Introduction
Lung cancer is the second most common cancer and the
leading cause of cancer-associated mortality, with an overall
5-year survival rate of 15% (1–3).
Non-small cell lung cancer (NSCLC) accounts for ~85% of all lung
cancer cases and ~70% of patients with NSCLC have locally advanced
disease or distant metastases at the time of diagnosis (4). The prognosis of patients with NSCLC
remains poor and most patients die from hematogenous dissemination,
whereas certain patients die from local organ failure. Therefore,
improving the understanding of the molecular mechanisms underlying
NSCLC development and progression is necessary to design effective
therapies for this disease.
Hepatocyte growth factor (HGF) and its receptor
tyrosine kinase c-Met are overexpressed in a variety of types of
human cancer, including NSCLC, renal, breast, ovarian cancer and
pleural mesothelioma (5). High
levels of expression of HGF represent an unfavorable prognostic
factor (6,7). The activation of the HGF/c-Met
signaling pathway promotes invasiveness and distant metastasis in
cancer (5,8) and increases tumor radioresistance
(9). In the cell microenvironment,
HGF is produced from the same cell (autocrine loop), neighboring
cells (paracrine loop), or distant organs (endocrine loop)
(10,11). By targeting its receptor c-Met, HGF
activates auto-phosphorylation in the catalytic domain. c-Met in
turn activates a series of signaling pathways including
Ras/extracellular signal regulated kinase and phosphoinositol 3
kinase/protein kinase B. Therefore, HGF and c-Met interaction and
expression are precisely monitored under physiological and
pathological conditions. Aberrant HGF/Met signaling activates
phosphorylation cascades, resulting in a comprehensive rewiring of
gene expression patterns and promoting tumor migration, invasion
and metastasis (12,13).
MicroRNAs (miRNAs/miRs), a class of small non-coding
RNAs, post-transcriptionally and/or translationally regulate gene
expression by binding to the 3′-untranslated region (3′-UTR) of
their target genes (14,15). Approximately one third to one half
of human genes are regulated by miRNAs, each of which has a number
of target transcripts (16). miRNAs
are involved in a wide range of human physiological and
pathological processes, including tumorigenesis (17,18).
miRNAs can function as oncogenes or tumor suppressors (19,20).
Studies have demonstrated that miR-200c, miR-193a-3p and
miR-193a-5p inhibit the proliferation, migration and invasion of
NSCLC cell lines (21,22). Furthermore, re-expression of miR-451
significantly reverses the radioresistance of docetaxel-resistant
lung adenocarcinoma cells by promoting apoptosis and DNA double
stranded breaks (23). miRNAs are
suggested as potential targets to enhance radiosensitivity in
cancer. In the present study, miRNAs capable of enhancing the
radiosensitivity of NSCLC cells by negatively regulating HGF
expression were investigated. miR-200a, located on chromosome
1p36.33, is involved in cell proliferation and migration by
targeting Kelch-like ECH-associated protein 1, Gαi1 and β-catenin.
miR-200a downregulation is associated with hepatic fibrosis, human
retinoblastomas, human glioma and breast cancer (24–26).
In the present study, the expression of HGF and
miRNAs was analyzed in NSCLC and normal tissues. Analysis of
clinicopathological data confirmed that HGF and miR-200a are
associated with tumor progression and the response to therapy.
Bioinformatics analysis and biological experiments demonstrated
that miR-200a negatively regulated HGF expression, thereby
decreasing NSCLC cell invasion and metastasis. The miR-200a/HGF
pathway also affected the radiosensitivity of NSCLC. These results
indicated that the miR-200a-mediated negative regulation of HGF is
involved in NSCLC invasion and metastasis at the cellular and
clinical levels.
Materials and methods
Tissue collection
Fresh tumor samples and adjacent normal tissues were
obtained from 11 patients who had undergone surgery at the Second
Affiliated Hospital of Soochow University (Suzhou, China) between
September 2016 and June 2017. Patients were aged between 45–65, of
whom 4 were female and 7 male. The pathological diagnosis of all
patients was NSCLC prior to surgery and no patients received
anticancer treatment prior to the operation. All tissue samples
were stored at −80°C. The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Soochow University
and written informed consent was provided by all patients.
Immunohistochemical (IHC)
staining
IHC staining was performed by a two-step procedure.
