SMURF1 and
LV5 control (LV5-NC) vectors were, respectively, co-transfected
with shuttle plasmid A1104 and packaging vectors pGag/Pol, pRev and
pVSV-G (Shanghai GenePharma Co., Ltd.) into 293T cells (Shanghai
Institutes for Biological Sciences, Chinese Academy of Sciences,
Shanghai, China) using the Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Following culturing for 72 h, the supernatants
of the transfected cells were harvested. The lentiviral titers were
determined by fluorescence microscopy analysis for green
fluorescence protein (GFP) expressed via the viral vectors. A total
of 1×105 OVCAR3 cells/well were transduced with
LV5-SMURF1 and LV5-NC vectors, respectively, at a multiplicity of
infection of 30. Following culturing for 48 h, western blot
analysis was performed to detect SMURF1 expression in transduced
OVCAR3 cells.
The lentiviral vector pGLV3/H1/GFP+Puro (Shanghai
GenePharma Co., Ltd.) was used to construct the human SMURF1 short
hairpin RNA (shRNA) plasmids; 3 different SMURF1 targeting and
silencing constructs were ligated into the LV3 vector via the
BamHI and EcoRI cloning sites. The purified LV3-shRNA
plasmids and packaging plasmids pGag/Pol, pRev and pVSV-G were
co-transfected into 293T cells using 300 µl RNAi-Mate (Shanghai
GenePharma Co., Ltd.). Following incubation at 37°C for 72 h, the
supernatant was collected and concentrated. For lentiviral
infection, SKOV3 cells (1×105 cells/well) were cultured
in 6-well plates. Concentrated viral particles were used to
transduce cells in an adherent monolayer culture at ~50% confluence
in 1 ml complete medium supplemented with 5 µg/ml
Polybrene®. Following overnight incubation at 37°C, the
viral particles were removed and the medium was changed to one for
normal growth conditions. Cells expressing non-targeting control
shRNA (NC) or SMURF1 targeting shRNA (shSMURF1) were monitored for
GFP expression. To determine the transfection efficiency of
LV3-shSMURF1, western blot analysis was performed to detect SMURF1
expression levels in transduced OVCAR3 cells.
RNA isolation and reverse
transcription-quantitative polymerase chain (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols
and reverse transcribed for quantification using a PrimeScript RT
reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.,
Dalian, China). qPCR reactions were performed using SYBR Premix Ex
Taq II (cat. no. DRRO81A; Takara Bio, Inc., Otsu Japan) on an ABI
7500 Fast Real-Time System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with a 50-µl reaction consisting of 5 ng of cDNA,
10 µM of each of the forward and reverse primers, and 25 µl of 2X
SYBR Premix. The qPCR conditions were 95°C for 10 sec, followed by
40 cycles at 95°C for 5 sec and 60°C for 34 sec, and a final stage
of dissociation analysis. The primers employed in the present study
were as follows: SMURF1 forward, 5′-ACCAGTGCCAACTCAAGGAG-3′ and
reverse, 5′-CGACAGTTCGTGTCTGAGGA-3′. The expression levels were
normalized to the endogenous control, GAPDH forward,
5′-GGGAAACTGTGGCGTGATGG-3′ and reverse,
5′-GTGTGGAAGTGGGAGACTCAAC-3′ and the relative expression was
calculated using the comparative 2−∆∆Cq method (16).
Immunohistochemistry (IHC)
Formalin-fixed paraffin embedded OC sections were
incubated with an anti-SMURF1 rabbit polyclonal antibody (dilution
1:100; cat. no. ab38866; Abcam, Cambridge, UK) and anti-cluster of
differentiation (CD) 34 rabbit polyclonal antibody (dilution 1:100;
cat. no. ab185732; Abcam). Immunohistochemical staining was
performed according to the manufacturer's protocol of the SPlink
Detection kit (Biotin-Streptavidin-HRP Detection system; cat. no.
SP-9001; ZSGB-BIO, Inc., Beijing, China). The deparaffinized
sections were placed in a pressure cooker at 100°C for 20 min for
antigen retrieval. A 3,3′-diaminobenzidine kit and hematoxylin were
used for staining. The methods of IHC scoring were performed as
described in the study by Wang et al (17).
Protein extraction and western blot
analysis
Cells were harvested 48 h following transduction and
total proteins were extracted using radioimmunoprecipitation assay
buffer (cat. no. R0010; Beijing Solarbio Science and Technology
Co., Ltd., Beijing China) according to the manufacturer's
protocols. Antibodies for SMURF1 (1:1,000; cat. no. 2174),
Ras-related C3 botulinum toxin substrate 1 (Rac1)/cell division
control protein 42 (cdc42) and phosphorylated Rac1/cdc42 (1:1,000;
cat. nos. 4651 and 2461, respectively) and β-actin (1:1,000; cat.
no. 8475) were purchased from Cell Signaling Technology, Inc.
(Danvers, MA, USA). Antibodies for Ras homolog family member A
(RhoA; 1:500; cat. no. ab86297), phosphorylated RhoA (1:1,000; cat.
no. ab41435), Rho-associated protein kinase (ROCK1; 1:1,000; cat.
no. ab97592) and phosphorylated ROCK1 (1:500; cat. no. ab203273)
were obtained from Abcam and incubated overnight (at 4°C) at a
1:500 dilution. Proteins were separated via 12% SDS-PAGE and then
transferred onto polyvinylidene difluoride (PVDF) membranes, which
were blocked with 5% non-fat dry milk. The blocked membranes were
incubated with primary antibodies in Tris-buffered saline with
Tween-20. The protein-antibody complexes were detected on the
membranes using an enhanced chemiluminescence detection system
(Amersham™ ECL™ Prime Western Blotting Reagents; GE Healthcare,
Chicago, IL, USA).
