Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is a
common type of blood cancer (1).
With advances in medical technologies, there has been a great
improvement in the survival rate of children with B-ALL. However,
certain children experience recurrence and a poor prognosis
following treatment (2,3). Previous studies have confirmed that
the onset of B-ALL is associated with aberrant gene expression
(4,5). Genes that have been identified to be
involved in the pathogenesis of ALL include ABL proto-oncogene 1,
non-receptor tyrosine kinase (ABL1), ABL2, protein tyrosine kinase
2β, Janus kinase (JAK)2, cytokine receptor-like factor 2 (CRLF2)
and colony stimulating factor 1 receptor (6). CRLF2 is located in the Xp22.3/Yp11.3
autosomal regions and is able to interact with interleukin-7
receptor-α. This protein not only serves an important role in the
growth and allergic and inflammatory responses of dendritic cells
and T cells, but is also able to regulate the proliferation and
apoptosis of B cells, which affects the progression of B-ALL
(7–9). Certain studies have reported that high
CRLF2 expression is associated with recurrence and poor prognosis
in children with B-ALL (10). It
has been documented that the activation of the RAC-α
serine/threonine-protein kinase (AKT)/serine/threonine-protein
kinase mTOR (mTOR) signaling pathway is closely associated with the
incidence of B-ALL, and targeted inhibition of AKT/mTOR is of great
significance to the treatment of this disease (11). Certain researchers have reported
that the CRLF2 gene is able to induce the incidence of B-ALL by
affecting the proliferation and differentiation of lymphocytes, via
activation of the JAK signaling pathway (12). However, the association between
CRLF2 and the AKT/mTOR signaling pathway has not been thoroughly
examined. Therefore, the present study aimed to investigate the
effects of CRLF2 on the invasion, migration and apoptosis of the
BaF3 leukemia cell line via the AKT/mTOR pathway.
Materials and methods
Study participants
The present study was approved by the Ethics
Committee of the Yichang Central People's Hospital (Yichang,
China). A total of 128 children (72 males and 56 females) who were
newly diagnosed with B-ALL and treated in Yichang Central People's
Hospital between June 2005 and January 2012 were enrolled in the
study. A further 26 healthy children, who were matched for sex and
age, were included as controls. Children with positive BCR-ABL1 or
Down's syndrome were excluded from the study. With the consent of
the patients and their families, bone marrow specimens and
peripheral blood were collected prior to the initial treatment.
Subsequently, the children with B-ALL were graded and treated
according to the Associazone Italiana Ematologia Oncologia
Pediatrica-Berlin-Frankfurt-Münster ALL-2000 protocol (13). Follow-ups were conducted in the form
of inpatient and outpatient visits and telephone calls. The 4-year
overall survival (OS) and the 3-year event-free survival (EFS)
rates of the children were documented during the follow-ups.
Immunohistochemistry
Frozen bone marrow specimens extracted from children
with B-ALL and healthy normal children were embedded in paraffin at
12–26°C for 10 h, and cut into sections at 5 µm thickness. The
paraffin sections were dried in an oven at 60°C for 30 min and
underwent routine dewaxing and hydration (treated with toluene I,
xylene II, 100 alcohol, 95 alcohol, 85 alcohol and 80% alcohol for
5 min, respectively) followed by rinsing with water for 2 min.
Subsequently, antigen retrieval was performed in 1 mM Tris-EDTA (pH
8.0) with microwave irradiation at 100°C. The sections were cooled
to room temperature and rinsed with PBS three times (5 min/wash).
Following this, 3% H2O2-methanol was added to
blocking endogenous peroxidase activity at room temperature for 10
min, followed by washing twice with PBS (5 min/wash). The primary
antibody against CRLF2 (rabbit anti-human; 5 µg/ml; cat. no.
ab109626; Abcam, Cambridge, UK) was added, while IgG was used as a
negative control. Samples were placed in a refrigerator at 4°C
overnight. On the following day, samples were washed in 0.1% PBS
with Tween-20 (PBST) three times (5 min/wash) and were incubated at
room temperature for 20 min following the addition of streptavidin
biotin-peroxidase complex reagent (SABC). The sections were washed
in 0.1% PBST three times (5 min/wash) and horseradish peroxidase
(HRP)-conjugated secondary IgG antibodies (goat anti-rabbit; cat.
no. ab205718; Abcam) were added for a 30-min incubation at room
temperature, prior to washing again in PBS three times.
Subsequently, the samples were incubated with SABC for 20 min and
were rinsed with 0.1% PBST three times (5 min/wash).
3,3′-Diaminobenzidine staining was conducted for 5 min at room
temperature, followed by a rinse with distilled water to stop the
reaction. The samples were counterstained with hematoxylin at room
temperature for 3 min, rinsed, differentiated in 1% hydrochloric
acid alcohol for 20 sec, and rinsed again until the samples turned
blue. Dehydration was conducted and neutral balsam was used for
mounting. A total of 500 cells were counted under the light
microscope (Olympus CX41; Olympus Corporation, Tokyo, Japan); brown
cells were identified as positive, and samples were graded
according to the percentage of positive cells in all tumor cells.
The criteria for immunohistochemical scoring were as follows:
Negative expression, with a positive cell percentage of <10% (0
point); a positive cell percentage of 10–25% (1 point); a positive
cell percentage of 25–50% (2 points); and a positive cell
percentage of >50% (3 points). A total score of 0–1 indicated
negative expression, 2–4 indicated low expression, 5–8 indicated
moderate expression and 9–12 indicated high expression. Children
with B-ALL were subdivided into a high expression group and a low
expression group (including children with negative, low and
moderate CRRF2 expression), and the CRLF2 protein expression was
compared among B-ALL patients with high and low CRRF2 expression
and healthy normal children.
Clinical data
The white blood cell count (WBC), hemoglobin (HGB)
and platelet count (PLT) in the peripheral blood of patients (prior
to the initial treatment) were measured using an automatic blood
analyzer (cat. no. SK9000; Shenzhen Sinothinker Technology, Co.,
Ltd., Shenzhen, China). The concentration of lactate dehydrogenase
(LDH) was determined using an LDH kit (cat. no. MAK066-1KT;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). MRI was utilized to
examine the degree of tumor infiltration in the liver and spleen.
All these indices were analyzed and compared between the children
with high CRLF2 expression and children with low CRLF2
expression.
Cell culture
BaF3 leukemia cell lines (cat. no. YB-ATCC-2781)
were purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The BaF3 cells were first thawed and
resuscitated, and subsequently placed into a centrifuge tube
containing RPMI-1640 medium without fetal bovine serum (FBS) (cat.
no. PM150110; Procell Life Science and Technology Co., Ltd., Wuhan,
China) for resuspension and centrifugation for 5 min (377 × g;
4°C). The supernatant was removed, and the cells were rinsed once
or twice and seeded in RPMI-1640 cell culture medium containing 15%
FBS (Sigma-Aldrich; Merck KGaA). The cells were incubated at 37°C
in an atmosphere of 5% CO2. The cells were routinely
sub-cultured. Only cells in the exponential phase were selected for
the experiments.
Cell treatment and grouping
BaF3 cells were subdivided into the following five
groups: The Blank group (untreated), the NC group (transfected with
a negative control sequence), the short hairpin (sh)CRLF2 group
(transfected with a CRLF2 shRNA plasmid), the LY294002 group
(treated with an AKT/mTOR pathway-specific inhibitor) and the
siCRLF2 + LY294002 group. The CRLF2 shRNA sequence
(5′-CACCGGAGTTCCGTTATGGTACTGGCGAACCAGTACCATAACGGAACTCC-3′) was
designed online with the BLOCK-iT™ RNAi Designer
(https://rnaidesigner.thermofisher.com/rnaiexpress/design.do).
