Introduction
As the leading cause of cancer-related deaths
worldwide, lung adenocarcinoma (LAD), which is the most prevalent
subtype of lung cancer, accounts for ~14% of all neoplasms and is
estimated to have produced >150,000 deaths in the last year
(1). It is common to for distant
metastasis to occur in patients with LAD, including bone
metastasis, cutaneous metastasis, thyroid metastasis and brain
metastasis (2–5). Despite comprehensive treatment
approaches, including chemotherapy, radiotherapy, surgical
resection and molecular targeted therapy, the 5-year survival rate
of LAD remains unsatisfactory (6,7).
Therefore, understanding the molecular mechanisms and pathways of
LAD metastasis is important to increase the treatment efficacy and
improve the prognosis of patients with LAD.
Long non-coding RNAs (lncRNAs) are RNA transcripts
with a length of >200 nucleotides. lncRNAs are involved in
multiple cancer-related biological progresses, including
proliferation, apoptosis, drug resistance, epithelial-mesenchymal
transition and metastasis (8–12). The
lncRNA differentiation antagonizing non-protein coding RNA (DANCR)
is 1,189-bp nucleotides in length and is located at chromosome
4q12. DANCR has been reported to act as an oncogene in various
types of cancer (13–16). Currently, few studies have
investigated DANCR in LAD. Lu et al reported that DANCR
contributed to LAD progression by sponging microRNA (miR)-496 to
modulate mTOR expression (17). It
is well established that lncRNAs act via regulation of different
downstream genes. The detailed mechanism of how DANCR functions in
LAD requires further exploration.
High mobility group AT-hook 2 (HMGA2) is part of the
HMGA protein family and is encoded by the HMGA2 gene located at
chromosome 12q13-15. HMGA1a, HMGA1b and HMGA2 have analogous
structures, and HMGA2 is well preserve evolutionarily (18,19).
HMGA2 has been widely reported as a key regulator in multiple
malignant tumor types, including gastric, thyroid and colorectal
cancer, as well as esophageal squamous cell carcinoma and lung
cancer (20–25). Meyer et al reported that
HMGA2 was overexpressed in non-small cell lung cancer (NSCLC) and
served as a molecular marker for lung cancer (26). Xu et al reported that
angiogenin promoted the migration, invasion and proliferation
capacity of squamous cell carcinoma of lung cells by directly
upregulating HMGA2 (27). Li et
al demonstrated that lncRNA nuclear paraspeckle assembly
transcript 1 facilitated cell growth and invasion by upregulation
of HMGA2 in breast cancer (28).
Presently, whether DANCR regulates HMGA2 to mediate metastasis
remains unclear.
In the present study, DANCR and HMGA2 were
overexpressed in LAD and were involved in the invasion of LAD
cells. Additionally, HMGA2 was revealed to be a downstream gene of
DANCR. DANCR promoted invasion via upregulation of HMGA2 in LAD
cells.
Materials and methods
Patients and tissue samples
Specimens, 45 LAD tissue and paired para-tumor
samples, were collected during tumorectomy at the Central Hospital
Affiliated to Shenyang Medical College (Shenyang, China) between
August 2012 and August 2017. Written informed consent was provided
by all patients whose tissue was used in the present study. The
Institute Research Medical Ethics Committee of Central Hospital
Affiliated to Shenyang Medical College granted approval for this
study. All 45 cases were diagnosed based on a definite pathological
diagnosis and the clinical stages of these patients were determined
according to the tumor node metastasis (TNM) classification (8th
edition) of the International Union Against Cancer (UICC).
Cell culture
Human normal bronchial epithelial cell line 16HBE,
and human LAD cell lines, SPCA1, A549, H1299 and H1975, were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China). 16HBE cells were
cultured in Airway Epithelial Cell Basal Medium (American Type
Culture Collection, Manassas, VA, USA) A549 cells were cultured in
F-12K medium (ATCC), and SPCA1, H1299 and H1975 cells were cultured
in RPMI-1640 medium (ATCC). All culture media were supplemented
with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), 100 IU/ml penicillin (Baomanbio, Shanghai,
China) and 100 mg/ml streptomycin (Baomanbio). All cell lines were
cultured at 37°C in a humidified atmosphere containing 5%
CO2.
Reverse transcription and quantitative
real-time PCR (qRT-PCR)
The procedure was performed as previously described
(11). Total RNAs from tissue
specimens and cell lines were extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). A
Takara RNA PCR kit (Takara Biotechnology Co., Ltd., Dalian, China)
was use to synthesize cDNA according to the manufacturer's
instructions. The condition for reverse transcription was 37°C for
15 min and 85°C for 5 sec. PCR reactions containing SYBR Premix Ex
Taq II (Takara Biotechnology Co., Ltd.) were performed according to
the manufacturer's instructions. PCR amplification condition was
shown as below: 95°C for 5 min, 38 cycles of 95°C for 5 sec, 61°C
for 30 sec. GAPDH was used as an internal control to assess the
expression levels of the DANCR and HMGA2. The following primer
sequences were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China): DANCR forward, 5′-GCGCCACTATGTAGCGGGTT-3′ and
reverse, 5′-TCAATGGCTTGTGCCTGTAGTT-3′; HMGA2 forward,
5′-TCTCCTGAGCAGGCTTCTTC-3′ and reverse, 5′-AAGGCAGCAAAAACAAGAGC-3′;
GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse,
5′-TGGTGAAGACGCCAGTGGA-3′.
Oligonucleotide transfection
Effective small interfering RNA (siRNA)
oligonucleotides that targeted DANCR (accession no. NR_024031.2;
siDANCR-01 and siDANCR-02) and HMGA2 (accession no. NM_001300918;
siHMGA2-01 and siHMGA2-02) and a corresponding control siRNA
(si-con) were synthesized by Guangzhou RiboBio Co., Ltd. Full
length DANCR and HMGA2 fragments were amplified and cloned into the
pcDNA3.1 vector to create DANCR and HMGA2 overexpression plasmids
(oe-DANCR and oe-HMGA2) synthesized by Guangzhou RiboBio Co., Ltd.