Transwell invasion and migration
assays
Cell migration and invasion assays using ovarian
cell lines were conducted as previously described (6,17). A
total of 1×105 or 1×107 cells/ml that were
serum-starved overnight were seeded in the upper chamber in 200 µl
DMEM/F-12 medium without FBS; 500 µl DMEM/F-12 medium supplemented
with 10% FBS was added to the lower chamber (pore size, 8-µm;
24-well plates; cat. no. 3422; Corning Inc., Corning, NY, USA).
Cells were incubated at 37°C for 48 h (migration assay) or 72 h
(invasion assay). For the invasion assay, the membrane inserts were
coated with Matrigel (BD Biosciences, San Jose, CA, USA) on the
upper side for 30 min. Cells on the upper membrane surface were
removed with a cotton swab. Cells on the lower membrane surface
were fixed with 4% paraformaldehyde for 20 min and stained with
0.1% crystal violet for 30 min. Cells were counted in five randomly
selected fields under an inverted light microscope at an ×200
magnification (Olympus Corp., Tokyo, Japan). Each assay was
performed 3 times.
Statistical analysis
Experimental data were presented as the mean ±
standard error of the mean. The Student's two tailed unpaired
t-test or one-way analysis of variance (ANOVA) followed by the
Tukey's multiple comparison was used to determine statistical
significance of in vitro experiments and analyze the
association of clinical pathology features and SMURF1 expression.
Statistical data analyses were conducted using GraphPad Prism
software (version 5; GraphPad Software, Inc., La Jolla, CA, USA).
Survival statistical analyses were performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Kaplan-Meier curves were
constructed to determine patient overall survival (OS) and
relapse-free survival (RFS) rates. Patients who were not located
for follow-up or who died from causes unrelated to OC were treated
as censored events. The statistical differences in survival among
subgroups were compared using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of SMURF1 in OC cell lines
with different invasive abilities
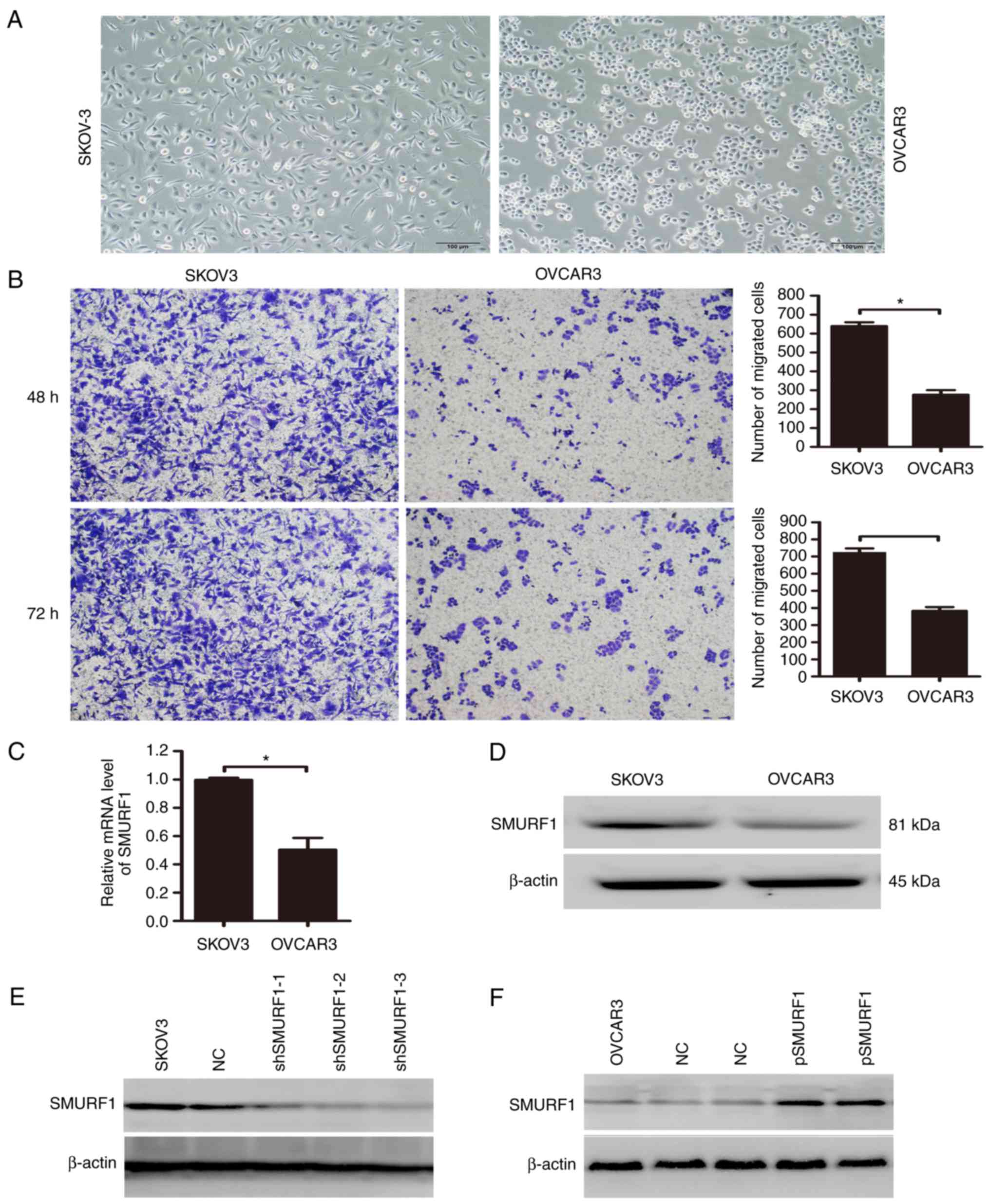
Previous studies have demonstrated that the OC cell
lines, SKOV3 and OVCAR3, possess varying invasive abilities. OVCAR3
cells demonstrated an elliptical, less invasive morphology, while
SKOV3 cells exhibited an elongated and more invasive morphological
phenotype, and have been reported to be significantly more
metastatic than OVCAR3 cells in xenograft mouse models (18,19).
