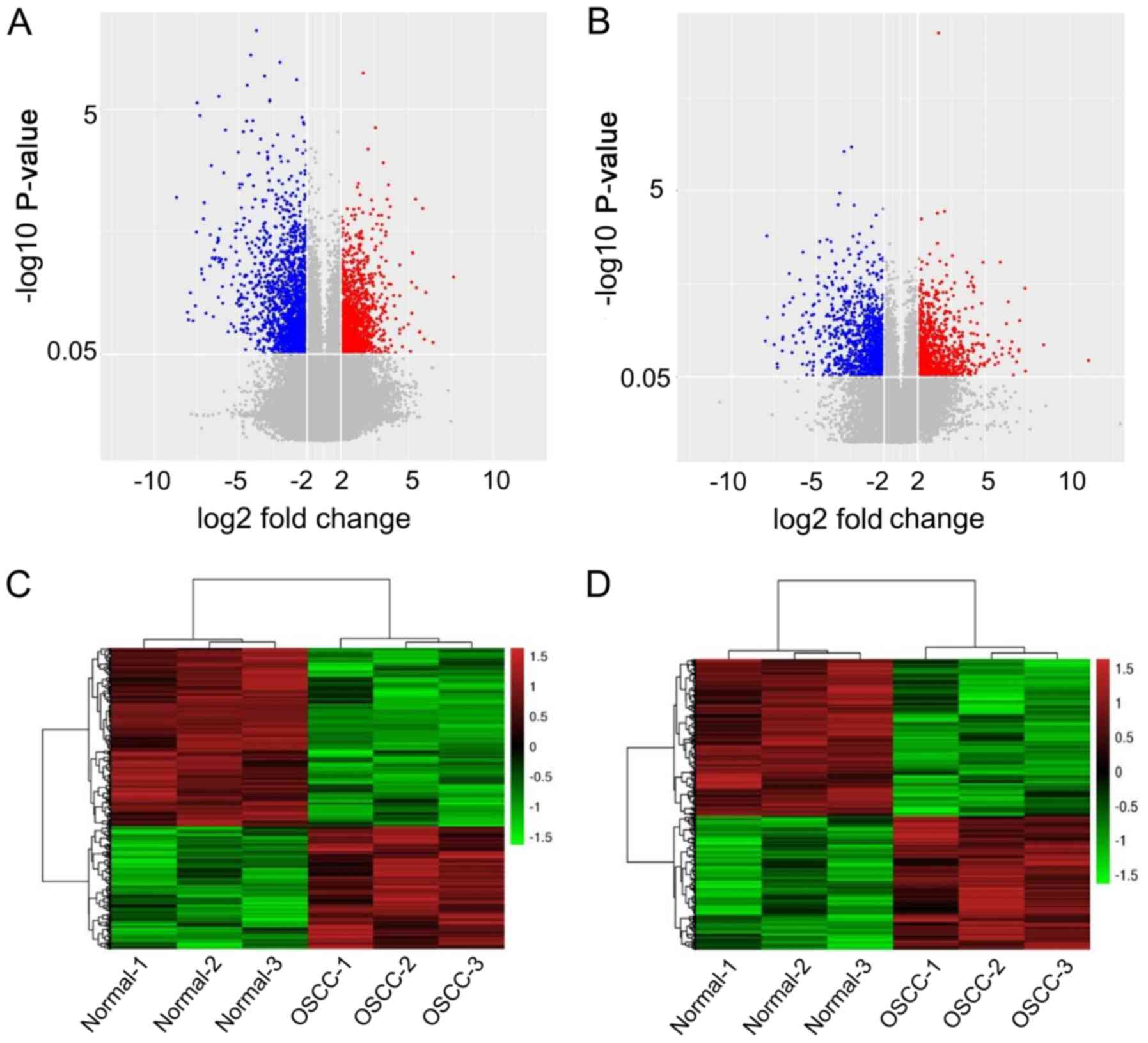
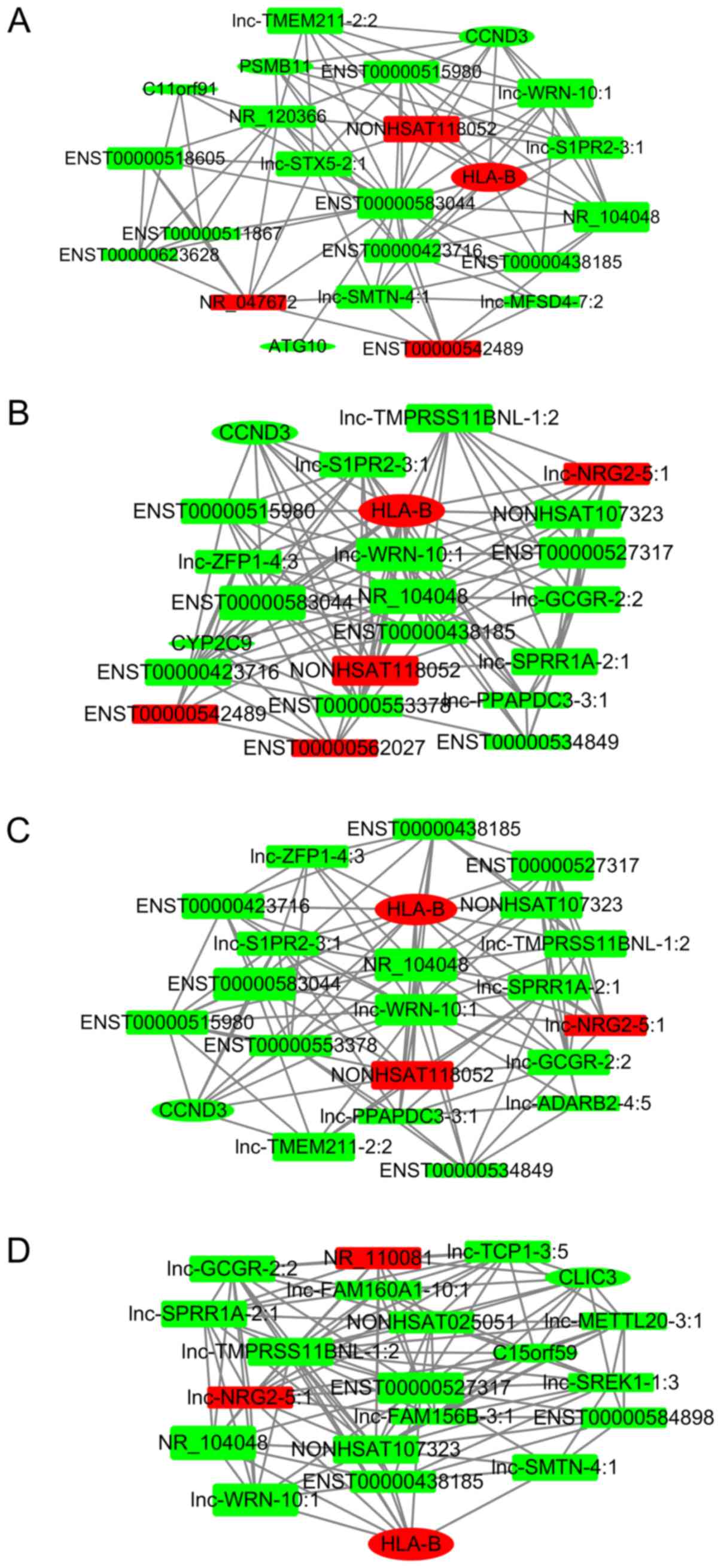
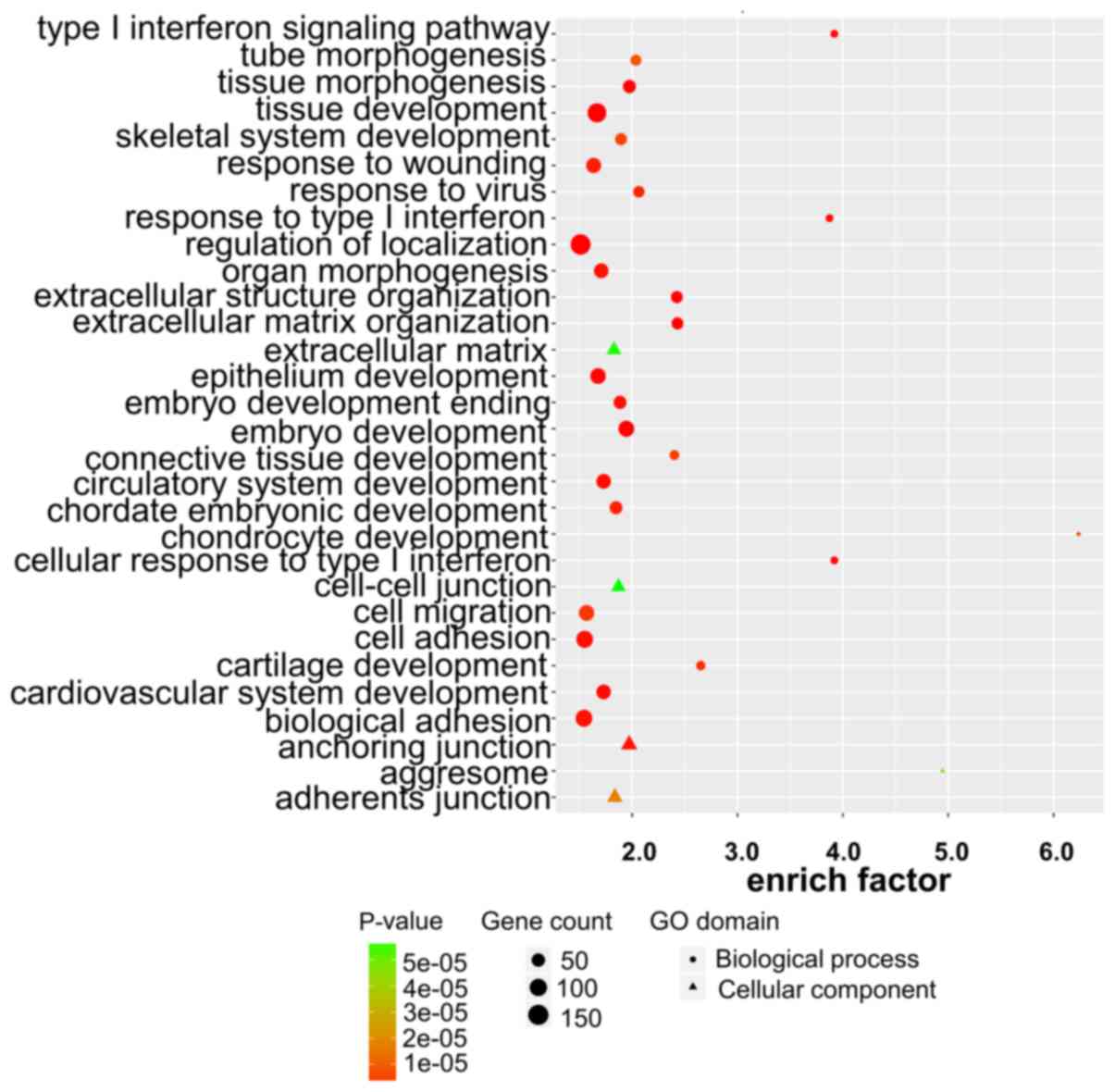
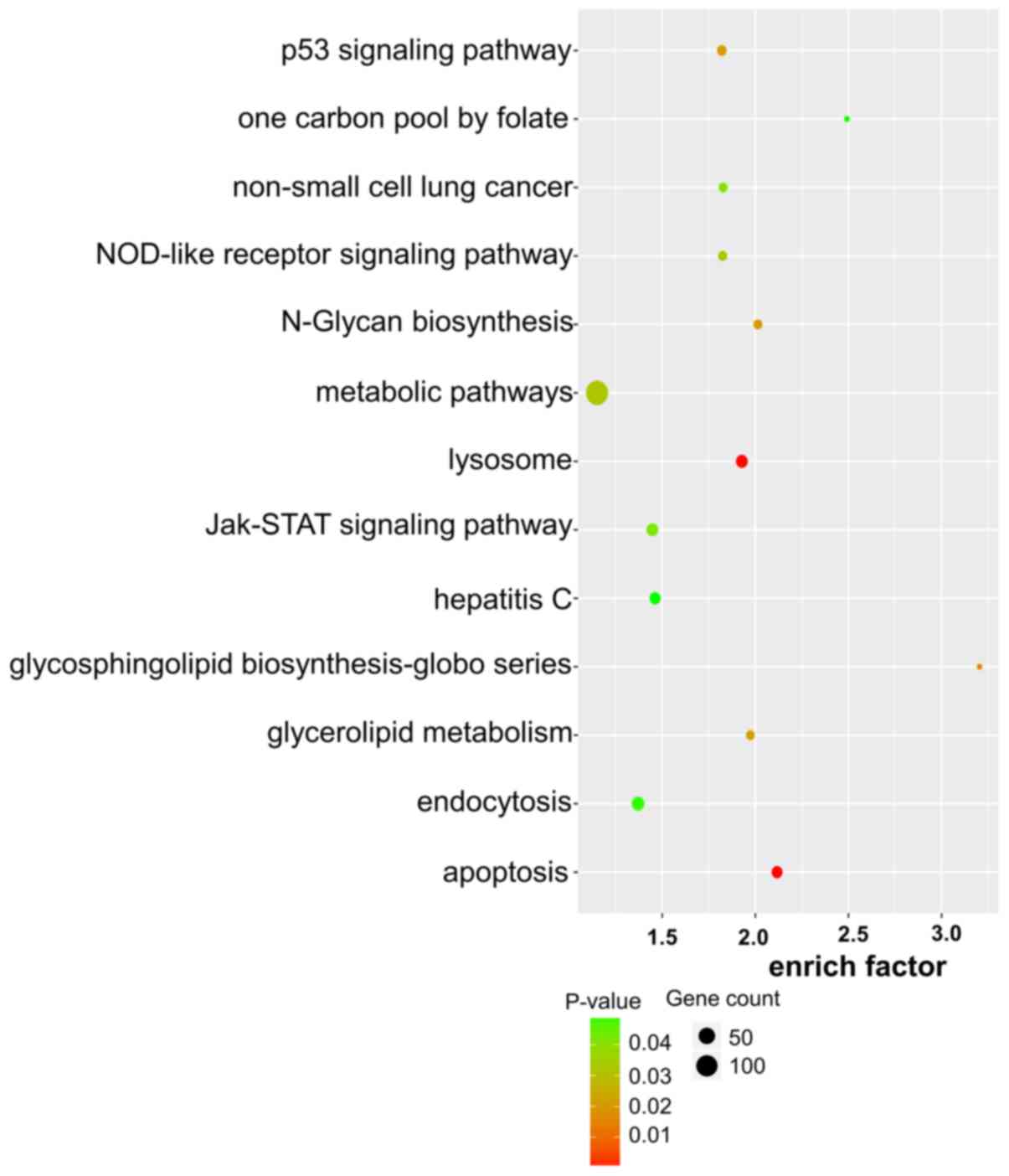