The sequences of the siRNAs were as follows: siDANCR-01,
GGUAAAGUUAAUUGACUAA; siDANCR-02, GGUAUUUCAAUUGACUUAA; siHMGA2-01,
GGGCAAUCUUAUAUAUCUA; siHMGA2-02, GGAAAGUGUCUUCAAACAA. When SPCA1
and A549 cells reached 70% confluence, the plasmids were
transfected into the cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions, as previously reported (29).
Transwell assay
The invasion assay was performed as previously
described (30). Briefly, SPCA1 and
A549 cells were seeded on the Matrigel-coated upper chambers of
Transwell inserts (BD Biosciences, Franklin Lakes, NJ, USA).
Culture medium with and without 10% FBS was supplemented into the
lower and upper chambers, respectively, and incubated for 24 h. The
subsequent day, the non-invaded cells were wiped from the membrane.
Then the membranes were fixed in 90% alcohol and crystal violet
staining followed. Five random fields were counted per chamber
using an inverted microscope (Olympus Corp., Tokyo, Japan).
Western blot analysis
Total proteins were isolated using
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck
KGaA) and qualified using a bicinchoninic acid assay detecting kit
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) according to the
manufacturer's protocol. Proteins samples (50 µg/well) were
subjected to 10% SDS-PAGE and transferred onto a polyvinylidene
difluoride membranes. Anti-HMGA2 and anti-GAPDH antibodies
(dilution 1:1,000 for anti-HMGA2; cat. no. ab97276; and dilution
1:500 for anti-GAPDH; cat. no. ab205718; Abcam, Cambridge, UK) were
applied and incubated with the membranes at 4°C overnight. The
following day, the membranes were incubated with secondary
antibodies (dilution 1:2,000; cat. no. ab205718; Abcam) for 1 h at
room temperature. Protein bands were detected on X-ray film using
an enhanced chemiluminescence detection system.
Immunohistochemical (IHC)
staining
The IHC procedure was performed as previously
described (31). The LAD tissue
specimens were processed as follows: 4% paraformaldehyde fixation,
paraffin-embedding, sectioned to 4-µm thickness, deparaffinization,
rehydration, hydrogen peroxide incubation, antigen retrieval,
blocked in 10% goat serum (Bioworld Technology, Inc., St. Louis
Park, MN, USA), primary antibody incubation (anti-HMGA2 and
anti-GAPDH) at 4°C overnight, secondary antibody incubation (goat
anti-rabbit IgG H&L; Abcam) at 37°C for 20 min,
streptavidin-horseradish peroxidase complex incubation,
diaminobenzidine tetrahydrochloride (MedChemExpress, Monmouth
Junction, NJ, USA) staining, hematoxylin (Amresco, LLC, Solon, OH,
USA) and counterstaining. All sections were assessed individually
by two experienced pathologists.
Statistical analysis
All experiments were repeated in triplicate and all
data from three independent experiments are presented as the mean ±
standard deviation. GraphPad Prism software v5.0 (GraphPad
Software, Inc., La Jolla, CA, USA) and SPSS 19.0 statistical
software (IBM Corp., Armonk, NY, USA) were used for statistical
analysis. Association between DANCR and clinicopathological
features of patients with LAD was analyzed using the Pearson's
Chi-square test. Survival analysis was performed using the log-rank
test in GraphPad Prismdv5.0. Differences between two groups were
analyzed using the Student's t-test or Student-Newman-Keuls method
(S-N-K) method. P<0.05 was considered to indicate a
statistically significant difference.
Results
DANCR is upregulated and associated
with poor prognosis in patients with LAD
The expression of DANCR in the collected LAD tissue
specimens was determined using qRT-PCR. DANCR was upregulated in
the majority of LAD tissue specimens (39/45, 86.67%) compared with
that in para-tumor tissue specimens (Fig. 1A and B). Additionally, DANCR was
notably higher in patients with lymph node metastasis (N1 and N2)
compared to patients without lymph node metastasis (N0; Fig. 1C). Furthermore, the association
between the elevated DANCR expression and the clinicopathological
features in patients with LAD was analyzed. As displayed in
Fig. 1D and Table I, elevated DANCR was associated with
a shorter survival rate (determined by Kaplan-Meier analysis), an
advanced TNM stage (IIIa; P=0.013), lymph node metastasis (P=0.001)
and a larger tumor size (P=0.023). Finally, the expression level of
DANCR was assessed in a normal human bronchial epithelial cell
line, 16HBE, and in four human LAD cell lines, SPCA1, A549, H1299
and H1975. DANCR was significantly upregulated in the four LAD cell
lines (particularly in SPCA1 and A549) compared with 16HBE cells
(Fig. 1E). Collectively, the
results indicated that DANCR may act as an oncogene in LAD.