Consistent with these studies, the results of the present study
revealed that OVCAR3 cells exhibited reduced cell migration
compared with SKOV3 cells (Fig. 1A and
B). To investigate the potential role of SMURF1 in the
regulation of OC metastasis, the mRNA and protein expression levels
of SMURF1 in SKOV3 and OVCAR3 cell lines were determined. The
results revealed that the mRNA and protein levels of SMURF1 were
increased in SKOV3 cells compared with in OVCAR3 cells (Fig. 1C and D). To assess the function of
SMURF1 in OC cells, we determined whether expression within these
cells could be manipulated. The LV3-shSMURF1 and LV5-SMURF1
lentiviral vectors were transduced into SKOV3 and OVCAR3 cells,
respectively. To select the cell line with the most effective and
stable transfection, the expression levels of SMURF1 protein in
stably transfected SKOV3 and OVCAR3 cells cultured with puromycin
were analyzed by western blotting. The results indicated that the
expression levels of SMURF1 were significantly decreased in all
three SMURF1 shRNA-transduced SKOV3 cell lines (Fig. 1E). Furthermore, compared with the
non-transduced control cells, ectopic expression of SMURF1 in
OVCAR3 cells was significantly increased at the protein level
(Fig. 1F).
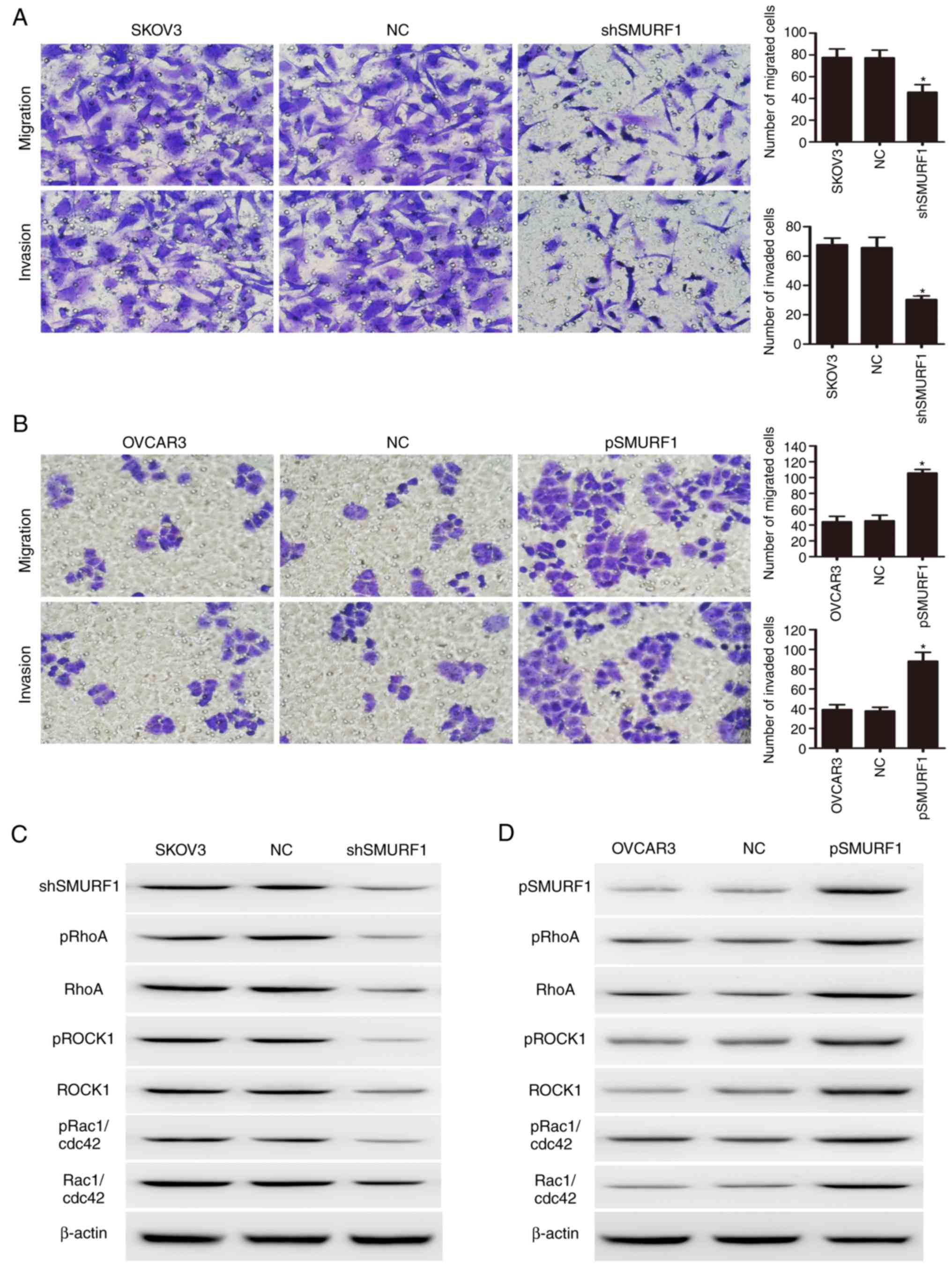
Effects of SMURF1 in OC cell migration
and invasion
To determine the role of SMURF1 in OC metastasis,
the effects of SMURF1 on the migration and invasion of OC cells
were analyzed in vitro. As presented in Fig. 2, inhibition of SMURF1 in SKOV3 cells
significantly reduced cell migration as detected by the number of
cells that had migrated via the Transwell membrane; reduced
invasion was also reported as observed by the number of cells
invading via the Matrigel-coated membrane (Fig. 2A). Conversely, the stable ectopic
expression of SMURF1 in OVCAR3 cells significantly enhanced cell
migration and invasive abilities (Fig.