BaF3 cells were seeded in 6-well plates at a density of
3×105/ml. Once the cells reached 90% confluence, they
were transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). During this
process, 4 µg target plasmid (negative control plasmid and CRLF2
shRNA plasmid; Shanghai GenePharma Co., Ltd., Shanghai, China) and
10 µl Lipofectamine 2000 were separately diluted in 250 µl
serum-free Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) and
mixed gently. After being placed at room temperature for 5 min, the
above two solutions were mixed. The mixture was subsequently added
to the culture wells after 20 min and incubated at 37°C in an
atmosphere of 5% CO2. A total of 6 h subsequently, the
medium was replaced, and the mixture was cultured for a further 48
h prior to collecting the cells. CRLF2 shRNA and negative control
double-stranded RNA were synthesized by Shanghai GenePharma Co.,
Ltd. In addition, the cells were cultured in RPMI-1640 cell culture
medium containing LY294002 (50 µmol/l) when they reached 80%
confluence.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from BaF3 cells in each
group using TRIzol® reagent (cat. no. 15596026; Thermo
Fisher Scientific, Inc.). Ultraviolet spectrophotometry and agarose
gel electrophoresis were performed to detect the RNA concentration,
purity and integrity. One-step RT-qPCR was performed using a
SuperScript® III Platinum® One-Step RT-qPCR
kit (cat. no. 11732088; Thermo Fisher Scientific, Inc.). The
reaction system included SuperScript™ III
RT/Platinum™ Taq Mix (1 µl), 2X Reaction Mix (25 µl),
forward primer (10 µM), reverse primer (10 µM), fluorogenic probe
(10 µM), template (1 µl) and diethyl pyrocarbonate-treated water
(50 µl). Running parameters were set as follows: One cycle at 50°C
for 15 min; one cycle at 95°C for 2 min; and 40 cycles at 95°C for
15 sec and 60°C for 30 sec. GAPDH was used as an internal control
and the mRNA expression was quantified using the 2−ΔΔCq
method (14). Primer sequences are
listed in Table I.
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Gene |
| Primer sequence
(5′→3′) |
|---|
| CRLF2 | Forward |
GTTCAGTAGCGGAGCCCCTTC |
|
| Reverse |
GTTTGGGAGGCGTTGGTGTC |
| AKT | Forward |
TTTATTGGCTACAAGGAACG |
|
| Reverse |
AGTCTGAATGGCGGTGGT |
| mTOR | Forward |
ATGACCAGACCCAGGCTAAG |
|
| Reverse |
GCCAGTCCTCTACAATACGC |
| 4EBP1 | Forward |
GACCTGCCAACCATTCCAG |
|
| Reverse |
CACCTGCCCGCTTATCTTC |
| S6K1 | Forward |
CGTGGAGTCTGCGGCGGGTC |
|
| Reverse |
CGCTCTGCTTTCGTGTGGGC |
| GAPDH | Forward |
GACATCAAGAAGGTGGTGAAGC |
|
| Reverse |
TGTCATTGAGAGCAATGCCAGC |
Western blotting
BaF3 cells from each group were washed with PBS,
resuspended and centrifuged. The supernatant was collected and
radioimmunoprecipitation lysis buffer was added (cat. no. P0013B;
Beyotime Institute of Biotechnology, Haimen, China) and 0.1 mM
phenylmethylsulfonyl fluoride. Following gentle shaking and
resuspension, the cells were incubated on ice for 30 min and
centrifuged for 10 min (241 × g; 4°C). The total cellular protein,
which remained in the supernatant, was subsequently collected. The
protein concentration was determined using a Bicinchoninic Acid
Protein Assay kit (Beyotime Institute of Biotechnology) and the
protein concentration was adjusted to 4 µg/µl. Total cellular
protein (30 µg) was extracted for 10% SDS-PAGE, followed by
transferring to the nitrocellulose (NC) membranes by 50 V wet
electrotransfer and blocked in 5% skim milk [TBS with Tween-20
(TBST) formulation] for 1.5 h. at room temperature. Antibodies were
diluted in primary antibody diluents, according to the
manufacturer's protocol. Primary antibodies included CRLF2 (rabbit
anti-human; 1 µg/ml; cat. no. ab109626; Abcam, Cambridge, UK),
phosphorylated (p)-AKT (rabbit anti-human; 1:1,000; cat. no.
ab52192; Abcam), p-mTOR (rabbit anti-human; 1:1,000; cat. no.
ab109268; Abcam), eukaryotic translation initiation factor
4E-binding protein 1 (4EBP1; rabbit anti-human; 1:5,000; cat. no.
ab32024; Abcam), ribosomal protein S6 kinase β-1 (S6K1; rabbit
anti-human; 1:5,000; cat. no. ab32529; Abcam), and GAPDH (rabbit
anti-human; 1:10,000; cat. no. ab181602; Abcam). The blocked NC
membranes were placed in a plastic container into which the above
antibodies were added, followed by agitation and storage at 4°C for
24 h. The membranes were washed in TBST three times (15 min/wash)
and were subsequently incubated at room temperature for 2 h
following the addition of diluted HRP-labeled secondary antibodies
IgG (goat anti-rabbit; 1:5,000; cat. no. ab205718; Abcam). The
membranes were washed in TBST three times (15 min/wash) and treated
with enhanced chemiluminescence (ECL western blotting substrate
kit; Thermo Fisher Scientific, Inc.). Samples were photographed
using SmartView Pro 2000 (UVCI-2100; Major Science, Saratoga, CA,
USA). Protein banding grayscale analysis was performed using
Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Cell Counting Kit-8 (CCK-8) assay
BaF3 cells from each group were washed with PBS
twice, digested with 0.25% trypsin and dissociated into single-cell
suspensions. Following cell counting, the cells were seeded in
triplicate in 96-well plates at a density of 1×104
cells/well. At 24, 48 and 72 h of incubation, respectively, 10 µl
CCK-8 reagent (cat. no. 40203ES60; Shanghai Yeasen Biotechnology
Co., Ltd., Shanghai, China) was added to each well for a 4-h
incubation. The optical density (OD) of each well was measured at
450 nm with an automated quantitative microplate reader (MK3;
Thermo Fisher Scientific, Inc.). The cell viability curves were
plotted with time on the x-axis and the OD value on the y-axis.
Each test was repeated three times.
Wound healing assay
A total of 48 h after transfection, culture was
resumed with BaF3 cells extracted from each group. When the cells
reached 80–90% confluence, scratches were made with a 200 µl
pipette tip using a ruler. The cells were rinsed with PBS three
times to remove detached cells, and RPMI-1640 medium containing 10%
FBS was added. Samples were subsequently incubated at 37°C in an
atmosphere of 5% CO2. The cells were observed and
photographed at 0 and 48 h under a phase contrast microscope
(Olympus Corporation) at ×100 magnification. The wound healing was
observed dynamically and the scratch width of the cells in each
group was measured. The migration of cells was calculated as
follows: Migration rate = (scratch width at 0 h - scratch width at
48 h)/scratch width at 0 h.
Transwell invasion assay
The Transwell invasion assay was performed according
to the instruction of the Costar 24-Well Transwell™ (EMD
Millipore, Billerica, MA, USA). Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA) which was previously stored at −20°C was defrosted
at 4°C. The Matrigel was placed on ice and diluted 1:1 using
serum-free medium. Following mixing, the mixture was added into the
Transwell chamber (Costar; EMD Millipore.) with 15 µl per well and
coated at 37°C for 1 h, followed by three washes with serum-free
medium. Following complete digestion, BaF3 cells were washed twice
in serum-free medium and counted. The cell suspensions (containing
1×105 cells), along with serum-free DMEM were added into
the upper compartment and diluted to 400 µl, and triplicate wells
were set up for each group. Complete DMEM (600 µl) containing 15%
FBS was added into the lower compartment. Samples were incubated at
37°C in an atmosphere of 5% CO2 for 24 h. Subsequently,
the liquid in the compartment was removed and the cells in the
compartment were wiped away using cotton swabs. The compartment was
immersed in fixation solution (50% methanol) at 4°C for 15 min and
washed with PBS three times. Following staining with crystal violet
for 30 min at room temperature and air-drying, six fields were
randomly selected in each well for photographing and cell counting
under an inverted phase contrast microscope (Olympus Corporation)
at ×200 magnification. The mean cell count in each field was
calculated.