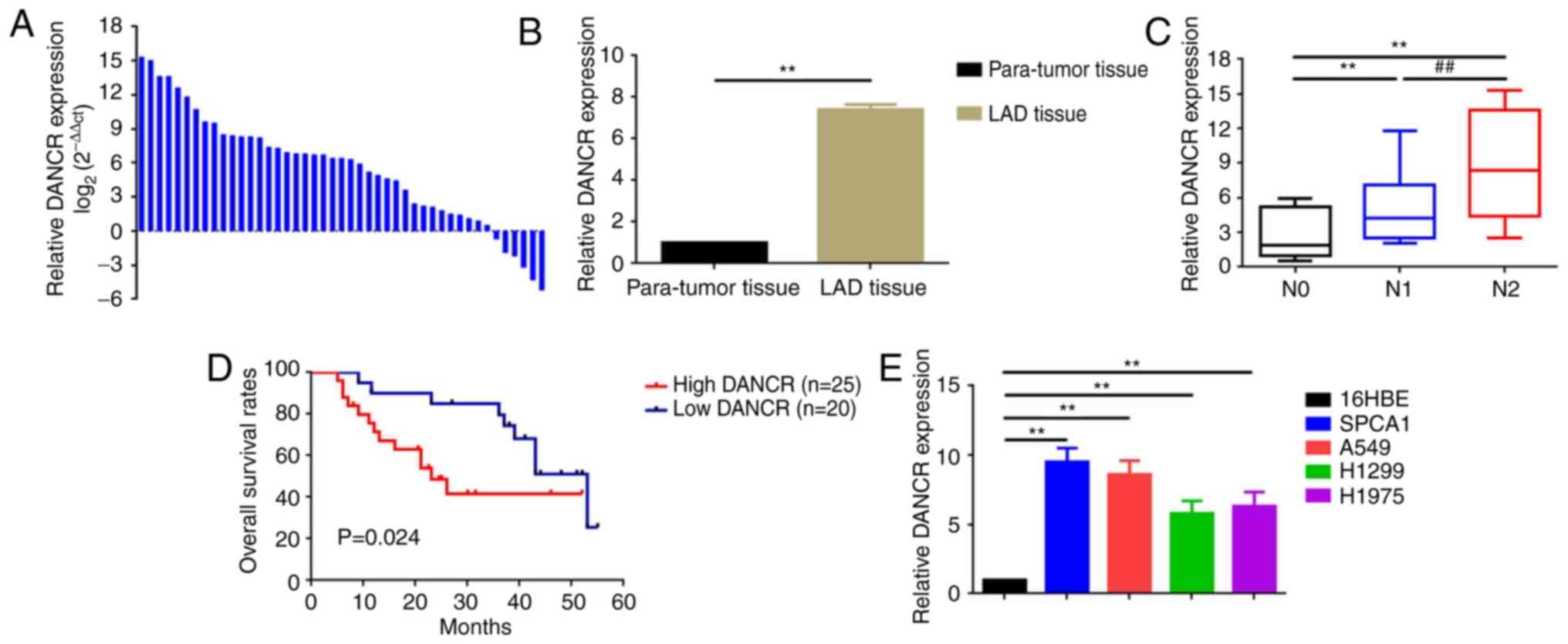 | Figure 1.DANCR is upregulated and correlated
with poor prognosis in patients with LAD. (A and B) Expression of
DANCR is elevated in LAD tissue specimens compared to para-tumor
tissue specimens as determined by qRT-PCR. Data is presented as (A)
log2 (2−ΔΔCt) and (B) ΔCt. **P<0.01 vs.
the para-tumor tissue group. (C) DANCR is upregulated in patients
with lymph node metastasis as detected by qRT-PCR. **P<0.01 vs.
the N0 group and ##P<0.01 vs. the N1 group. (D) OS in
the patients with high DANCR (n=25) was significantly shorter than
that of patients with low DANCR (n=20), P=0.024 as determined by
log-rank test. (E) DANCR expression is upregulated in LAD cell
lines, SPCA1, A549, H1299 and H1975, compared with normal human
bronchial epithelial cell line, 16HBE. **P<0.01 vs. the 16HBE
group. Data are presented as the mean ± standard deviation from
three independent experiments. DANCR, differentiation antagonizing
non-protein coding RNA; LAD, lung adenocarcinoma; qRT-PCR,
quantitative real-time polymerase chain reaction; OS, overall
survival. |
 | Table I.Association of DANCR expression with
clinicopathological features of LAD. |
Table I.
Association of DANCR expression with
clinicopathological features of LAD.
|
|
| DANCR |
|
|---|
|
|
|
|
|
|---|
| Features | No. of cases | High | Low |
P-valuea |
|---|
| Age (years) |
|
|
| 0.841 |
|
≤65 | 24 | 13 | 11 |
|
|
>65 | 21 | 12 | 9 |
|
| Sex |
|
|
| 0.787 |
|
Male | 26 | 14 | 12 |
|
|
Female | 19 | 11 | 8 |
|
| TNM stage |
|
|
| 0.013 |
| I +
II | 20 | 7 | 13 |
|
| III +
IV | 25 | 18 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.001 |
|
Negative | 17 | 4 | 13 |
|
|
Positive | 28 | 21 | 7 |
|
| Tumor size
(cm) |
|
|
| 0.023 |
| ≤5 | 23 | 9 | 14 |
|
|
>5 | 22 | 16 | 6 |
|
| Smoking
history |
|
|
| 0.463 |
|
Smokers | 23 | 14 | 9 |
|
|
Non-smokers | 22 | 11 | 11 |
|
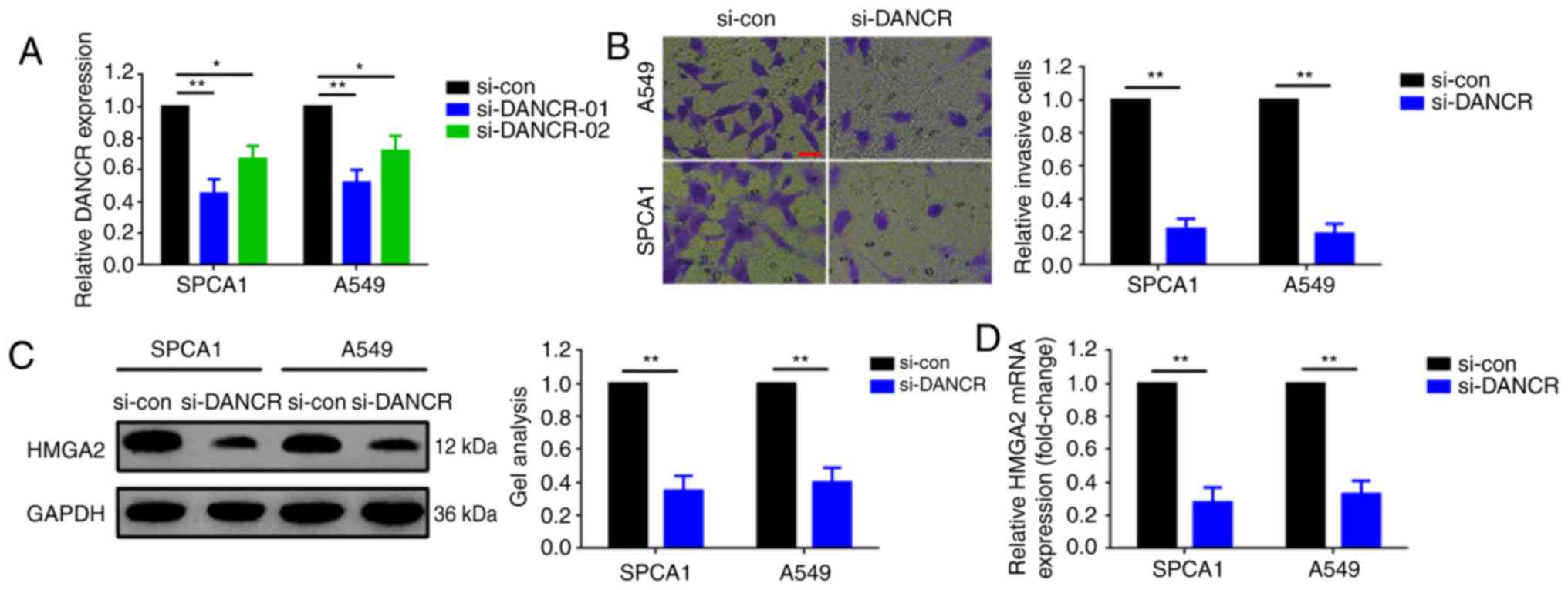
Downregulation of DANCR inhibits
metastasis and HMGA2 expression in SPCA1 and A549 cells