2B).
Enhancement of RhoA/ROCK signaling in
OC cells by SMURF1
The present study further investigated the molecular
mechanism by which SMURF1 promotes OC metastasis. It has been
demonstrated that SMURF1 promoted the degradation of the small GTP
protein, RhoA, by increasing its ubiquitination (5). RhoA is one of the most extensively
investigated members of the Rho GTPase family of proteins; the
RhoA/ROCK signaling pathway has been associated with malignant
transformation, as well as tumor invasion and metastasis (20,21).
To determine whether SMURF1 affects OC cell migration and invasion
via this particular signaling pathway, the effects of SMURF1 on the
activation of several key downstream signaling molecules were
investigated via western blotting. As presented in Fig. 2, compared with SKOV3 control cells,
inhibition of SMURF1 within SKOV3 cells significantly decreased the
phosphorylation of RhoA, ROCK1, Rac1 and cdc42 (Fig. 2C). Conversely, overexpression of
SMURF1 in OVCAR3 cells was observed to enhance RhoA/ROCK signaling
in these OC cells (Fig. 2D). The
findings of the present study indicated that SMURF1 may promote
metastasis by activating the RhoA/ROCK-associated signaling
pathways in OC cells.
Overexpression of SMURF1 in OC tissues
is associated with poor prognosis
Previously, we observed SMURF1 overexpression in
human OC tissues (6). To
investigate the biological significance of this finding, the
association between the SMURF1 expression levels and the clinical
features of patients with OC was investigated in the present study.
Real-time qPCR results demonstrated that the expression levels of
SMURF1 in OC tissue samples were greater than the levels in the
normal ovarian tissue samples (median, 3.195±0.102 vs. 1.971±0.115;
P<0.0001). Since the median SMURF1 mRNA expression level in OC
tissues was 3.195±0.102, we chose a cut-off value of 3.0 to divide
the patients into those with high and low expression levels of
SMURF1. According to this cut-off value, 42 patients had a SMURF1
level ≥3.0 and 38 patients had a SMURF1 level <3.0. The
associations between the SMURF1 mRNA expression levels and the
clinicopathological features are shown in Table I. Statistical analyses revealed that