Cell sensitivity to chemotherapeutic
agents
The concentration of BaF3 cells in each group was
adjusted to 0.5×1010/l by adding RPMI-1640 medium. Cells
were cultured in vitro for 24 h following the addition of 25
µmol/l imatinib in each group. Subsequently, the cells were
harvested and the apoptosis of BaF3 cells in each group was
detected using an Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) double staining kit (cat. no. 556547;
Shanghai FuShen Biotechnology Co., Ltd., Shanghai, China). The
procedure for the test was as follows: 10X binding buffer was
diluted 10 times using deionized water and the cells from each
group were centrifuged for 5 min (670 × g; 4°C); subsequently, the
cells were harvested and resuspended with pre-cooled 1X PBS,
followed by centrifugation at for 5–10 min (670 × g; 4°C); next,
the cells were washed and suspended with 1X binding buffer (300
µl); 5 µl Annexin V-FITC was added and mixed, and the mixture was
incubated away from light at room temperature for 15 min; 5 min
prior to the cell analysis by flow cytometry (Cube 6; Sysmex Partec
GmbH, Görlitz, Germany), 5 µl PI was added and the cells were
placed in an ice bath away from light for 5 min. The excitation
wavelength was 480 nm; FITC was detected at 530 nm and PI was
detected at a wavelength >575 nm.
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used for
the statistical analysis. Each experiment was run in triplicate.
Measurement data are expressed as mean ± standard deviation.
Comparisons between the two groups were conducted by t-test and
comparisons across multiple groups were conducted by one-way
analysis of variance with the Bonferroni post hoc test. The OS and
EFS in children with high CRLF2 expression and children with low
CRLF2 expression were compared using a Kaplan-Meier survival curve
and the log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
CRLF2 expression levels in bone
marrow
The immunohistochemical results in bone marrow
tissues from healthy normal children and children with B-ALL are
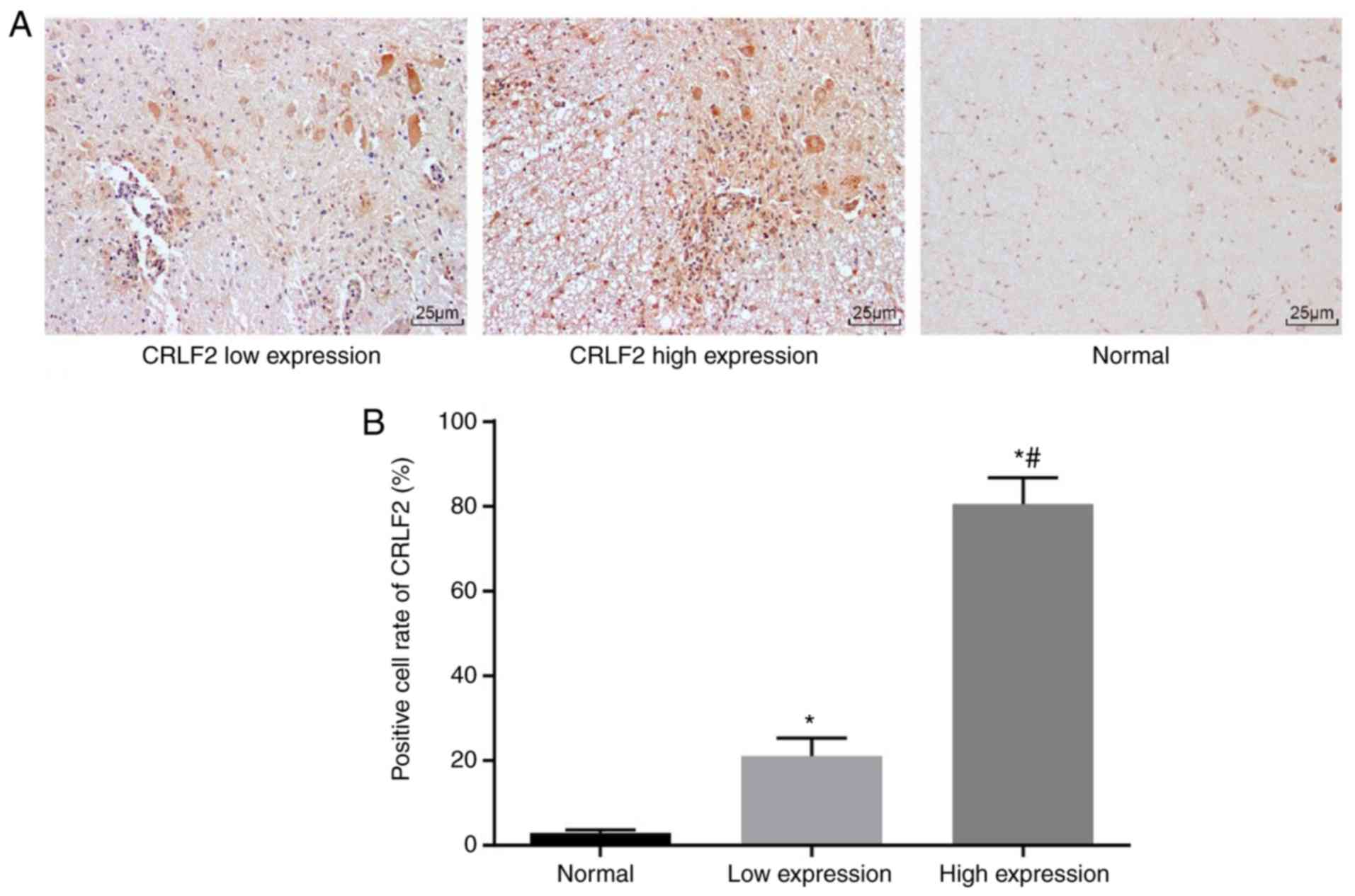
presented in Fig. 1A. The CRLF2
expression was hardly visible in the normal bone marrow tissues. By
contrast, the CRLF2 expression was increased in the bone marrow
tissues from children with B-ALL. The histogram of positive cell
rates of CRLF2 in each group is displayed in Fig. 1B. The CRLF2 expression in the bone
marrow of children with B-ALL was significantly upregulated
compared with that in healthy normal children (P<0.05).
Likewise, among all the children with B-ALL, the upregulation of
CRLF2 expression in the high expression group (n=95) was greater
compared with that in the low expression group (n=33)
(P<0.05).
Factors associated with CRLF2 high
expression
It was identified that the age, sex, PLT count, HGB
concentration, liver and lymph node infiltration indexes, as well
as risk stage were similar between children with low and high CRLF2
expression (all P>0.05), whereas WBC count, LDH concentration
and spleen infiltration indexes were different between these two
groups (all P<0.05; Table
II).
 | Table II.Correlation between CRLF2 expression
and patients' characteristics. |
Table II.