Upon rehydration in anhydrous ethanol for 5 min, 95% ethanol for 5
min, 85% ethanol for 5 min, 75% ethanol for 5 min, the slides were
washed 3 times in tap water, and then a phosphate-buffered saline
(PBS) solution was used 3 times for 5 min each time. A three-way
dip wax was used as a fixative, at 56–58°C, for ~3 h, and finally
embedded in a wax block casette, the continuous slice thickness is
3 µm. The slides were subjected to antigen retrieval by
pressure-cooking at 92–98°C for 15 min. Endogenous peroxidase
activity was neutralized using peroxide block placement on the
slides for 10 min at room temperature. The slides were then
incubated with anti-HGF polyclonal antibody (dilution 1:200; cat.
no. ab83760; Abcam, Cambridge, MA, USA) at 4°C overnight. This was
followed by incubation with peroxidase-conjugated polymer (ChemMate
EnVision/horseradish peroxidase; Gene Tech Biotechnology Co., Ltd.,
Shanghai, China) for 30 min at room temperature. The chromogen
reaction was developed in 3′-diaminobenzidine (DAB; Gene Tech
Biotechnology Co., Ltd.) tetrahydrochloride for 5 min at room
temperature. Finally, hematoxylin was performed for 30 sec at room
temperature used as a light nuclear counterstain.
The percentage of positive-staining cells was graded
on a scale of 0–3, with <5% positive-staining cells as grade 0,
5–25% as grade 1, 26–50% as grade 2 and >50% as grade 3. The
intensity of staining was also graded on a scale of 0–2, with
negative to weak intensity as grade 0, weak-moderate intensity as
grade 1 and moderate to strong intensity as grade 2. Finally, the
percentage and intensity scores were multiplied. The final score
between 0–2 was determined as low expression and a score higher
than 2 was determined as high expression.
miRNA screening
The targetscan.org and microrna.org
websites were used to screen miRNAs. Firstly, human species was
selected and HGF was entered as the target gene in TargetScan. This
identified eight broadly conserved 8-mer predicted target sites,
including miR-200a-3p, miR-141-3p, miR-26-5p, miR-199-5p,
miR-19-3p, miR-101-3p.1, miR-204-5p and miR-211-5p. Then, in the
mircorna.org website, after entering HGF as target
mRNA and selecting Homo sapiens, 29 miRNAs were displayed
and ordered by the sum of mirSVR scores, including miR-200a,
miR-141, miR-495 and miR-1297 among others. The results of the two
sites were then matched, finally selecting miR-200a and miR-141 for
subsequent experiments.
Cell culture
The parental human lung adenocarcinoma cell lines
(A549 and H1299) were obtained from the Cell Bank of Shanghai
Institute of Cell Biology (Chinese Academy of Medical Sciences,
Shanghai, China). A549 and H1299 cells were grown in Dulbecco's
modified Eagle's medium (DMEM; Corning, Inc., Corning, NY, USA)
containing 10% certified fetal bovine serum (FBS)-heat inactivated
(Biological Industries, Kibbutz Beit Haemek, Israel), penicillin
(100 U/ml), and streptomycin (100 U/ml) and maintained in an
incubator at 37°C with 5% CO2 in a humidified
atmosphere.