SMURF1 expression in OC specimens was positively associated with
advanced clinical FIGO stages (III and IV stage) (P=0.0043;
Table I) and tumor lymph node
metastasis (P=0.0032; Table I). Of
note, higher SMURF1 expression levels were associated with
increased microvessel density and the presence of metastasis
(Fig. 3), indicating that SMURF1
overexpression may contribute to the progression of OC by promoting
tumor metastasis.
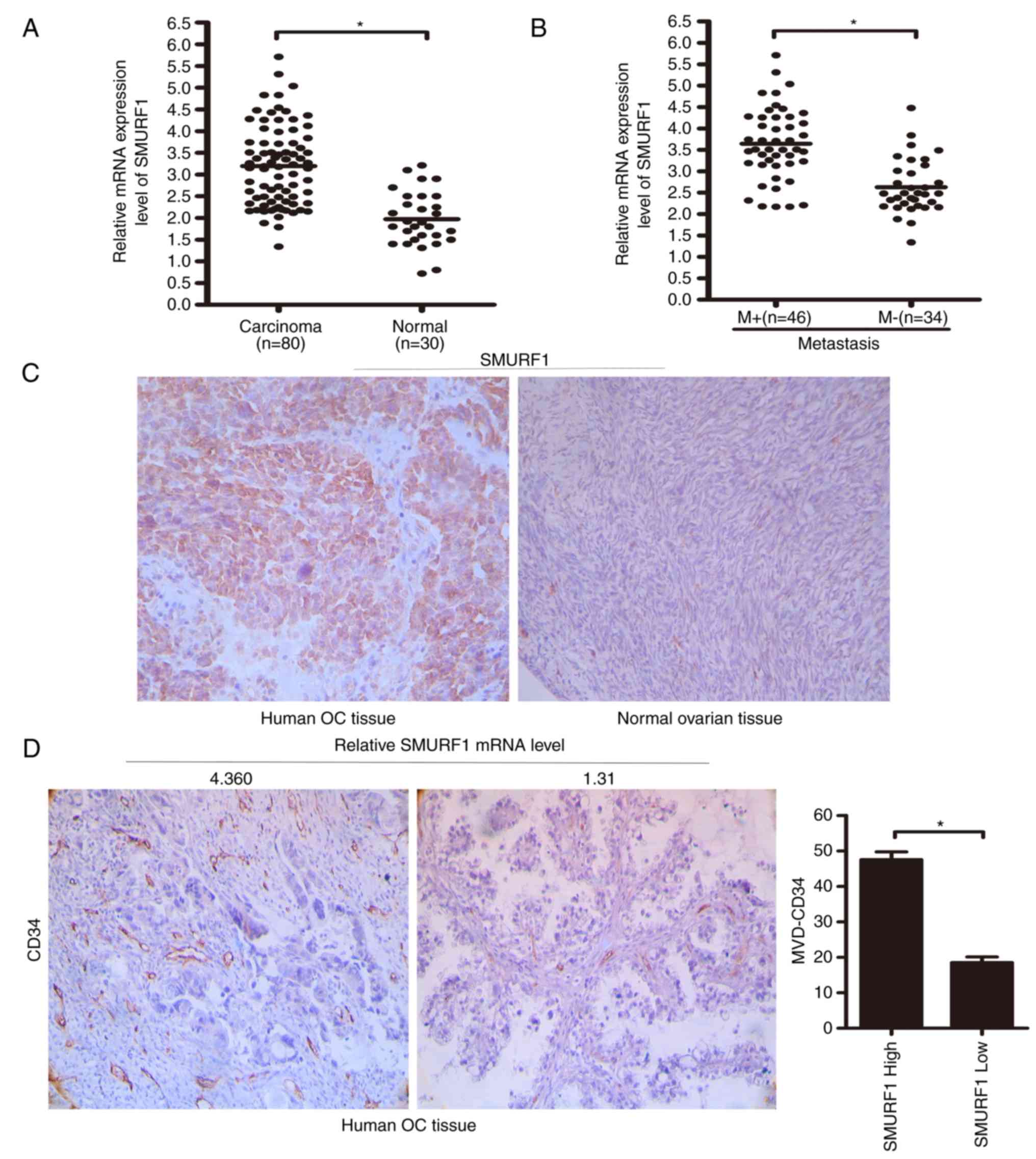 | Figure 3.SMURF1 is upregulated in human OC
tissues. (A) RT-qPCR analysis of SMURF1 expression levels in human
ovarian tissues (n=80) and normal ovarian tissues (n=30). The
central horizontal line represents the mean value. (B) RT-qPCR
analysis of SMURF1 expression levels in patients with or without
lymph node metastasis. The central horizontal line represents the
mean value. (C) Expression of SMURF1 in human ovarian tissues was
analyzed via immunohistochemical staining. (Original magnification,
×400). (D) MVD was positively associated with SMURF1 expression in
human ovarian cancer tissues. (Original magnification, ×400). M+,
patients with lymph node metastasis; M-, patients without lymph
node metastasis; SMURF1 high and SMURF1 low: the patients were
divided into groups of high and low SMURF1 mRNA expression, with a
cut-off set at 3.0 according to the median SMURF1 mRNA expression
level in ovarian cancer tissues. High group, SMURF1 mRNA level
≥3.0; low group, SMURF1 mRNA level <3.0; *P<0.05. SMURF1,
SMAD specific E3 ubiquitin protein ligase 1; OC, ovarian cancer;
MVD, microvessel density. |
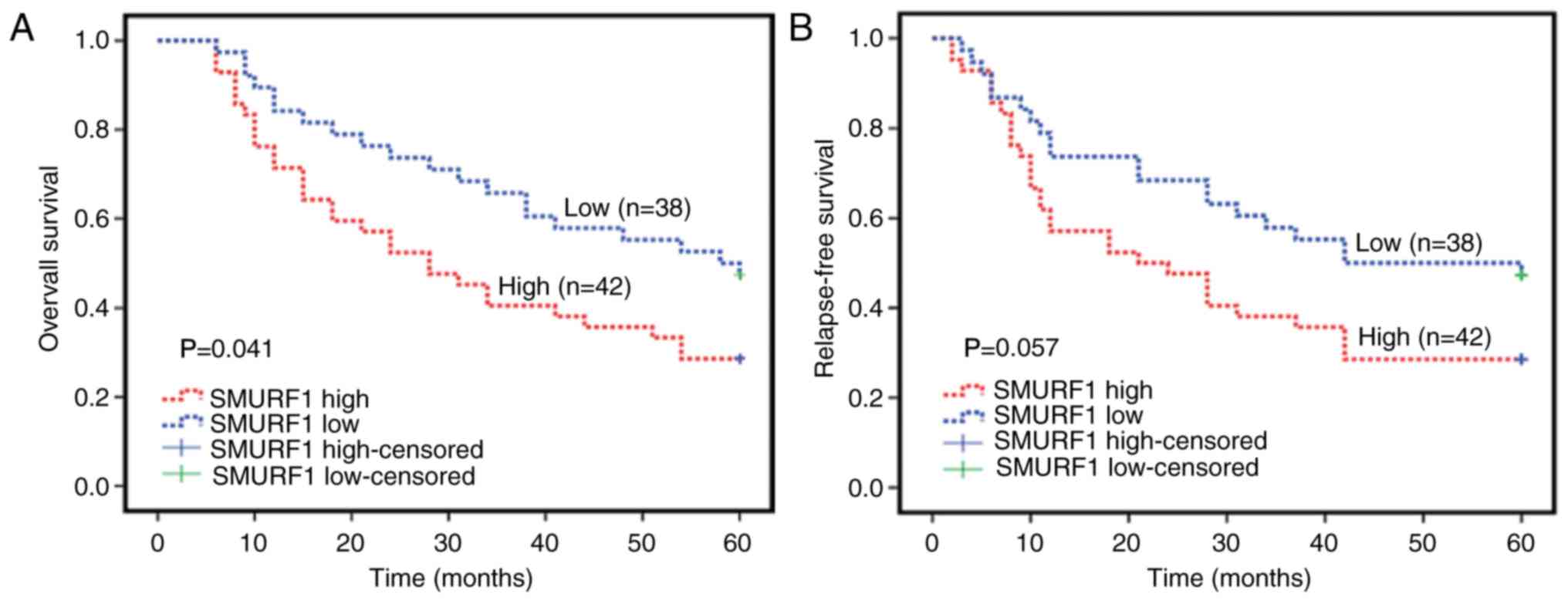
In the present study, Kaplan-Meier survival analyses
were performed to further investigate whether SMURF1 expression is
associated with the 5-year overall survival (OS) and relapse-free
survival (RFS) in patients with OC. The results revealed that
patients with lower SMURF1 expression levels exhibited longer
durations of survival, whereas the patients with higher SMURF1
expression levels had shorter duration of survival (P=0.041;
Fig. 4A). Additionally, the present
study reported an association between higher SMURF1 expression
levels with shorter RFS; however, statistical significance was not
observed (P=0.057; Fig. 4B).