Correlation between CRLF2 expression
and patients' characteristics.
| Clinical
characteristic | High expression of
CRLF2, n=33 (mean ± standard deviation) | Low expression of
CRLF2, n=95 (mean ± standard deviation) | P-value |
|---|
| Age, years | 8±2 | 9±4 | 0.1715 |
| Sex |
|
Male | 21 | 53 | 0.4320 |
|
Female | 12 | 42 |
|
| WBC,
109/l | 47.4±6.3 | 31.2±2.1 | <0.0001 |
| LDH, µg/l | 565±87 | 845±59 | <0.0001 |
| PLT,
109/l | 38.4±9.2 | 36.1±8.1 | 0.1771 |
| HGB, g/dl | 11.2±3.2 | 10.9±1.6 | 0.4858 |
| Extramedullary
infiltration, % |
|
Liver | 15.2±4.7 | 14.8±5.4 | 0.7058 |
|
Spleen | 66.4±5.2 | 30.5±4.3 | <0.0001 |
| Lymph
node | 48.2±6.1 | 45.7±7.5 | 0.3055 |
| Risk stage |
|
Standard risk | 19 | 63 | 0.3670 |
|
High-risk | 14 | 32 |
|
Association between CRLF2 expression
and patient prognosis
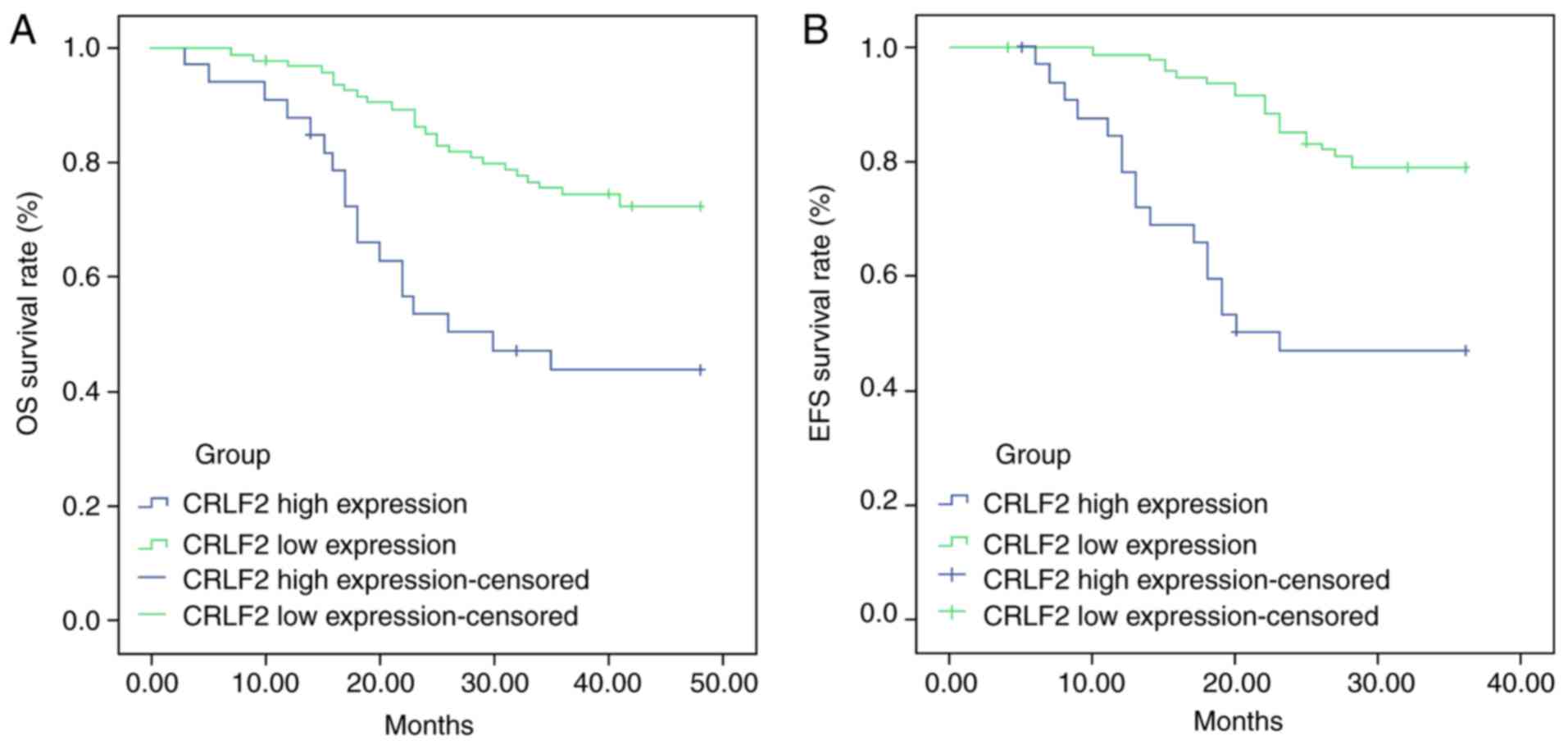
The comparison of OS and EFS in patients with high
CRLF2 expression and low CRLF2 expression is displayed in Fig. 2A and B, respectively. The results of
the analysis indicated that B-ALL patients with high CRLF2
expression had lower OS and EFS rates compared with those with low
expression (45.45 vs. 72.63%, P<0.001; 48.48 vs. 78.95%,
P<0.001, respectively), suggesting that CRLF2 overexpression was
associated with poor prognosis (P<0.001).
Detection of mRNA and protein
expression in BaF3 cell by RT-qPCR and western blotting
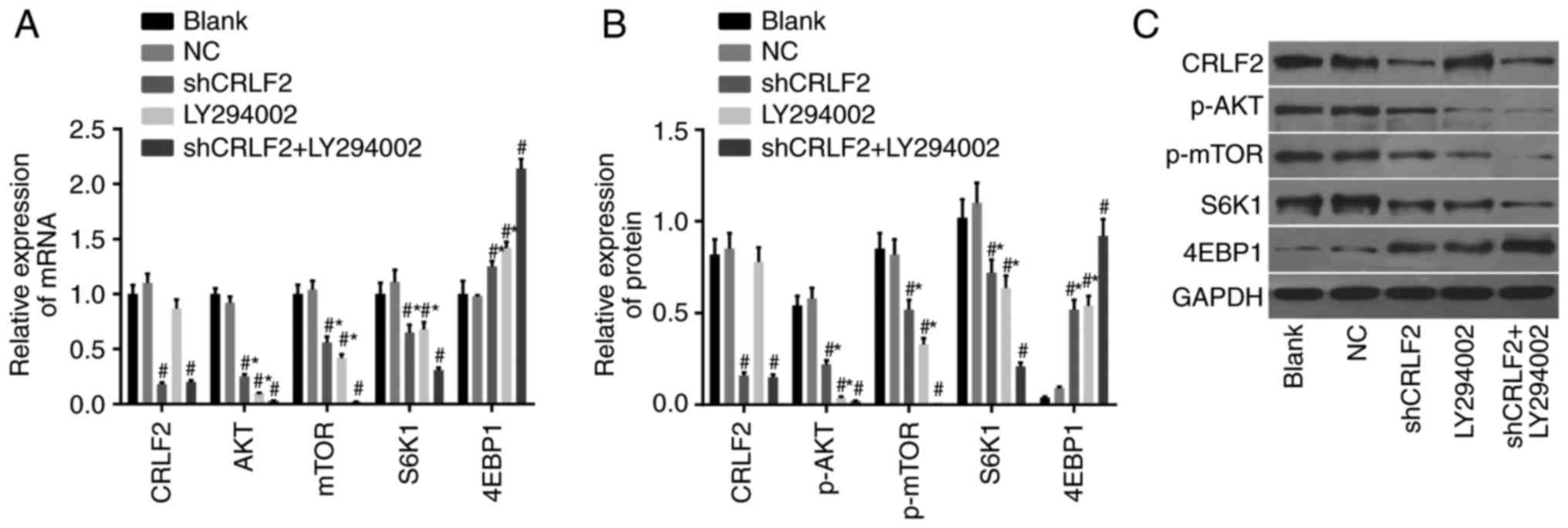
The results of the relative mRNA expression in each
group as assessed by RT-qPCR are presented in Fig. 3A; results of the relative protein
expression in each group, assessed by RT-qPCR, are presented in
Fig. 3B; results of the protein
bands in each group, assessed by western blotting, are displayed in
Fig. 3C. The findings revealed that
compared with the blank group, the expression of CRLF2 protein and
mRNA was significantly downregulated in the shCRLF2 and the shCRLF2
+ LY294002 groups (both P<0.05), whereas the expression levels
in the LY94002 group and the NC group were similar to those in the
blank group (both P>0.05).