The aforementioned results indicated that elevated
DANCR was associated with lymph node metastasis. Loss-of-function
experiments were then used to elucidate the potential role of DANCR
in SPCA1 and A549 cell invasion. DANCR was knocked down using
si-DANCR in SPCA1 and A549 cells (Fig.
2A). A Transwell assay was then performed to determine the
changes in invasive ability. Downregulation of DANCR significantly
inhibited the invasion of SPCA1 and A549 cells in the Transwell
assay (Fig. 2B).
HMGA2 has been commonly reported as a metastatic
gene in lung cancer. The expression changes of HMGA2 following
DANCR knockdown were also evaluated in the present study. As
presented in Fig. 2C and D,
downregulation of DANCR also inhibited HMGA2 expression at the mRNA
and protein levels.
HMGA2 is overexpressed in LAD and is
involved in SPCA1 and A549 cell metastasis
To further explore the function of HMGA2 in LAD, IHC
was performed to detect HMGA2 expression in LAD tissue specimens.
As presented in Fig. 3A, HMGA2
gradually increased with increasing pathological staging of LAD.
Additionally, the expression of HMGA2 was determined in LAD cell
lines. As revealed in Fig. 3B and
C, compared with 16HBE, HMGA2 was overexpressed in LAD cell
lines, SPCA1, A549, H1299 and H1975. Furthermore, knockdown of
HMGA2 using si-HMGA2 inhibited the invasion of SPCA1 and A549 cells
in a Transwell assay (Fig. 3D and
E).
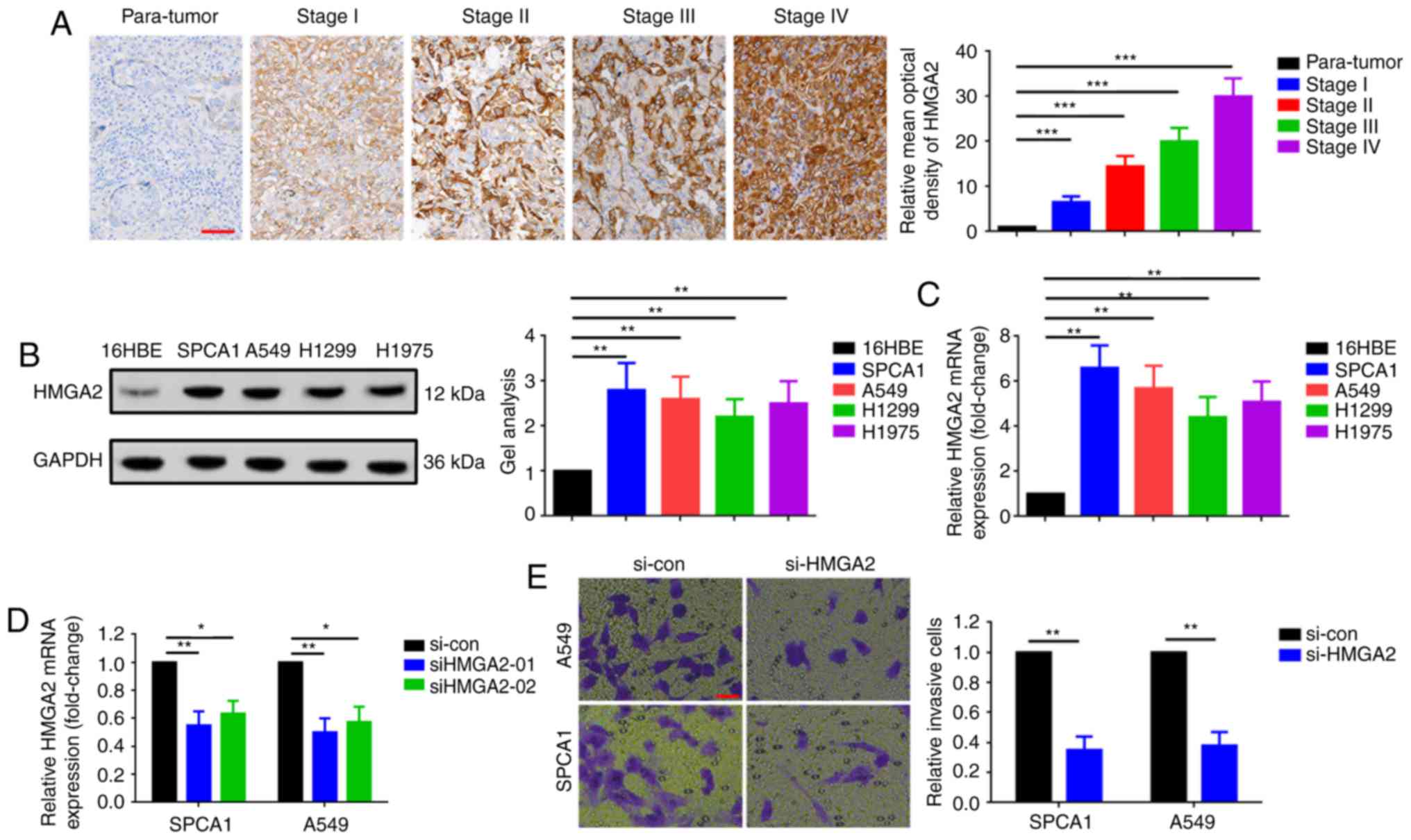 | Figure 3.HMGA2 is overexpressed in LAD and
involved in SPCA1 and A549 cell invasion. (A) HMGA2 was gradually
elevated with advanced staging, as shown in the representative
immunohistochemistry images at each stage. Scale bars, 20 µm;
magnification, ×400. ***P<0.001 vs. para-tumor tissue. (B and C)
Upregulated HMGA2 in SPCA1, A549, H1299 and H1975 cells compared
with 16HBE illustrated by (B) western blotting and (C) qRT-PCR
assay. **P<0.01 vs. 16HBE. (D) HMGA2 in SPCA1 and A549 cells
knocked down by siHMGA2-01 and siHMGA2-02 as confirmed by qRT-PCR.