Discussion
SMURF1, a member of the HECT family of E3 ubiquitin
ligases, is involved in the regulation of numerous pathological
processes (22,23). The findings of the present study
indicated that SMURF1 may promote ovarian cancer (OC) cell
migration and invasion, and that overexpression of SMURF1 in OC
tissues may facilitate the progression of OC.
SMURF1 has also been suggested as a promoter of
metastasis in numerous human malignancies. In breast cancer,
overexpression of the E3 ubiquitin ligase SMURF1 led to RhoA
ubiquitination and degradation (7).
Degradation of the small GTPase RhoA in tumor cells disrupted
F-actin cytoskeletal organization, reduced cell adhesion, increased
cell migration and invasion and promoted breast cancer progression
and metastasis (7). In prostate
cancer, SMURF1 was associated with androgen-induced cell migration
and invasion. Downregulating SMURF1 completely inhibited the
invasive ability of C4-2 cells (24). In the cervical cancer cell line,
HeLa and the breast cancer cell line MCF-7, SMURF1 was reported to
function as an upstream oncogenic factor, which negatively
regulated the antimetastatic factor, DAB2IP for the control of
aberrant tumor cell growth and migration (12). In gastric cancer, SMURF1 promoted
cancer cell migration and invasion by suppressing the expression of
double C2 protein/DAB2IP (25). Our
previous study reported SMURF1 as a direct target of microRNA
(miR)-497, in which miR-497 inhibited OC cell migration and
invasion (6). These findings
indicated a potential role of SMURF1 in cancer metastasis. In the
present study, it was observed that the expression of SMURF1 was
significantly increased in OC cells, which exhibited high invasive
potentials. The inhibition of SMURF1 expression suppressed the
invasion and migration of OC cells. In human OC specimens, SMURF1
was significantly overexpressed in the present study. Furthermore,
high expression levels of SMURF1 were associated with aggressive
tumor characteristics and poor OS of patients with OC. These
findings support the role of SMURF1 in promoting the progression of
OC; however, the regulatory mechanism of constitutive activation of
SMURF1 in OC remains unknown.
RhoA, a member of the Rho homolog family of small
GTPases, promotes the reorganization of the actin cytoskeleton and
regulates cell shape, attachment and motility (26). Increased activation of RhoA has been
associated with tumor cell proliferation and metastasis (27,28).
In OC, the activation of RhoA GTPase and downstream ROCK led to
enhanced cell invasion and migration (29,30).
The suppressive effects of ROCK inhibitors, as well as macitentan,
have demonstrated that RhoA may be involved in the cell migration
of epithelial OC (31). Via
interactions with a variety of factors, RhoA can modulate the
activities of various signaling pathways, including that of
phosphoinositide 3-kinase (PI3K)/protein kinase B, Ras/Raf and
mitogen-activated protein kinase/extracellular signal-regulated
kinase; loss of RhoA can further dysregulate proliferation and
metastasis-associated pathways, inducing tumor development
(12,32,33).
It was previously demonstrated that SMURF1-mediated ubiquitination
of RhoA was responsible for the precise temporal and spatial
regulation of RhoA, which is required for optimal cell migration
(34). Thus, the present study
investigated whether SMURF1 was involved in the regulation of OC
cell migration and invasion via activation of the RhoA signaling
pathway. Consistent with these previous studies, the present study
reported that reductions in SMURF1 expression decreased OC cell
migration and invasion, and attenuated RhoA-mediated activation of
the RhoA/ROCK signaling pathway. Recent studies have demonstrated
that SMURF1 regulated a variety of signaling networks, including
transforming growth factor β family signaling and the Wnt signaling
pathway (34); however, we only
investigated the role of SMURF1 in the promotion of OC metastasis
via the RhoA/ROCK oncogenic pathway in the present study. We
anticipate that future studies will reveal that SMURF1 may regulate
other signalling pathways within OC cells. The identification of
these pathways may provide novel insight into the function of
SMURF1 in the progression and metastasis of OC.