A downregulation of the expression levels of AKT,
mTOR and S6K1 mRNAs, and phosphorylated AKT and mTOR proteins, in
addition to an evident upregulation of the expression levels of
4EBP1 mRNA and protein, were observed in the shCRLF2, LY294002 and
shCRLF2 + LY294002 groups compared with the blank group (all
P<0.05). The same patterns were observed in both the shCRLF2 and
the LY294002 group compared with the shCRLF2 + LY294002 group (all
P<0.05). No intergroup differences were observed between the
blank group and the NC group for these indices (all P>0.05;
Fig. 3).
BaF3 cell viability in each group
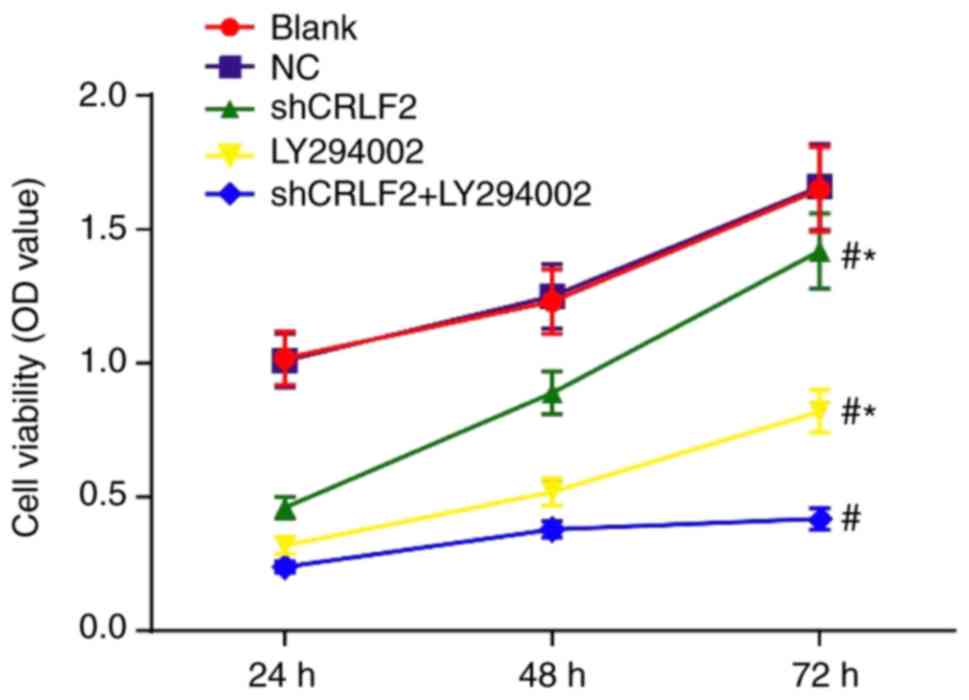
measured by CCK-8 assay
The viability of BaF3 cells at different time points
was measured by CCK-8 assay. The results indicated that the cell
viability in each group increased with the prolongation of culture
time. The cell viability in the shCRLF2, LY294002 and shCRLF2 +
LY294002 groups was inhibited at 24, 48 and 72 h compared with that
in the blank group (all P<0.05). The cell viability was greatly
increased at all time points in the shCRLF2 group and the LY294002
group compared with that in the shCRLF2 + LY294002 group (all
P<0.05). No intergroup difference was noted between the blank
group and the NC group (P>0.05; Fig.
4).
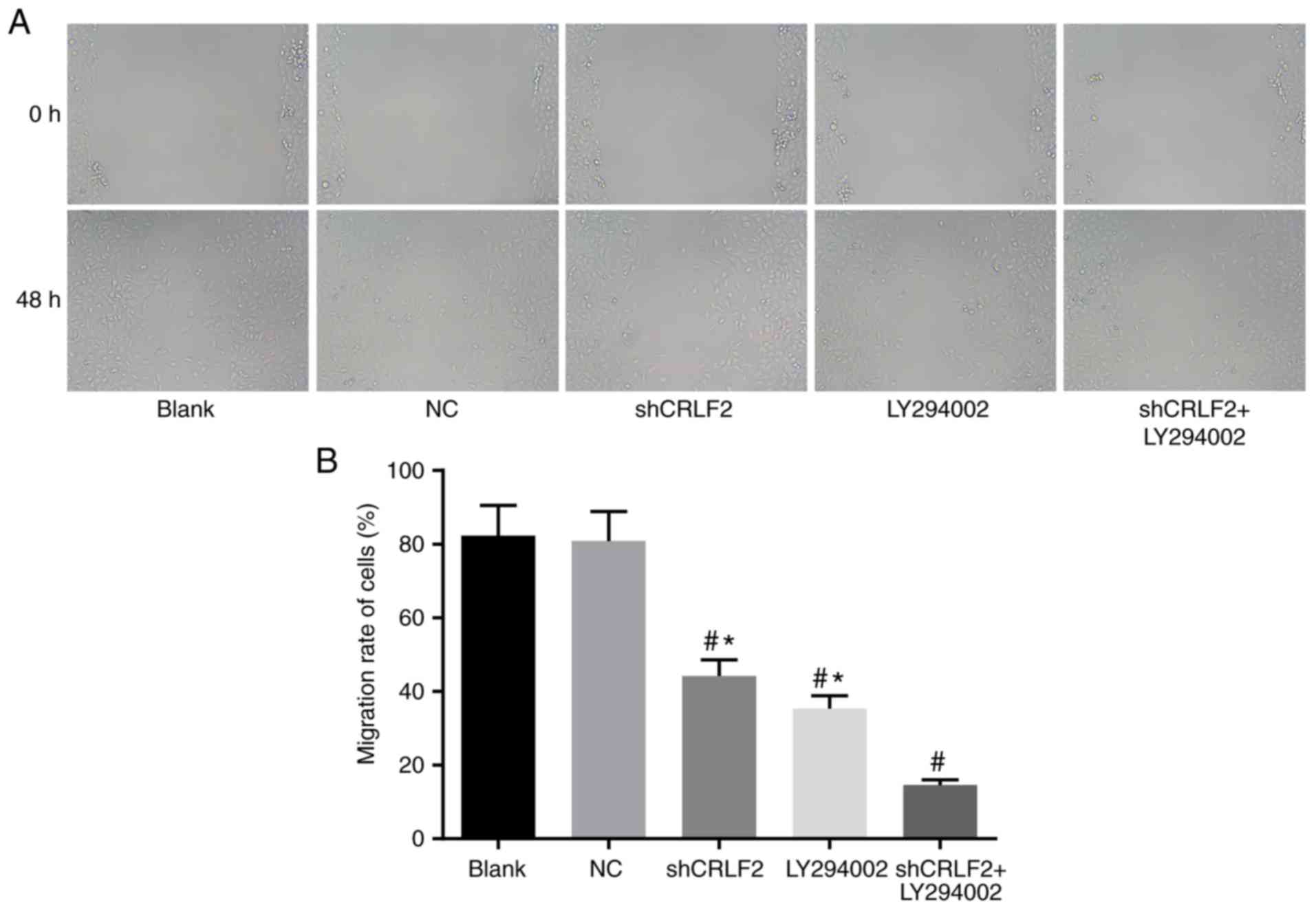
BaF3 cell migration in each group
measured by wound healing assay
The migration of BaF3 cells in each group was
measured by wound healing assay. The results indicated that the
migration of the cells was considerably inhibited in the shCRLF2
(44.20±3.41%), LY294002 (35.40±2.51%) and shCRLF2 + LY294002
(14.60±1.52%) groups compared with the blank group (82.30±8.10%;
all P<0.05). In addition, the migration of the cells was
increased considerably in the shCRLF2 and LY294002 groups compared
with the shCRLF2 + LY294002 group (all P<0.05). No intergroup
difference was observed between the blank group and the NC group
(P>0.05; Fig. 5).