siHMGA2-01 presented a more effective silencing efficacy and was
used as the silencing tool in subsequent experiments. *P<0.05
and **P<0.01 vs. the si-con group. (E) Knockdown of HMGA2
suppressed the invasive abilities of SPCA1 and A549 cells as
determined by a Transwell assay. Scale bars, 500 µm; magnification,
×4. **P<0.01 vs. the si-con group. Data are presented as the
mean ± standard deviation from three independent experiments.
HMGA2, high mobility group AT-hook 2; LAD, lung adenocarcinoma; si,
small interfering RNA; qRT-PCR, quantitative real-time polymerase
chain reaction. |
HMGA2 is a downstream effector in
DANCR-facilitated metastasis in SPCA1 and A549 cells
Since DANCR and HMGA2 were revealed to be involved
in SPCA1 and A549 cell invasion, the relationship between them was
further explored. An increase and decrease of DANCR positively
regulated HMGA2 expression at the mRNA and protein levels, which
indicated that HMGA2 is a downstream effector of DANCR (Fig. 4A-C). Furthermore, siRNA and
expression vectors were used to knockdown HMGA2 in
DANCR-overexpressed A549 cells and overexpress HMGA2 in
DANCR-silenced SPCA1 cells, as confirmed by qRT-PCR (Fig. 4D and E). Ultimately, a Transwell
assay was used determine the role of HMGA2 in DANCR-mediated
invasion. As revealed in Fig. 4F,
upregulation of DANCR promoted the invasion ability of A549 cells,
but the facilitative effect was attenuated by knockdown of HMGA2
(co-transfection of oe-DANCR and si-HMGA2). Conversely, knockdown
of DANCR inhibited the invasion of SPCA1 cells, however, the
suppressive effect was reversed by HMGA2 overexpression
(co-transfection of si-DANCR and oe-HMGA2; Fig. 4G). The findings strongly indicated
that DANCR promoted the invasion of LAD cells via positive
regulation of HMGA2.
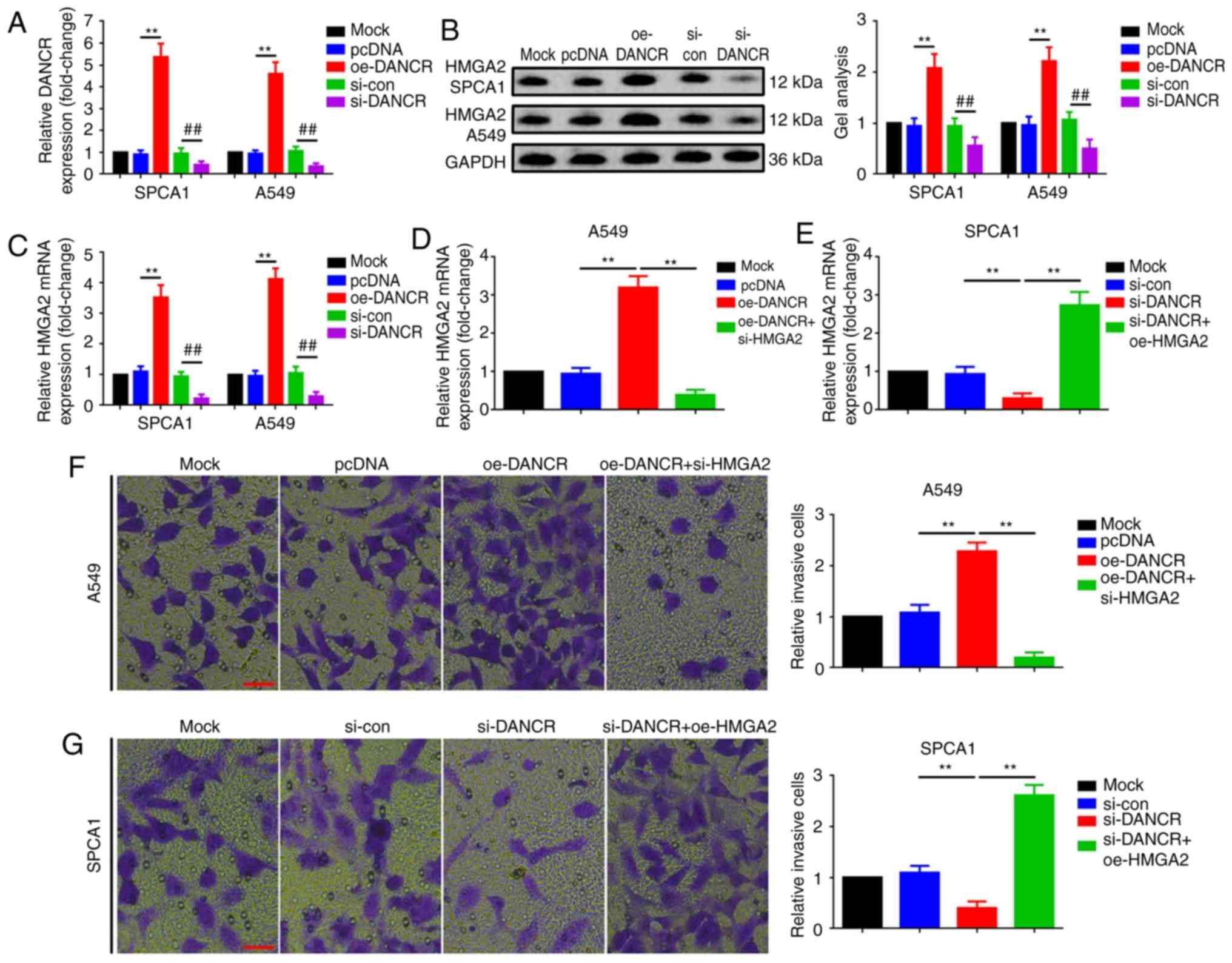 | Figure 4.HMGA2 is a downstream effector in
DANCR-mediated invasion in SPCA1 and A549 cells. (A) DANCR
expression in SPCA1 and A549 cells was up- and downregulated by
oe-DANCR and si-DANCR, as illustrated by qRT-PCR. **P<0.01 vs.
the pcDNA group and ##P<0.01 vs. the si-con group,
individually. (B and C) Up- and downregulation of DANCR positively
regulated HMGA2 at the (B) protein and (C) mRNA level. **P<0.01
vs. the pcDNA group and ##P<0.01 vs. the si-con
group, individually. (D) Upregulation of DANCR promoted HMGA2
expression at the mRNA level and the facilitative effect was
attenuated by si-HMGA2 in A549 cells. **P<0.01 vs. the oe-DANCR
group. (E) Knockdown of DANCR reduced HMGA2 mRNA expression and the
suppressive effect was reversed by a transfection with oe-HMGA2 in
SPCA1 cells, as revealed by qRT-PCR. **P<0.01 vs. the si-DANCR
group. (F) Upregulation of DANCR promoted the invasion of A549
cells and the facilitative effect was attenuated with transfection
of si-HMGA2 in a Transwell assay. **P<0.01 vs. the oe-DANCR
group. Scale bars, 500 µm; magnification, ×4. (G) Knockdown of
DANCR reduced the invasion of SPCA1 cells and the suppressive
effect was reversed by oe-HMGA2 in a Transwell assay. **P<0.01
vs. the si-DANCR group. Scale bars, 500 µm; magnification, ×4. All
data are normalized to the Mock group and presented as the mean ±
standard deviation from three independent experiments. HMGA2, high
mobility group AT-hook 2; DANCR, differentiation antagonizing
non-protein coding RNA; oe, overexpression; si, small interfering