At present, a few oncogenes have been reported to
possess pro-metastatic functions in OC. Yu et al (35) identified a functional kinase, spleen
tyrosine kinase (SYK), which was associated with OC cell motility
and invasiveness. In addition, the inhibition of SYK significantly
decreased the invasive ability of OC cells. Dorayappan et al
(36) reported that patient-derived
exosomes from ascites-associated OC cells cultured under hypoxic
conditions, exhibited an increased abundance of potent oncogenic
proteins, including signal transducer and activator of
transcription 3 and Fas, which are capable of significantly
increasing cell migration/invasion. Vascular endothelial growth
factor (VEGF)-C and VEGF-D were proposed to contribute to
tumor-associated lymphatic vessel growth, enhancing the metastatic
spread of tumor cells to lymph nodes. Secreted protein acidic and
rich in cysteine functions as a tumor suppressor by inhibiting
lymph node metastasis via the regulation of VEGF-C/D expression in
OC cells (37). Formyl peptide
receptor 2 (FPR2) expression levels were upregulated in OC cells.
The inhibition of FPR2 was observed to reduce the migration and
invasion of OC cells (38). Park
et al (39) reported that
Toll-like receptor 5/7-mediated PI3K activation induced
epithelial-mesenchymal transition and the metastasis of OC cells
via the expression of Wiskott-Aldrich syndrome protein family
member 3-dependent mesothelin or POU domain class 5 transcription
factor 1/SRY-box 2. The results of the present study revealed that
SMURF1 promoted OC cell metastasis via the activation of the
RhoA/ROCK signaling pathway. This finding indicated that SMURF1 may
be a promising molecular target for the optimization of individual
therapeutic approaches for patients with OC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Natural Science Foundation of Henan Province of China (grant no.
182300410358).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WW, HD, HL and FH conducted the data acquisition and
the patient follow-up. WW and GL contributed to the conception and
design, the data analysis and the interpretation of the results. WW
and GL wrote, reviewed and edited the manuscript. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The present study was approved by the Life Sciences
Ethics Committee of Zhengzhou University (Zhengzhou, China) and
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ng JS, Low JJ and Ilancheran A: Epithelial
ovarian cancer. Best practice & research. Clin Obstet Gynaecol.
26:337–345. 2012.
|
|
3
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wen M, Ma X, Cheng H, Jiang W, Xu X, Zhang
Y, Zhang Y, Guo Z, Yu Y, Xu H, et al: Stk38 protein kinase
preferentially inhibits TLR9-activated inflammatory responses by
promoting MEKK2 ubiquitination in macrophages. Nat Commun.
6:71672015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee MG, Jeong SI, Ko KP, Park SK, Ryu BK,
Kim IY, Kim JK and Chi SG: RASSF1A directly antagonizes RhoA
activity through the assembly of a smurf1-mediated destruction
complex to suppress tumorigenesis. Cancer Res. 76:1847–1859. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang W, Ren F, Wu Q, Jiang D, Li H, Peng
Z, Wang J and Shi H: MicroRNA-497 inhibition of ovarian cancer cell
migration and invasion through targeting of SMAD specific E3
ubiquitin protein ligase 1. Biochem Biophys Res Commun.
449:432–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu L, Liu X, Cui K, Di Y, Xin L, Sun X,
Zhang W, Yang X, Wei M, Yao Z, et al: SND1 acts downstream of TGFβ1
and upstream of Smurf1 to promote breast cancer metastasis. Cancer
Res. 75:1275–1286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khammanivong A, Gopalakrishnan R and
Dickerson EB: SMURF1 silencing diminishes a CD44-high cancer stem
cell-like population in head and neck squamous cell carcinoma. Mol
Cancer. 13:2602014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang C, Rajfur Z, Yousefi N, Chen Z,
Jacobson K and Ginsberg MH: Talin phosphorylation by Cdk5 regulates
Smurf1-mediated talin head ubiquitylation and cell migration. Nat
Cell Biol. 11:624–630. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu C, Billadeau DD, Abdelhakim H, Leof E,
Kaibuchi K, Bernabeu C, Bloom GS, Yang L, Boardman L, Shah VH, et
al: IQGAP1 suppresses TβRII-mediated myofibroblastic activation and
metastatic growth in liver. J Clin Invest. 123:1138–1156. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kwon A, Lee HL, Woo KM, Ryoo HM and Baek
JH: SMURF1 plays a role in EGF-induced breast cancer cell migration
and invasion. Mol Cells. 36:548–555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Dai X, Wan L, Inuzuka H, Sun L and
North BJ: Smurf1 regulation of DAB2IP controls cell proliferation
and migration. Oncotarget. 7:26057–26069. 2016.PubMed/NCBI
|
|
13
|
Wang Z, Wang J, Li X, Xing L, Ding Y, Shi
P, Zhang Y, Guo S, Shu X and Shan B: Bortezomib prevents
oncogenesis and bone metastasis of prostate cancer by inhibiting
WWP1, Smurf1 and Smurf2. Int J Oncol. 45:1469–1478. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kwei KA, Shain AH, Bair R, Montgomery K,
Karikari CA, van de Rijn M, Hidalgo M, Maitra A, Bashyam MD and
Pollack JR: SMURF1 amplification promotes invasiveness in
pancreatic cancer. PLoS One. 6:e239242011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Javadi S, Ganeshan DM, Qayyum A, Iyer RB
and Bhosale P: Ovarian cancer, the revised FIGO staging system, and
the role of imaging. AJR Am J Roentgenol. 206:1351–1360. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Liang WJ, Min GT, Wang HP, Chen W
and Yao N: LTBP2 promotes the migration and invasion of gastric
cancer cells and predicts poor outcome of patients with gastric
cancer. Int J Oncol. 52:1886–1898. 2018.PubMed/NCBI
|
|
18
|
Chen J, Wang L, Matyunina LV, Hill CG and
McDonald JF: Overexpression of miR-429 induces
mesenchymal-to-epithelial transition (MET) in metastatic ovarian
cancer cells. Gynecol Oncol. 121:200–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shaw TJ, Senterman MK, Dawson K, Crane CA
and Vanderhyden BC: Characterization of intraperitoneal,
orthotopic, and metastatic xenograft models of human ovarian
cancer. Mol Ther. 10:1032–1042. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kataoka K and Ogawa S: Variegated RHOA
mutations in human cancers. Exp Hematol. 44:1123–1129. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Chu S, Yao S, Li Y, Fan S, Sun X,
Su L and Liu X: CD74 interacts with CD44 and enhances tumorigenesis
and metastasis via RHOA-mediated cofilin phosphorylation in human
breast cancer cells. Oncotarget. 18:68303–68313. 2016.