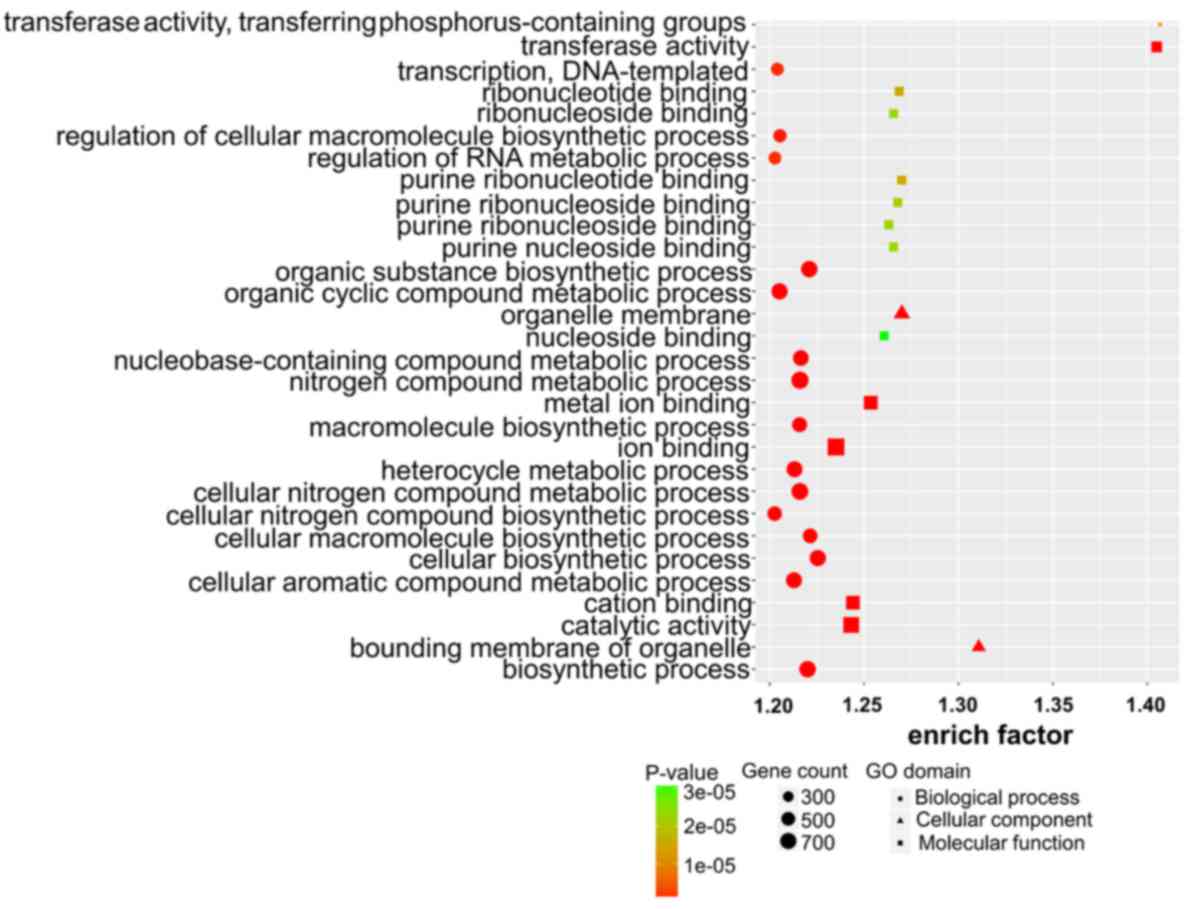
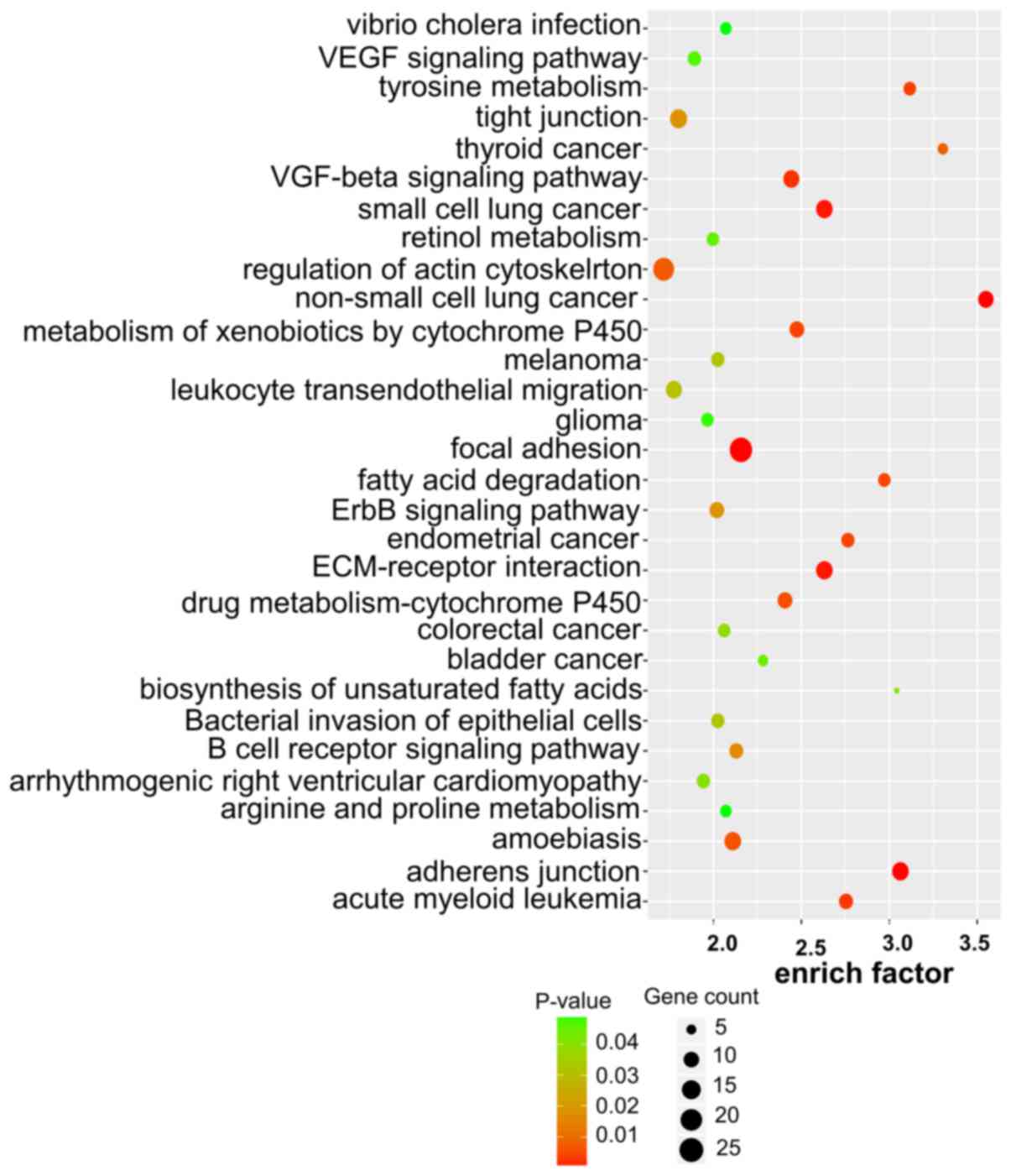
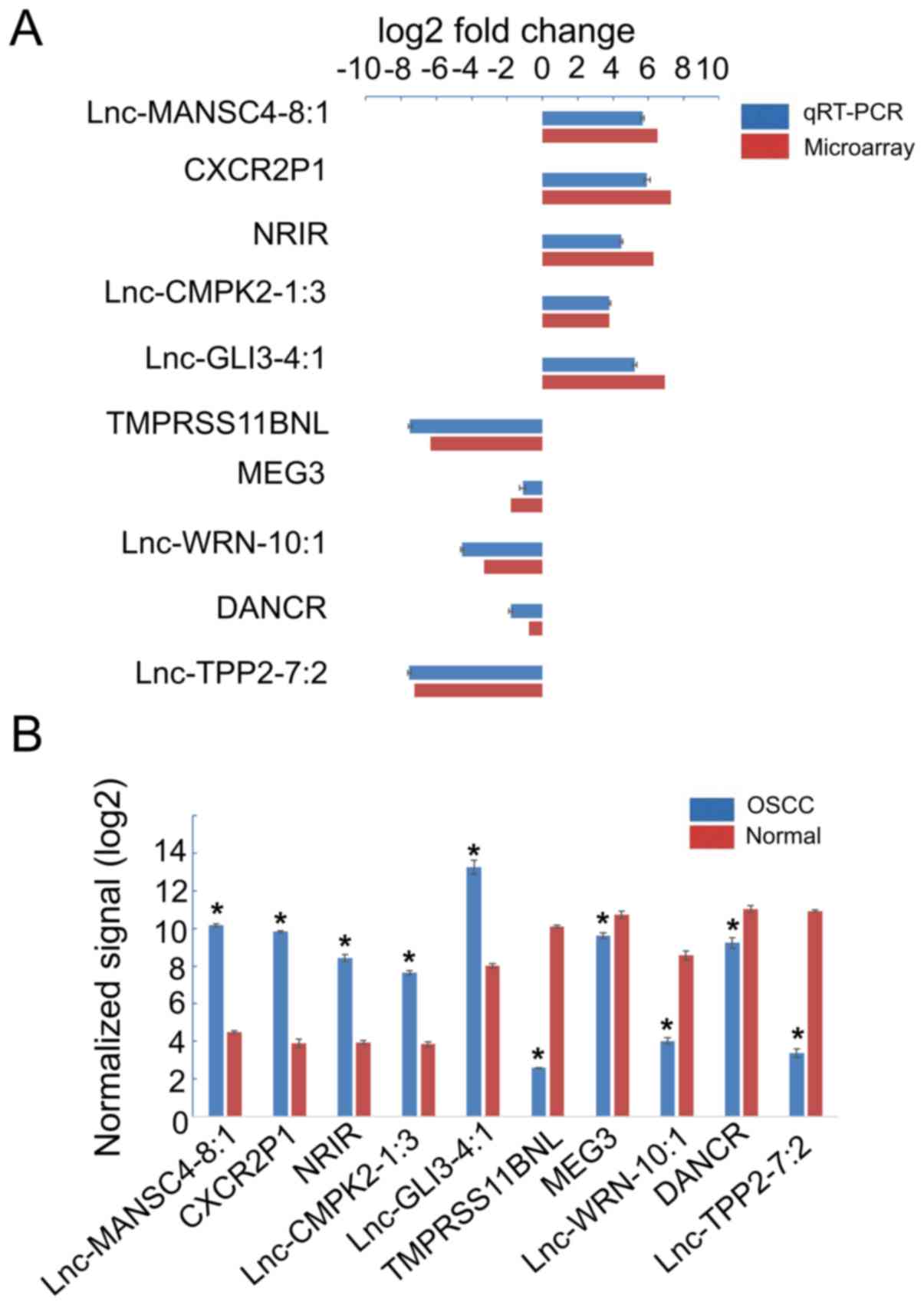