RNA; qRT-PCR, quantitative real-time polymerase chain reaction. |
Discussion
A growing body of evidence has revealed that lncRNAs
have important regulatory roles in various cellular behaviors and
processes (32). DANCR, also termed
ANCR, was initially identified as a non-coding RNA required to
enforce the undifferentiated cell state within the epidermis
(33–35). Currently, increasing evidence
indicates that DANCR is involved in multiple biological processes,
including stem cell differentiation, cell proliferation and cancer
progression (13,16,29,36).
Wang et al reported that DANCR promoted the proliferation,
migration and invasion of NSCLC cell lines, SPC-A1 and H1299, via
regulation of the tumor suppressor miR-758-3p (37). Zhen et al demonstrated that
ectopic DANCR expression induced the proliferation and colony
formation of lung cancer cells, whereas DANCR silencing promoted
the opposing effect (38). In the
present study, it was demonstrated that DANCR was overexpressed in
LAD tissue specimens and in LAD cell lines compared with para-tumor
tissue and a normal lung cell line, respectively. Additionally,
elevated DANCR was associated with more progressive malignant
phenotypes, such as advanced staging (IIIa, P=0.013), larger tumor
size (P=0.023), lymph node metastasis (P=0.001) and shorter
survival time (P=0.024). This phenomenon indicated that DANCR may
be a tumor initiator in LAD. Furthermore, the role of DANCR in the
invasive abilities of SPCA1 and A549 cells was explored in
loss-of-function experiments, which revealed that DANCR promoted
the invasion of LAD cells.
HMGA2 protein is encoded by the HMGA2 gene that has
at least five exons. HMGA2 participates in multiple nuclear
processes including long-range chromatin interactions, chromosome
condensation, inhibition of nucleotide excision repair and
regulation of gene transcription (39). HMGA2 is commonly reported as a
transcriptional modulator that mediates motility and self-renewal
in cancer stem cells (40). Through
in vitro and in vivo study, Fan et al
demonstrated that miR-543 inhibited the proliferation and
metastasis of colorectal cancer cells by targeting KRAS, metastasis
associated 1 and HMGA2 (41). Yang
et al reported that HMGA2 mRNA and protein were highly
expressed in metastatic breast cancer cells and that an inhibition
of protease-activated receptor 1 suppressed HMGA2-driven invasion
in breast cancer cells (42). In a
lung cancer study, Gao et al demonstrated that HMGA2 is a
target of miR-195 and that ectopic expression of HMGA2 increased
the proliferation and migration ability of A549 cells (43). The expression and function of HMGA2
in LAD was also investigated in the present study. HMGA2 was highly
expressed and promoted the invasion of the LAD cell lines, SPCA1
and A549. Knockdown of HMGA2 using siRNA inhibited the invasive
abilities of SPCA1 and A549 cells. Additionally, the relationship
between DANCR and HMGA2 was explored. DANCR regulated the
expression of HMGA2 in a positive manner, and upregulation of DANCR
promoted the invasion LAD cells, but the facilitative effect was
attenuated by HMGA2 silencing. In opposing experiments, elevation
of HMGA2 reversed the suppressive effect of DANCR silencing on LAD
cell line invasion. This phenomenon indicated that HMGA2 is a key
downstream effector in DANCR-mediated invasion of LAD cells.