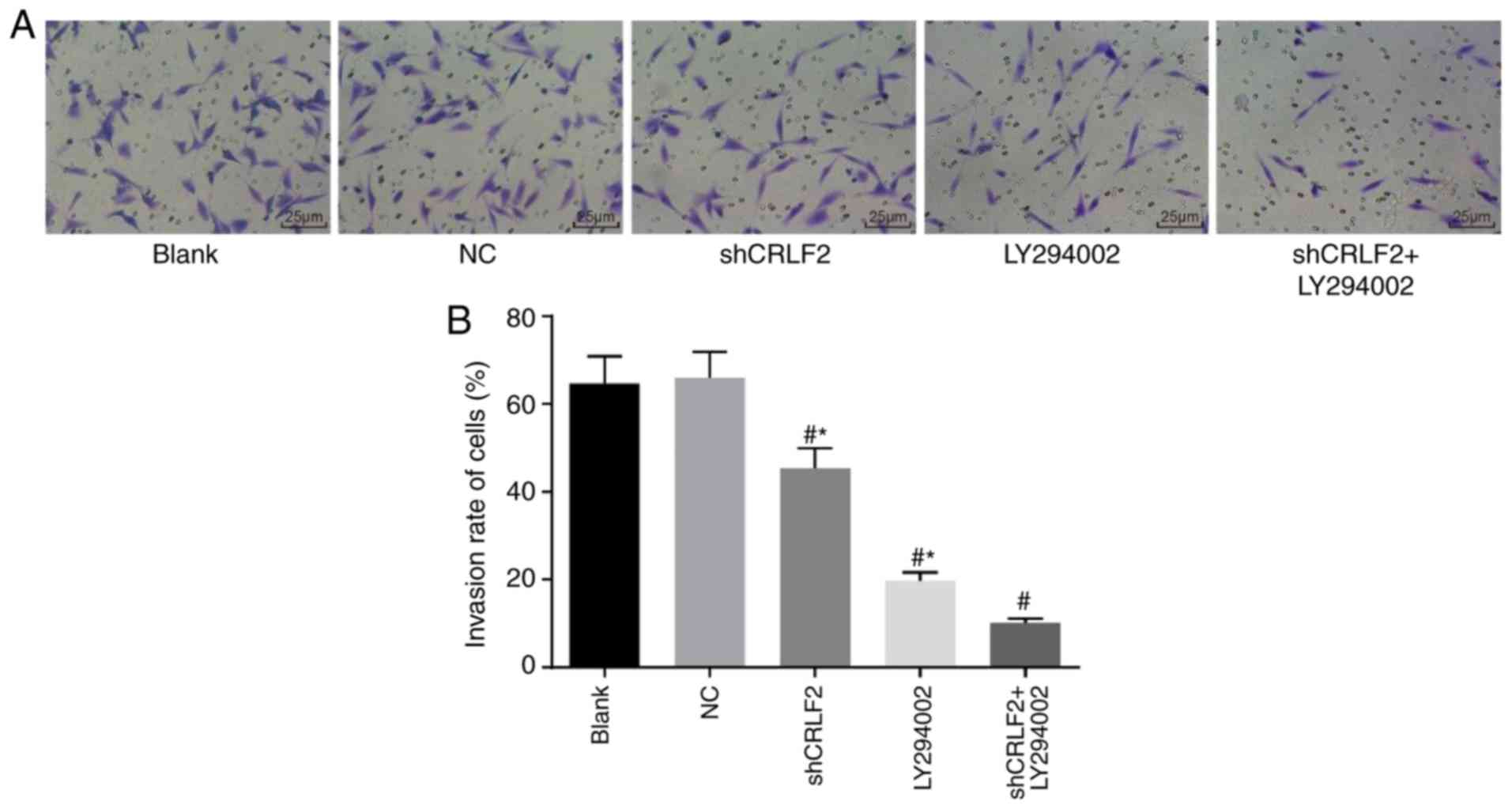
BaF3 cell invasion in each group
detected by Transwell invasion assay
The invasion of BaF3 cells in each group may be
observed in Fig. 6A and the
histogram of the BaF3 cell invasion rate in each group is presented
in Fig. 6B. The results documented
that the invasion of BaF3 cells was markedly suppressed in the
shCRLF2 (45.40±5.28%), LY294002 (19.70±2.41%) and shCRLF2 ±
LY294002 (10.20±1.66%) groups compared with the blank group
(64.70±5.25%; all P<0.05). The invasion of cells was
significantly enhanced in the shCRLF2 and LY294002 groups compared
with the shCRLF2 + LY294002 group (all P<0.05; Fig. 6).
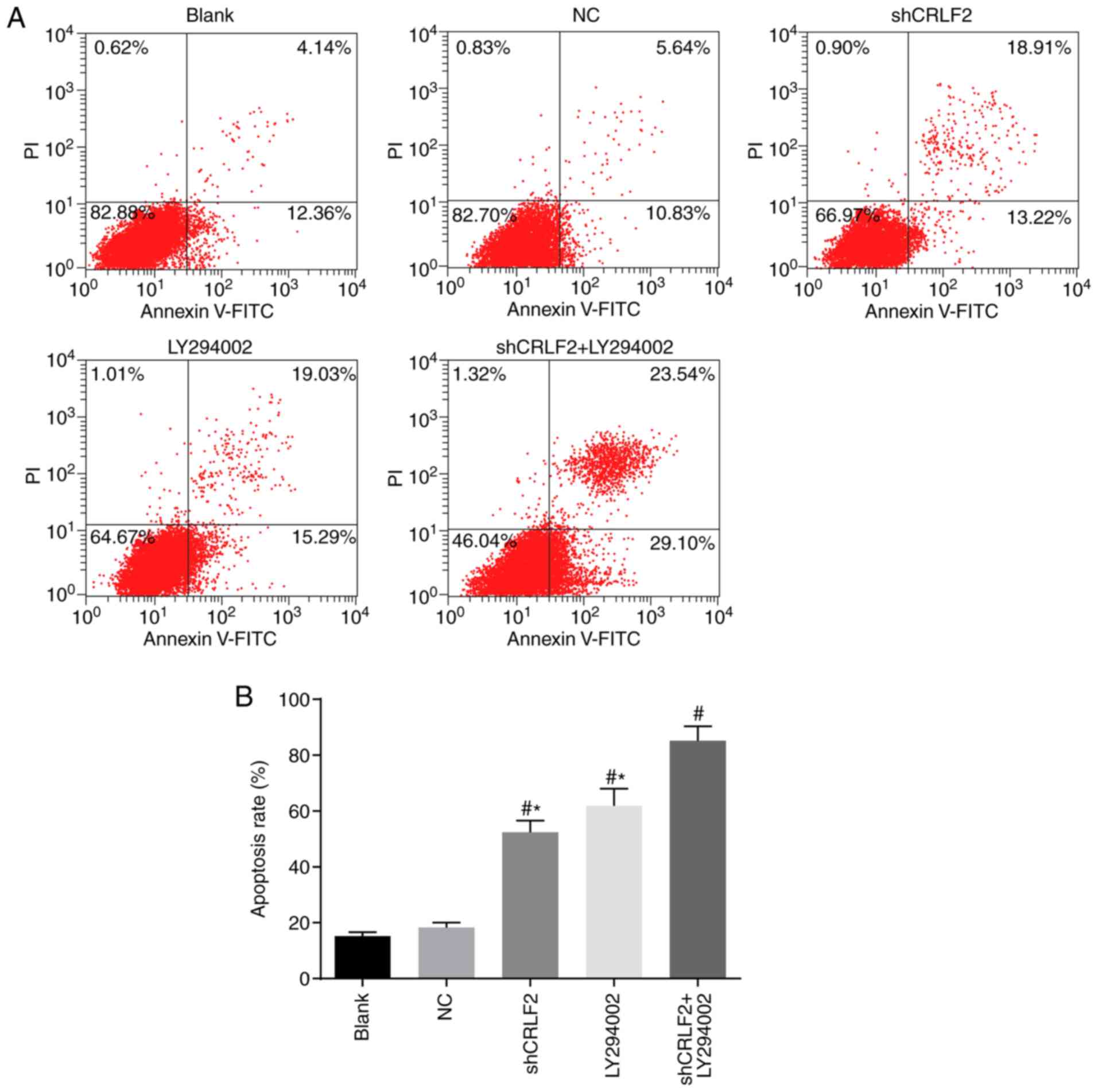
Cell apoptosis in each group, assessed
by Annexin V-FITC/PI double staining assay
The Annexin V-FITC/PI double staining assay was
performed to detect the drug-induced apoptosis of imatinib-treated
cells. The results of the BaF3 cell apoptosis and apoptosis rates
in each group are presented in Fig. 7A
and B, respectively. The results demonstrated in the shCRLF2
group, LY294002 group and shCRLF2 + LY294002 group there was an
increase in the sensitivity of BaF3 cells to chemotherapeutic
agents compared with the blank group. Furthermore, the shCRLF2 +
LY294002 group achieved even better results compared with the
shCRLF2 and LY294002 groups; the cell apoptosis rates in the
shCRLF2 and LY294002 groups were decreased compared with the
shCRLF2+LY294002 group (both P<0.05; Fig. 7).