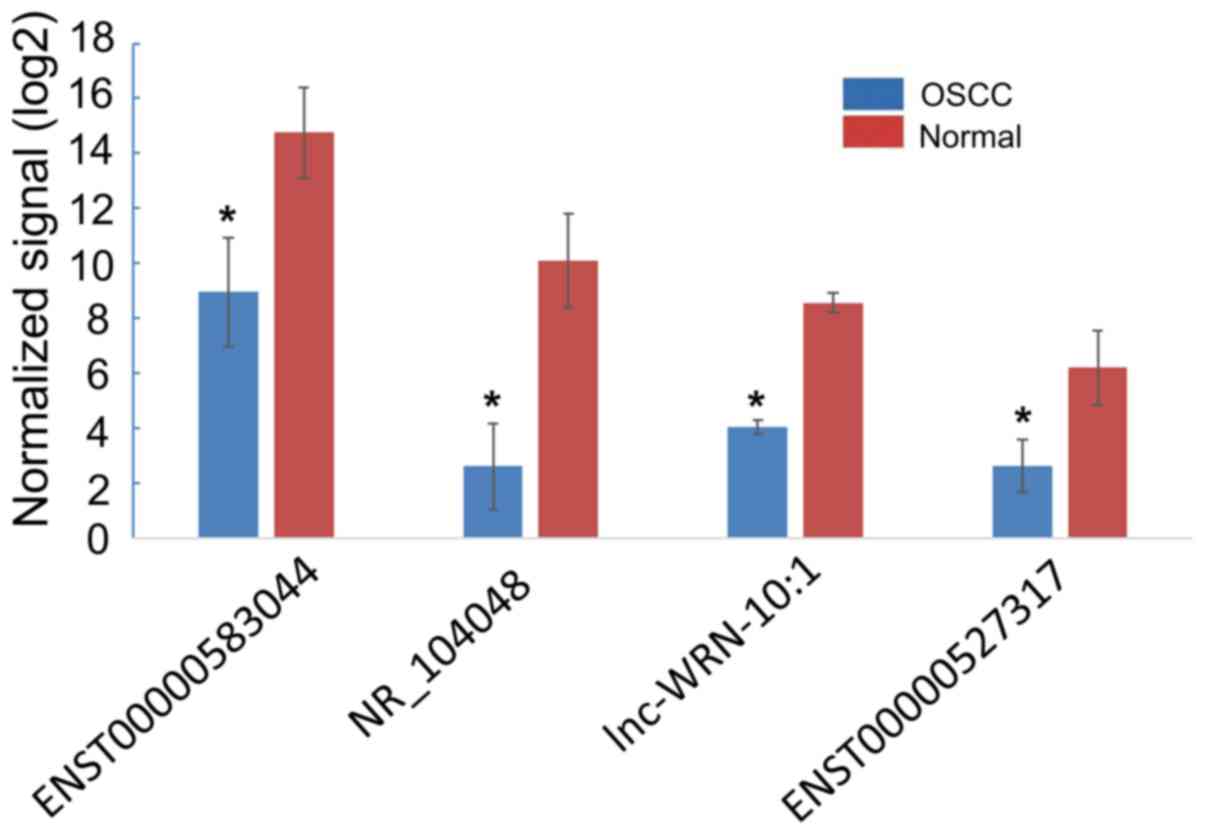
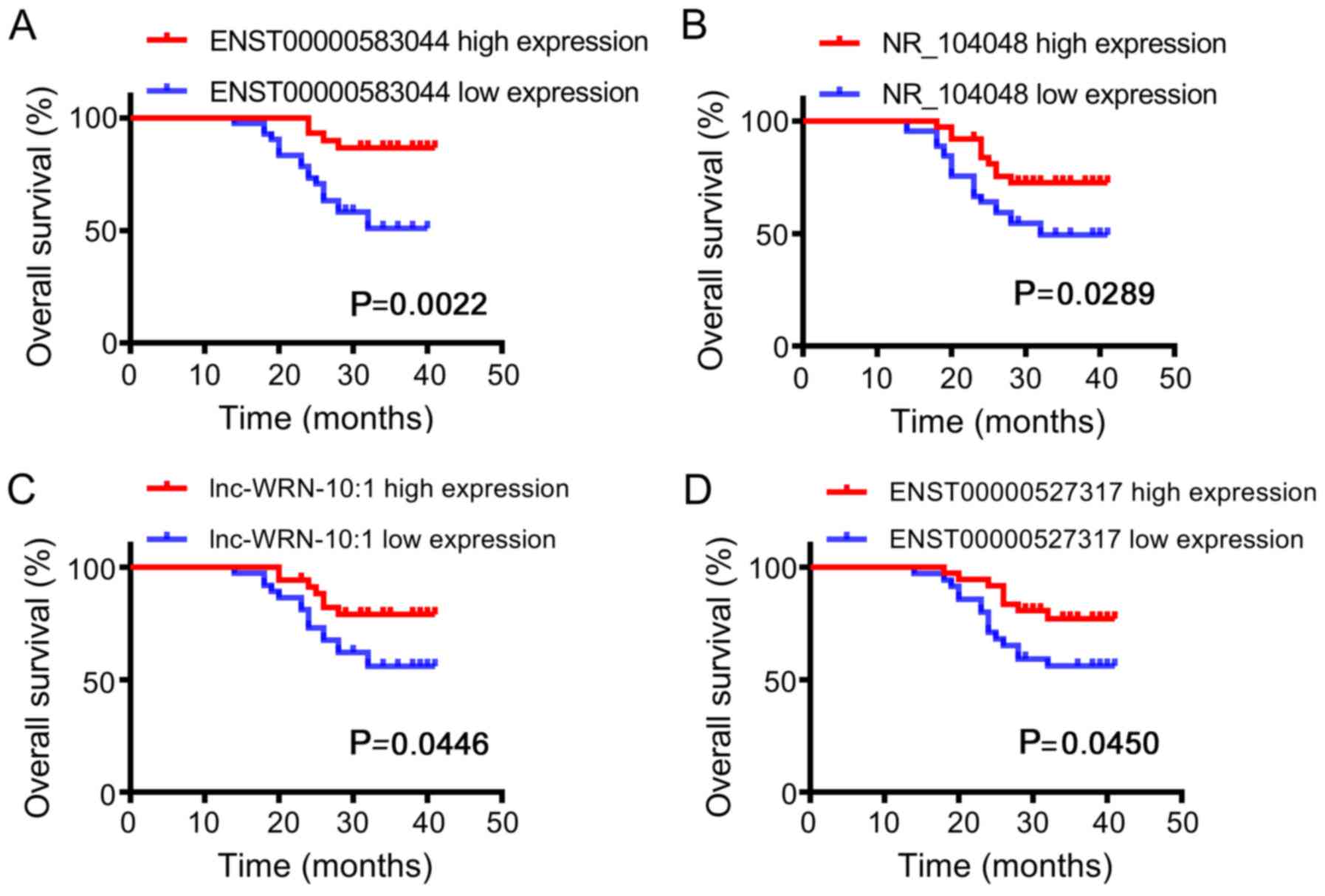
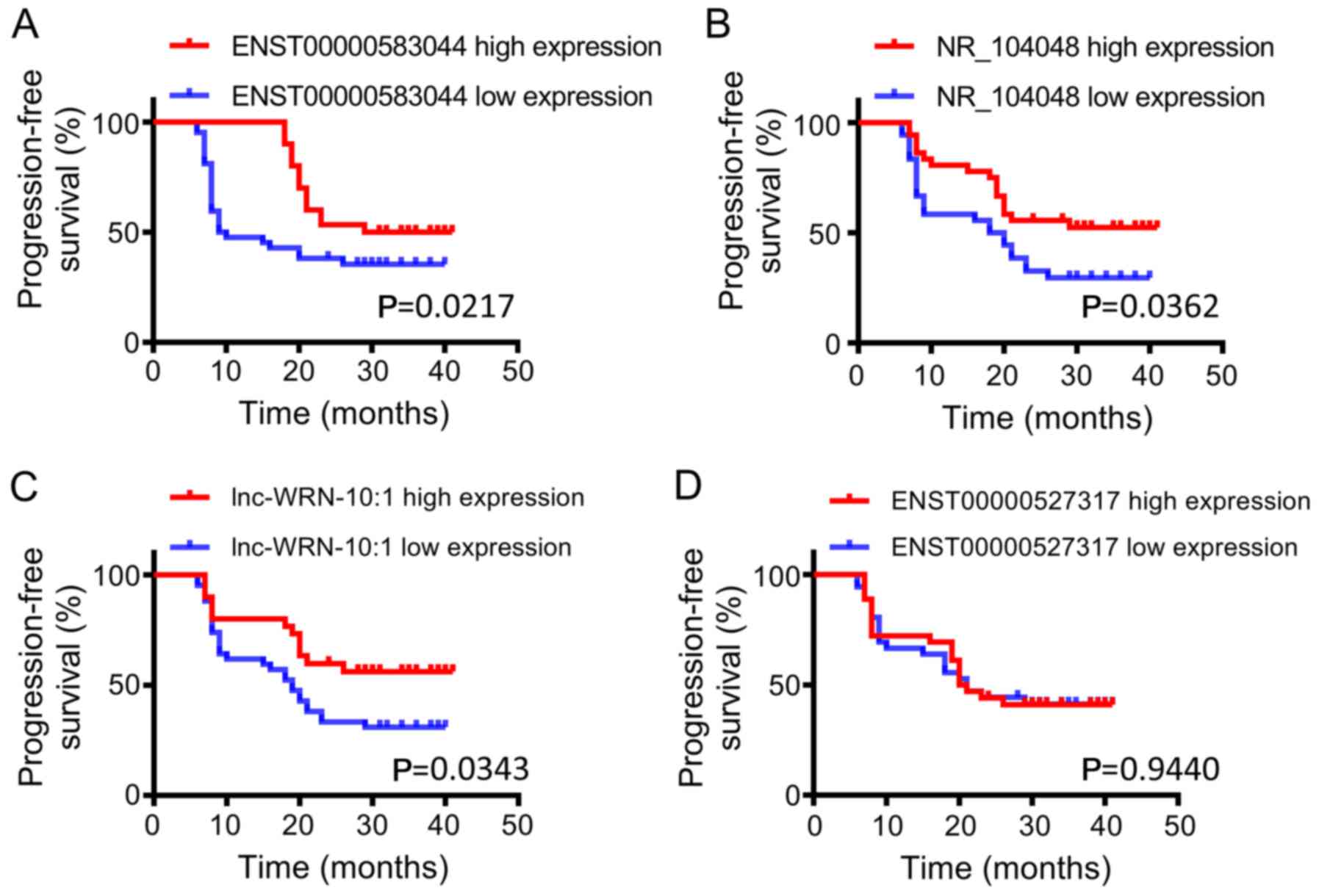
|
1
|
Poveda-Roda R, Bagán JV, Jiménez-Soriano
Y, Margaix-Muñoz M and Sarrión-Pérez G: Changes in smoking habit
among patients with a history of oral squamous cell carcinoma
(OSCC). Med Oral Patol Oral Cir Bucal. 15:e721–e726. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Messadi DV: Diagnostic aids for detection
of oral precancerous conditions. Int J Oral Sci. 5:59–65. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y,
Wei M, Chen J, Gao X, Xu C, et al: The prostate cancer-up-regulated
long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation
through reciprocal regulation of androgen receptor. Urol Oncol.