It is well established that lncRNAs exert their
functions via diverse mechanisms, including post-transcriptional
regulation, genomic imprinting, chromatin remodeling and regulation
of protein activity (44). The
present study, only focused on the expression levels of DANCR and
HMGA2, and the regulative effect that DANCR had on HMGA2. It was
demonstrated that HMGA2 is a downstream effector involved in
DANCR-induced invasion of LAD cells. However, the detailed
molecular mechanism and action sites between DANCR and HMGA2 still
require further exploration. The present study illustrated that a
DANCR/HMGA2 axis may be a novel target for treating LAD.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
SMC General Science Foundation (grant nos. 20181025 and
20181022).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WJ conceived the experiments. NZ performed the
experiments and analyzed the data. WJ wrote the manuscript. Both
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All LAD tissues and matched para-tumor tissues were
in accordance with the ethical guidelines of the Central Hospital
Affiliated to Shenyang Medical College and the Helsinki
declaration. The ethics consents were signed by each patient before
the study. All patients agreed that the data from their samples
could be used for experimental studies and paper presentations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdeen Y, Amireh S, Patel A, Al-Halawani
M, Shaaban H and Miller R: Cutaneous metastasis as a first
presentation for lung adenocarcinoma. N Am J Med Sci. 8:222–225.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dao A, Jabir H, Taleb A, Benchakroun N,
Bouchbika Z, Nezha T, Jouhadi H, Sahraoui S and Benider A: Lung
adenocarcinoma with thyroid metastasis: A case report. BMC Res
Notes. 10:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li H, Lian J, Han S, Wang W, Jia H, Cao J,
Zhang X, Song X, Jia S, Ren J, et al: Applicability of graded
prognostic assessment of lung cancer using molecular markers to
lung adenocarcinoma patients with brain metastases. Oncotarget.
8:70727–70735. 2017.PubMed/NCBI
|
|
5
|
Yu M, Su Y, Cui D, Sun Q, Luan B and Zhao
D: Chemotherapy effectively suppresses interleukin-20, receptor
activator of nuclear factor kappa-B ligand, and osteoprotegerin
levels in patients with lung adenocarcinoma and bone metastasis. J
Cancer Res Ther. 12:963–968. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Behera M, Owonikoko TK, Gal AA, Steuer CE,
Kim S, Pillai RN, Khuri FR, Ramalingam SS and Sica GL: Lung
adenocarcinoma staging using the 2011 IASLC/ATS/ERS classification:
A pooled analysis of adenocarcinoma in situ and minimally invasive
adenocarcinoma. Clin Lung Cancer. 17:e57–e64. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borczuk AC: Prognostic considerations of
the new World Health Organization classification of lung
adenocarcinoma. Eur Respir Rev. 25:364–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen B, Ma J, Li C and Wang Y: Long
noncoding RNA KCNQ1OT1 promotes proliferation and
epithelial-mesenchymal transition by regulation of SMAD4 expression
in lens epithelial cells. Mol Med Rep. 18:16–24. 2018.PubMed/NCBI
|
|
9
|
Wang L, Ma L, Xu F, Zhai W, Dong S, Yin L,
Liu J and Yu Z: Role of long non-coding RNA in drug resistance in
non-small cell lung cancer. Thorac Cancer. 9:761–768. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Lu Z, Wang N, Feng J, Zhang J,
Luan L, Zhao W and Zeng X: Long noncoding RNA DANCR promotes
colorectal cancer proliferation and metastasis via miR-577
sponging. Exp Mol Med. 50:572018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Zhang Y, Yang T, Zhao W, Wang N,
Li P, Zeng X and Zhang W: Long non-coding RNA MALAT1 for promoting
metastasis and proliferation by acting as a ceRNA of miR-144-3p in
osteosarcoma cells. Oncotarget. 8:59417–59434. 2017.PubMed/NCBI
|
|
12
|
Wu X, Zheng Y, Han B and Dong X: Long
noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin
resistance of gastric cancer via sponging miR-361. Biomed
Pharmacother. 99:832–838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma X, Wang X, Yang C, Wang Z, Han B, Wu L
and Zhuang L: DANCR Acts as a diagnostic biomarker and promotes
tumor growth and metastasis in hepatocellular carcinoma. Anticancer
Res. 36:6389–6398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017.PubMed/NCBI
|
|
16
|
Sha S, Yuan D, Liu Y, Han B and Zhong N:
Targeting long non-coding RNA DANCR inhibits triple negative breast
cancer progression. Biol Open. 6:1310–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu QC, Rui ZH, Guo ZL, Xie W, Shan S and
Ren T: lncRNA-DANCR contributes to lung adenocarcinoma progression
by sponging miR-496 to modulate mTOR expression. J Cell Mol Med.
22:1527–1537. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson KR, Lehn DA and Reeves R:
Alternative processing of mRNAs encoding mammalian chromosomal
high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol.
9:2114–2123. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pallante P, Sepe R, Puca F and Fusco A:
High mobility group a proteins as tumor markers. Front Med
(Lausanne). 2:152015.PubMed/NCBI
|
|
20
|
Chang HY, Ye SP, Pan SL, Kuo TT, Liu BC,
Chen YL and Huang TC: Overexpression of miR-194 Reverses
HMGA2-driven Signatures in Colorectal Cancer. Theranostics.