Discussion
Certain studies have confirmed that CRLF2 serves a
key role in the development of B lymphocytes, and mutations in the
CRLF2 gene may further activate the CRLF2/thymic stromal
lymphopoietin (TSLP) signaling pathway and induce B-ALL (15–17).
Moreover, CRLF2 is a marker for Ph-like ALL. The abnormal
expression of CRLF2 is closely associated with the occurrence,
recurrence and poor prognosis of ALL (18). There was a study in which CRLF2
expression was observed in TSLP humanized transplantation mouse
models following injection of human HS27 cells expressing the human
TSLP gene, and the results demonstrated that CRLF2 is able to
promote the differentiation and growth of B cells, in addition to
the proliferation of lymphocytes at the early stage in TSLP mouse
models (19). The AKT/mTOR
signaling pathway has been proven to serve an essential role in
cell proliferation and growth, and the abnormal activation of this
signaling pathway has been observed in studies on T-cell ALL
(20). Certain studies have
compared the effects of different signaling pathway inhibitors on
B-ALL cells and have reported that AKT/mTOR signaling pathway
inhibitors can significantly inhibit the B-ALL cell cycle and
reduce the cell survival rate (21). The present study performed a further
investigation based on these findings and identified that silencing
the CRLF2 gene may suppress the activation of the AKT/mTOR
signaling pathway, which may have protective effects on B-ALL.
In the present study, significant upregulation of
CRLF2 expression in the bone marrow of B-ALL patients was observed,
implying that CRLF2 may be implicated in the pathogenesis of B-ALL.
In order to investigate the association between the CRLF2 gene and
the pathogenesis of B-ALL, the present study divided BaF3 cell
lines into different groups and examined the expression levels of
mRNAs and proteins associated with the CRLF2 and AKT/mTOR pathways,
the cell viability and cell sensitivity to chemotherapeutic agents.
The results indicated that the mRNA and protein expression levels
of CLRF2 were downregulated in the shCRLF2 and shCRLF2 + LY294002
groups as compared with the Blank and NC groups, however, no
significant difference was noted in the LY294002 group. Moreover,
the expression levels of AKT, mTOR and S6K1 mRNAs and associated
phosphorylated proteins, cell viability, cell migration and cell
invasion in the shCRLF2, LY294002, and shCRLF2 + LY294002 groups
were significantly decreased compared with the Blank and NC groups,
indicating that CLRF2 gene could affect the protein expression,
cell migration and cell invasion in BaF3 cells via AKT, mTOR and
other signaling pathways. The results revealed that silencing CRLF2
expression or inhibiting the AKT/mTOR signaling pathway could
reduce the cell viability, migration and invasion in B-ALL and
promote cell apoptosis. Other studies have stated that >80% of
patients with ALL have abnormal activation of the AKT/mTOR pathway,
which further confirms the findings in the present study (22,23).
Gene rearrangement refers to the movement of a gene
from a site distal to the promoter to a site proximal to the
promoter, thereby influencing gene transcription and expression.
During the treatment of children with B-ALL, Raghunathan et
al (24) identified that
children with CRLF2 rearrangement had improved treatment responses
and 4-year recurrence-free survival rates compared with those
without CRLF2 rearrangement, suggesting that the abnormal
expression of CRLF2 induced by gene rearrangement may be highly
correlated with the prognosis of B-ALL. In the present study, the
Annexin V-FITC/PI double staining assay was used, and it was
identified that the silencing of the CRLF2 gene and the use of the
AKT/mTOR signaling pathway inhibitor significantly increased the
sensitivity of BaF3 cells to chemotherapeutic agents against B-ALL
compared with the blank group and NC group; furthermore, the
combined use of these two methods achieved an even a better result.
Survival analysis indicated that among all the children with B-ALL,
those with high CRLF2 expression had much lower OS and EFS levels
than those with low CRLF2 expression, implying that low CRLF2
expression may have a positive effect on the prognosis of B-ALL.
Useful findings have been obtained in the present study by
investigating the association between silencing of the
CRLF2-mediated AKT/mTOR pathway and the treatment efficacy and
prognosis of pediatric B-ALL. However, due to regional and
individual differences, further studies are required in the future
for verification.
The present study demonstrated that silencing the
CRLF2 gene may inhibit the activation of the AKT/mTOR pathway,
which is closely associated with the treatment effect and prognosis
in children with B-ALL. CRLF2 is a poor prognostic factor for B-ALL
and may be used as a key target in the management of this disease.
The suppression of CRLF2 expression or the use of an AKT/mTOR
signaling pathway inhibitor may be helpful in the treatment of
B-ALL, and the results of the present study may provide a basis for
the future study and treatment of this disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fujian
Province Medical Innovation Project (grant no. 2017-CXB-19); Xiamen
Science and Technology Plan Project (grant no. 3502Z20164066);
Fujian Province Medical Innovation Project (grant no. 2014-CXB-51);
Fujian Province Natural Science Foundation Project (grant no.
2017D006).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
MJ was the guarantor of the integrity of the entire
study. MJ and XZ contributed to the study concepts, the definition
of intellectual content, literature research, clinical studies,
experimental studies, manuscript preparation and manuscript
editing. XZ contributed to data acquisition, data analysis and
statistical analysis. LL contributed to the study design. MJ and LL
contributed to manuscript review. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Yichang Central People's Hospital (Yichang,
China), and consent was obtained from the patients and their
families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRLF2
|
cytokine receptor-like factor 2
|
|
B-ALL
|
B-cell acute lymphoblastic
leukemia
|
|
OS
|
overall survival
|
|
EFS
|
event-free survival
|
|
WBC
|
white blood cell count
|
|
HGB
|
hemoglobin
|
|
PLT
|
platelet count
|
|
LDH
|
lactate dehydrogenase
|
References
|
1
|
Buitenkamp TD, Izraeli S, Zimmermann M,
Forestier E, Heerema NA, van den Heuvel-Eibrink MM, Pieters R,
Korbijn CM, Silverman LB, Schmiegelow K, et al: Acute lymphoblastic
leukemia in children with down syndrome: A retrospective analysis
from the Ponte di Legno study group. Blood. 123:70–77. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tasian SK, Doral MY, Borowitz MJ, Wood BL,
Chen IM, Harvey RC, Gastier-Foster JM, Willman CL, Hunger SP,
Mullighan CG, et al: Aberrant STAT5 and PI3K/mTOR pathway signaling
occurs in human CRLF2-rearranged B-precursor acute lymphoblastic
leukemia. Blood. 120:833–842. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maude S, Tasian S, Vincent T, Hall J,
Roberts K, Collins R, Mullighan C, Hunger S, Willman C, Loh M, et
al: Targeting JAK2 and mTOR in xenograft models of
CRLF2-overexpressing Acute Lymphoblastic Leukemia (ALL). Pediat
Blood Cancer. 58:1014. 2012.