31:1117–1123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng L, Houck JR, Lohavanichbutr P and
Chen C: Transcriptome analysis reveals differentially expressed
lncRNAs between oral squamous cell carcinoma and healthy oral
mucosa. Oncotarget. 8:31521–31531. 2017.PubMed/NCBI
|
|
6
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eades G, Zhang YS, Li QL, Xia JX, Yao Y
and Zhou Q: Long non-coding RNAs in stem cells and cancer. World J
Clin Oncol. 5:134–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Xu HG and Lu C: A novel long
non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to
induce cellular apoptosis in T-cell acute lymphoblastic leukemia
cells. Leuk Lymphoma. 55:1373–1382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zang W, Wang T, Huang J, Li M, Wang Y, Du
Y, Chen X and Zhao G: Long noncoding RNA PEG10 regulates
proliferation and invasion of esophageal cancer cells. Cancer Gene
Ther. 22:138–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA
lncTCF7 promotes self-renewal of human liver cancer stem
cells through activation of Wnt signaling. Cell Stem Cell.
16:413–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan P, Luo S, Lu JY and Shen X: Cis- and
trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin
Genet Dev. 46:170–178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang
YH, Liu P, Hong M, Miao KR, Liu P, et al: miR-181a/b significantly
enhances drug sensitivity in chronic lymphocytic leukemia cells via
targeting multiple anti-apoptosis genes. Carcinogenesis.
33:1294–1301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista
DL, Gejman R, Ansell PJ, Zhao J, Weng C and Klibanski A: Activation
of p53 by MEG3 non-coding RNA. J Biol Chem. 282:24731–24742. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma N, Li S, Zhang Q, Wang H, Qin H and
Wang S: Long non-coding RNA GAS5 inhibits ovarian cancer cell
proliferation via the control of microRNA-21 and SPRY2 expression.
Exp Ther Med. 16:73–82. 2018.PubMed/NCBI
|
|
23
|
Lu MY, Liao YW, Chen PY, Hsieh PL, Fang
CY, Wu CY, Yen ML, Peng BY, Wang DP, Cheng HC, et al: Targeting
LncRNA HOTAIR suppresses cancer stemness and metastasis in oral
carcinomas stem cells through modulation of EMT. Oncotarget.
8:98542–98552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Arun G, Mao YS, Lazar Z, Hung G,
Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al: The lncRNA
Malat1 is dispensable for mouse development but its
transcription plays a cis-regulatory role in the adult. Cell
Rep. 2:111–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X,
Ma Z, Li X and Zhang Y: LincRNA-p21 enhances the sensitivity of
radiotherapy for human colorectal cancer by targeting the
Wnt/beta-catenin signaling pathway. Oncol Rep. 31:1839–1845. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roy R, Singh R, Chattopadhyay E, Ray A,
Sarkar N, Aich R, Paul RR, Pal M and Roy B: MicroRNA and target
gene expression based clustering of oral cancer, precancer and
normal tissues. Gene. 593:58–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zaid KW, Nhar BM, Ghadeer Alanazi SM,
Murad R, Domani A and Alhafi AJ: Lack of effects of recombinant
human bone morphogenetic protein2 on angiogenesis in oral squamous
cell carcinoma induced in the syrian hamster cheek Pouch. Asian Pac
J Cancer Prev. 17:3527–3531. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is
associated with meningioma pathogenesis and progression. Cancer
Res. 70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sasahira T and Kirita T: Hallmarks of
cancer-related newly prognostic factors of oral squamous cell
carcinoma. Int J Mol Sci. 19:E24132018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lala M, Chirovsky D, Cheng JD and Mayawala
K: Clinical outcomes with therapies for previously treated
recurrent/metastatic head-and-neck squamous cell carcinoma (R/M
HNSCC): A systematic literature review. Oral Oncol. 84:108–120.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wurdak M, Schneider M, Iftner T and
Stubenrauch F: The contribution of SP100 to cottontail rabbit
papillomavirus transcription and replication. J Gen Virol. Jan
24–2018.(Epub ahead of print). doi: 10.1099/jgv.0.001012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Berscheminski J, Brun J, Speiseder T,
Wimmer P, Ip WH, Terzic M, Dobner T and Schreiner S: Schreiner,
Sp100A is a tumor suppressor that activates p53-dependent
transcription and counteracts E1A/E1B-55K-mediated transformation.
Oncogene. 35:3178–3189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim T, Veronese A, Pichiorri F, Lee TJ,
Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, et al:
p53 regulates epithelial-mesenchymal transition through microRNAs
targeting ZEB1 and ZEB2. J Exp Med. 208:875–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tan BS, Tiong KH, Choo HL, Chung FF, Hii
LW, Tan SH, Yap IK, Pani S, Khor NT, Wong SF, et al: Mutant
p53-R273H mediates cancer cell survival and anoikis resistance
through AKT-dependent suppression of BCL2-modifying factor (BMF).
Cell Death Dis. 6:e18262015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Z, Bai J, Wu A, Wang Y, Zhang J, Wang
Z, Li Y, Xu J and Li X: Co-LncRNA: Investigating the lncRNA
combinatorial effects in GO annotations and KEGG pathways based on
human RNA-Seq data. Database. 2015:bav0822015. View Article : Google Scholar : PubMed/NCBI
|