7:3889–3900. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang C, Cao Y, Lei T, Wang Y, Fu J, Wang
Z and Lv Z: microRNA-363-3p inhibits cell growth and invasion of
non-small cell lung cancer by targeting HMGA2. Mol Med Rep.
17:2712–2718. 2018.PubMed/NCBI
|
|
22
|
Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J and
Shi ZZ: miR-125b-5p functions as a tumor suppressor gene partially
by regulating HMGA2 in esophageal squamous cell carcinoma. PLoS
One. 12:e01856362017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Šamija I, Mateša N, Kožaj S, Majstorović
I, Bolanča A and Kusić Z: HMGA2 gene expression in fine-needle
aspiration samples of thyroid nodules as a marker for preoperative
diagnosis of thyroid cancer. Appl Immunohistochem Mol Morphol. Feb
5–2018.(Epub ahead of print). doi: 10.1097/PAI.0000000000000637.
View Article : Google Scholar :
|
|
24
|
Zhu J, Wang H, Xu S and Hao Y:
Clinicopathological and prognostic significance of HMGA2
overexpression in gastric cancer: A meta-analysis. Oncotarget.
8:100478–100489. 2017.PubMed/NCBI
|
|
25
|
Zhuo HC, Song YF, Ye J, Lai GX and Liu DL:
MicroRNA-154 functions as a tumor suppressor and directly targets
HMGA2 in human non-small cell lung cancer. Genet Mol Res. 15:2016.
View Article : Google Scholar :
|
|
26
|
Meyer B, Loeschke S, Schultze A, Weigel T,
Sandkamp M, Goldmann T, Vollmer E and Bullerdiek J: HMGA2
overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu L, Liao WL, Lu QJ, Li CG, Yuan Y, Xu
ZY, Huang SD and Chen HZ: ANG promotes proliferation and invasion
of the cell of lung squamous carcinoma by directly up-regulating
HMGA2. J Cancer. 7:862–871. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Wang S, Li Z, Long X, Guo Z, Zhang
G, Zu J, Chen Y and Wen L: The lncRNA NEAT1 facilitates cell growth
and invasion via the miR-211/HMGA2 axis in breast cancer. Int J
Biol Macromol. 105:346–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Zeng X, Wang N, Zhao W, Zhang X,
Teng S, Zhang Y and Lu Z: Long noncoding RNA DANCR, working as a
competitive endogenous RNA, promotes ROCK1-mediated proliferation
and metastasis via decoying of miR-335-5p and miR-1972 in
osteosarcoma. Mol Cancer. 17:892018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Sun J, Wei X, Luan L, Zeng X, Wang
C and Zhao W: Decrease of miR-622 expression suppresses migration
and invasion by targeting regulation of DYRK2 in colorectal cancer
cells. OncoTargets Ther. 10:1091–1100. 2017. View Article : Google Scholar
|
|
31
|
Wang Y, Yang T, Liu Y, Zhao W, Zhang Z, Lu
M and Zhang W: Decrease of miR-195 promotes chondrocytes
proliferation and maintenance of chondrogenic phenotype via
targeting FGF-18 pathway. Int J Mol Sci. 18(pii): E9752017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kretz M, Webster DE, Flockhart RJ, Lee CS,
Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al:
Suppression of progenitor differentiation requires the long
noncoding RNA ANCR. Genes Dev. 26:338–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang
P, Li Z and Liu Y: PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop
regulates the biological behavior of glioma cells. Theranostics.
8:1084–1105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J,
Chen J, Liu L, Cai H, Li Z, et al: Knockdown of SOX2OT inhibits the
malignant biological behaviors of glioblastoma stem cells via
up-regulating the expression of miR-194-5p and miR-122. Mol Cancer.
16:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Ding R and Xu Y: Effects of long
non-coding RNA SPRY4-IT1 on osteosarcoma cell biological behavior.
Am J Transl Res. 8:5330–5337. 2016.PubMed/NCBI
|
|
36
|
Zhang L, Yang C, Chen S, Wang G, Shi B,
Tao X, Zhou L and Zhao J: Long noncoding RNA DANCR is a positive
regulator of proliferation and chondrogenic differentiation in
human synovium-derived stem cells. DNA Cell Biol. 36:136–142. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang S and Jiang M: The long non-coding
RNA-DANCR exerts oncogenic functions in non-small cell lung cancer
via miR-758-3p. Biomed Pharmacother. 103:94–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX,
Zhao XJ, Lv BL and Liu JB: lncRNA DANCR promotes lung cancer by
sequestering miR-216a. Cancer Control. 25:10732748187698492018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cleynen I and Van de Ven WJ: The HMGA
proteins: A myriad of functions (Review). Int J Oncol. 32:289–305.
2008.PubMed/NCBI
|
|
40
|
Sun J, Sun B, Zhu D, Zhao X, Zhang Y, Dong
X, Che N, Li J, Liu F, Zhao N, et al: HMGA2 regulates CD44
expression to promote gastric cancer cell motility and sphere
formation. Am J Cancer Res. 7:260–274. 2017.PubMed/NCBI
|
|
41
|
Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma
H, Yu D, Maitikabili A, Xiao H, Zhang C, et al: MicroRNA-543
suppresses colorectal cancer growth and metastasis by targeting
KRAS, MTA1 and HMGA2. Oncotarget. 7:21825–21839. 2016.PubMed/NCBI
|
|
42
|
Yang E, Cisowski J, Nguyen N, O'Callaghan
K, Xu J, Agarwal A, Kuliopulos A and Covic L: Dysregulated protease
activated receptor 1 (PAR1) promotes metastatic phenotype in breast
cancer through HMGA2. Oncogene. 35:1529–1540. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gao X, Dai M, Li Q, Wang Z, Lu Y and Song
Z: HMGA2 regulates lung cancer proliferation and metastasis. Thorac
Cancer. 8:501–510. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Akhade VS, Pal D and Kanduri C: Long
noncoding RNA: Genome organization and mechanism of action. Adv Exp
Med Biol. 1008:47–74. 2017. View Article : Google Scholar : PubMed/NCBI
|