|
|
4
|
Tasian SK, Maude SL, Hall J, Vincent T,
Mullighan CG, Willman CL, Hunger S, Loh ML, Teachey DT and Grupp
SA: In vivo monitoring of JAK/STAT and PI3K/mTOR signal
transduction inhibition in pediatric CRLF2-rearranged acute
lymphoblastic leukemia (ALL). J Clin Oncol. 9506:2012.
|
|
5
|
Krawczyk J, Haslam K, Lynam P, Kelly J,
Storey L, O'Marcaigh A, Langabeer SE and Smith OP: No prognostic
impact of P2RY8-CRLF2 fusion in intermediate cytogenetic risk
childhood B-cell acute lymphoblastic leukaemia. Br J Haematol.
160:555–556. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Francis OL: TSLP-induced mechanisms and
potential therapies for CRLF2 B-cell acute lymphoblastic leukemia.
Dissertations Theses-Gradworks. Loma Linda University Electronic
Theses, Dissertations & Projects. 282:2015.
|
|
7
|
Maude SL, Tasian SK, Vincent T, Hall JW,
Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, et
al: Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like
acute lymphoblastic leukemia. Blood. 120:3510–3518. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chiaretti S, Brugnoletti F, Messina M,
Paoloni F, Fedullo AL, Piciocchi A, Elia L, Vitale A, Mauro E,
Ferrara F, et al: CRLF2 overexpression identifies an unfavourable
subgroup of adult B-cell precursor acute lymphoblastic leukemia
lacking recurrent genetic abnormalities. Leuk Res. 41:36–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Konoplev S, Lu X, Konopleva M, Jain N,
Ouyang J, Goswami M, Roberts KG, Valentine M, Mullighan CG,
Bueso-Ramos C1, et al: CRLF2-positive B-cell acute lymphoblastic
leukemia in adult patients: A single-institution experience. Am J
Clin Pathol. 147:357–363. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dou H, Chen X, Huang Y, Su Y, Lu L, Yu J,
Yin Y and Bao L: Prognostic significance of P2RY8-CRLF2 and CRLF2
overexpression may vary across risk subgroups of childhood B-cell
acute lymphoblastic leukemia. Genes Chromosomes Cancer. 56:135–146.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Neri LM, Cani A, Martelli AM, Simioni C,
Junghanss C, Tabellini G, Ricci F, Tazzari PL, Pagliaro P, McCubrey
JA, et al: Targeting the PI3K/Akt/mTOR signaling pathway in
B-precursor acute lymphoblastic leukemia and its therapeutic
potential. Leukemia. 28:739–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tasian SK, Teachey DT, Li Y, Shen F,
Harvey RC, Chen IM, Ryan T, Vincent TL, Willman CL, Perl AE, et al:
Potent efficacy of combined PI3K/mTOR and JAK or ABL inhibition in
murine xenograft models of Ph-like acute lymphoblastic leukemia.
Blood. 129:177–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Möricke A, Zimmermann M, Valsecchi MG,
Stanulla M, Biondi A, Mann G, Locatelli F, Cazzaniga G, Niggli F,
Aricò M, et al: Dexamethasone vs prednisone in induction treatment
of pediatric ALL: Results of the randomized trial AIEOP-BFM ALL
2000. Blood. 127:2101–2112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B, Suer S, Livak F, Adediran S,
Vemula A, Khan MA, Ning Y and Hussain A: Telomere and microtubule
targeting in treatment-sensitive and treatment-resistant human
prostate cancer cells. Mol Pharmacol. 82:310–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jain N, Lu X, Daver N, Thakral B, Wang SA,
Konoplev S, Patel K, Kanagal-Shamanna R, Valentine M, Tang G, et
al: Co-occurrence of CRLF2-rearranged and Ph+ acute lymphoblastic
leukemia: A report of four patients. Haematologica. 102:e514–e517.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sadras T, Heatley SL, Kok CH, Dang P,
Galbraith KM, McClure BJ, Muskovic W, Venn NC, Moore S, Osborn M,
et al: Differential expression of MUC4, GPR110 and IL2RA defines
two groups of CRLF2-rearranged acute lymphoblastic leukemia
patients with distinct secondary lesions. Cancer Lett. 408:92–101.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Francis OL, Shiraz P, Milford TA, Baez I,
Coats JS, Mayagoitia K, Ginelli E, Salcedoconcepcion KR, Martinez
S, Zhang X, et al: Abstract 3295: A novel patient-derived xenograft
model for evaluating the role of TSLP in CRLF2 B-ALL. Cancer Res.
75:3295. 2015. View Article : Google Scholar
|
|
18
|
Herold T, Schneider S, Metzeler KH,
Neumann M, Hartmann L, Roberts KG, Konstandin NP, Greif PA, Bräundl
K, Ksienzyk B, et al: Adults with Philadelphia chromosome-like
acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2
mutations, persistence of minimal residual disease and poor
prognosis. Haematologica. 102:130–138. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stoian C, Mambo NG, Mccarthy P, Vidales V,
Coats JS, Baez I, Dovat S, Gohar SF, Desai D, Kamal M and Payne KJ:
Abstract 5829: Targeting TSLP-induce upregulation of Mcl-1 for the
treatment of Ph-like ALL with CRLF2 alterations. Cancer Res.
77:5829. 2017. View Article : Google Scholar
|
|
20
|
Shi C, Han L, Tabe Y, Mu H, Wu SC, Zhou J,
Zeng Z, Fruman DA, Tasian SK, Weinstock DM and Konopleva M: Dual
targeting of JAK2 signaling with a type II JAK2 inhibitor and of
mTOR with a TOR kinase inhibitor induces apoptosis in
CRLF2-rearranged Ph-like acute lymphoblastic leukemia. Blood.
124:37062014.
|
|
21
|
Zhang Q, Shi C, Han L, Jain N, Roberts KG,
Ma H, Cai T, Cavazos A, Tabe Y, Jacamo RO, et al: Inhibition of
mTORC1/C2 signaling improves anti-leukemia efficacy of JAK/STAT
blockade in CRLF2 rearranged and/or JAK driven Philadelphia
chromosome-like acute B-cell lymphoblastic leukemia. Oncotarget.
9:8027–8041. 2018.PubMed/NCBI
|
|
22
|
Francis OL, Milford TA, Martinez SR, Baez
I, Coats JS, Mayagoitia K, Concepcion KR, Ginelli E, Beldiman C,
Benitez A, et al: A novel xenograft model to study the role of
TSLP-induced CRLF2 signals in normal and malignant human B
lymphopoiesis. Haematologica. 101:417–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lane AA, Bodegom DV, Chapuy B, Alexe G,
Sullivan TJ, Tivey T, Day T, Crispino J, Fox E, Stegmaier K and
Weinstock D: Trisomy of the down syndrome critical region
suppresses precursor B-cell differentiation and promotes B-cell
transformation associated with altered expression of polycomb
repressor complex 2 targets. Blood. 120:1152012.
|
|
24
|
Raghunathan R, Mahesula S, Kancharla K,
Janardhanan P, Jadhav YL, Nadeau R, Villa GP, Cook RL, Witt CM,
Gelfond JA, et al: Anti-CRLF2 antibody-armored biodegradable
nanoparticles for childhood B-ALL. Part Part Syst Charact.
30:355–364. 2013. View Article : Google Scholar : PubMed/NCBI
|