|
|
22
|
David D, Nair SA and Pillai MR: Smurf E3
ubiquitin ligases at the cross roads of oncogenesis and tumor
suppression. Biochim Biophys Acta. 1835:119–128. 2013.PubMed/NCBI
|
|
23
|
Cao Y and Zhang L: A Smurf1 tale: function
and regulation of an ubiquitin ligase in multiple cellular
networks. Cell Mol Life Sci. 70:2305–2317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gang X, Wang G and Huang H: Androgens
regulate SMAD ubiquitination regulatory factor-1 expression and
prostate cancer cell invasion. Prostate. 75:561–572. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tao Y, Sun C, Zhang T and Song Y: SMURF1
promotes the proliferation, migration and invasion of gastric
cancer cells. Oncol Rep. 38:1806–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blangy A: Tensins are versatile regulators
of Rho GTPase signaling and cell adhesion. Biol Cell. 109:115–126.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang HR, Nam S, Lee J, Kim JH, Jung HR,
Park HS, Park S, Ahn YZ, Huh I, Balch C, et al: Systematic approach
identifies RHOA as a potential biomarker therapeutic target for
Asian gastric cancer. Oncotarget. 7:81435–81451. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Xu T, Zou H, Chen X, Sun D and Yang
M: Cell migration microfluidics for electrotaxis-based
heterogeneity study of lung cancer cells. Biosens Bioelectron.
89:837–845. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Semprucci E, Tocci P, Cianfrocca R,
Sestito R, Caprara V, Veglione M, Castro VD, Spadaro F, Ferrandina
G, Bagnato A, et al: Endothelin A receptor drives invadopodia
function and cell motility through the beta-arrestin/PDZ-RhoGEF
pathway in ovarian carcinoma. Oncogene. 35:3432–3442. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paul NR, Allen JL, Chapman A,
Morlan-Mairal M, Zindy E, Jacquemet G, Fernandez del Ama L,
Ferizovic N, Green DM, Howe JD, et al: α5β1 integrin recycling
promotes Arp2/3-independent cancer cell invasion via the formin
FHOD3. J Cell Biol. 210:1013–1031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tocci P, Caprara V, Cianfrocca R, Sestito
R, Di Castro V, Bagnato A and Rosanò L: Endothelin-1/endothelin A
receptor axis activates RhoA GTPase in epithelial ovarian cancer.
Life Sci. 159:49–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zandvakili I, Lin Y, Morris JC and Zheng
Y: Rho GTPases: Anti- or pro-neoplastic targets? Oncogene.
36:3213–3222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jansen S, Gosens R, Wieland T and Schmidt
M: Paving the Rho in cancer metastasis: Rho GTPases and beyond.
Pharmacol Ther. 183:1–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang C: Roles of E3 ubiquitin ligases in
cell adhesion and migration. Cell Adh Migr. 4:10–18. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu Y, Suryo Rahmanto Y, Lee MH, Wu PH,
Phillip JM, Huang CH, Vitolo MI, Gaillard S, Martin SS, Wirtz D, et
al: Inhibition of ovarian tumor cell invasiveness by targeting SYK
in the tyrosine kinase signaling pathway. Oncogene. 37:3778–3789.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dorayappan KD, Wanner R, Wallbillich JJ,
Saini U, Zingarelli R, Suarez AA, Cohn DE and Selvendiran K:
Hypoxia-induced exosomes contribute to a more aggressive and
chemoresistant ovarian cancer phenotype: A novel mechanism linking
STAT3/Rab proteins. Oncogene. 37:3806–3821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peng F, Zhong Y, Liu Y, Zhang Y, Xie Y, Lu
Y, Zhang X and Li D: SPARC suppresses lymph node metastasis by
regulating the expression of VEGFs in ovarian carcinoma. Int J
Oncol. 51:1920–1928. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie X, Yang M, Ding Y, Yu L and Chen J:
Formyl peptide receptor 2 expression predicts poor prognosis and
promotes invasion and metastasis in epithelial ovarian cancer.
Oncol Rep. 38:3297–3308. 2017.PubMed/NCBI
|
|
39
|
Park GB and Kim D: TLR5/7-mediated PI3K
activation triggers epithelial-mesenchymal transition of ovarian
cancer cells through WAVE3-dependent mesothelin or OCT4/SOX2
expression. Oncol Rep. 38:3167–3176. 2017. View Article : Google Scholar : PubMed/NCBI
|