Western blot analysis
Briefly, NSCLC tissues milled into powder in liquid
nitrogen or A549 and H1299 cells were extracted using
radioimmunoprecipitation assay cell lysis reagent containing
proteinase and phosphatase inhibitors (Beyotime Institute of
Biotechnology, Haimen, China) at 4°C for 30 min. Cell lysates were
centrifuged at 12,000 × g for 20 min at 4°C and the protein
concentrations of the supernatant were determined using the
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). The supernatants containing total protein were then
mixed with a corresponding volume of 5X SDS loading buffer and
heated at 100°C for 10 min. Then, the supernatant lysates were run
on 10% SDS-polyacrylamide gels (50 µg/lane) and proteins were
transferred to polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA) by semidry electroblotting (1.5
mA/cm2). PVDF membranes were then incubated in blocking
buffer [Tris-buffered saline (TBS) supplemented with 0.05%
(vol/vol) Tween-20; TBST] containing 5% (wt/vol) skimmed milk
powder for 2 h at room temperature followed by three 10-min washes
in TBST. The PVDF membranes were then incubated with anti-HGF
(dilution 1:1,000; cat. no. ab83760 Abcam) or anti-c-Met (dilution
1:1,000; cat. no. ab74217; Abcam) as internal normalizers in TBST
containing 5% (wt/vol) skimmed milk powder (antibody buffer)
overnight at 4°C on a three-dimensional rocking table. Then, the
membranes were washed three times for 10 min in TBST and incubated
with goat anti-rabbit IgG conjugated to horseradish peroxidase
(dilution 1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) in antibody buffer for 2 h. Finally, membranes were
washed three times for 10 min in TBST and exposed to ECL Advanced
reagent (EMD Millipore) for 2 min as described in the
manufacturer's protocol. Membranes were exposed to Hyperfilm-ECL
for 2–5 min and visualized using a Fluor S Multimager and Quantity
One 4.1 (Gene Company, Ltd., Hong Kong, China). The molecular
weights of the bands were calculated by comparison with prestained
molecular weight markers (molecular weight range, 6,500-250,000)
that were run in parallel with the samples. Semiquantitative
analysis of specific immunolabeled bands was performed using a
Fluor S image analyzer and Quantity One 4.1 (Gene Company,
Ltd.)
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA from cultured cells was extracted using
the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. miRNA
levels were measured by RT-qPCR. Total RNA was subsequently reverse
transcribed at 65°C for 5 min to cDNA with the stem-loop reverse
transcription primer (Beijing Genomics Institute, Beijing, China)
for HGF and miR-200a detection. Then PCR was performed out in a PCR
gene amplification apparatus: 42°C for 60 min, 70°C for 5 min, and
then the reaction was terminated, placed on ice for storage or
stored at −20°C. The primer sequences are: HGF qPCR-forward primer,
5′-CAACAAACTTAGCTCATCGCAA-3′ and HGF qPCR-reverse primer,
5′-GCCTGGGTGAAAGAATCCT-3′; GAPDH qPCR-forward primer,
5′-CAAGGTCATCCATGACAACTTTG-3′ and GAPDH qPCR-reverse primer,
5′-GTCCACCACCCTGTTGCTGTAG-3′; miR-200a RT:
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACATCGT-3′;
miR-200a qPCR-forward primer, 5′-GGGGTAACACTGTCTGGTAG-3′ and
miR-200a qPCR-reverse primer, 5′-TGCGTGTCGTGGAGTC-3′; U6 forward
primer, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse primer,
5′-CGCTTCACGAATTTGCGTGTCAT-3′.
qPCR was carried out using SYBR Premix Ex
Taq™ (Takara Biotechnology, Co., Ltd., Dalian, China).
qPCR thermocycling was performed according to the following:
Pre-deformation 95°C for 10 min, 1 cycle; denaturation 95°C for 15
sec and annealing 60°C for 60 sec, 40 cycles. The reactions were
placed in a 96-well plate using a preheated real-time instrument
(ABI 7500HT; Applied Biosystems Life Technologies; Thermo Fisher
Scientific, Inc.). The relative levels of expression were
quantified and analyzed using Bio-Rad iCycler iQ software 3.1
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cq values were
used to calculate the RNA expression levels. The amount of miR-200a
expression (2−ΔΔCq) (27) was normalized using the endogenous U6
reference.
In situ hybridization (ISH)
staining
Sections of 1–3 µm thickness were cut from
paraffin-embedded tissues to evaluate miR-200a expression by ISH.
In brief, the slides were incubated at 60°C for 2 h, deparaffinized
in xylene and rehydrated with graded alcohol washes. Slides were
then washed three times with diethyl pyrocarbonate-treated PBS,
digested with 5 µg/ml proteinase K at 37°C for 30 min and washed
and dehydrated in graded alcohol. Slides were hybridized at 55°C
for 2 h with 50 nmol/l locked nucleic acid-modified
digoxigenin-labeled probes for miR-200a (Wuhan Boster Biological
Technology, Ltd., Wuhan, China): miR-200a Probe sequence:
5′-ACATCGTTACCAGACAGTGTTA-3′.
Following stringency washes (5X, 1X and 0.2X SSC),
slides were placed in blocking solution for 1 h at room temperature
followed by incubation in alkaline phosphatase conjugated anti-DIG
Fab fragment solution at 37°C for 1 h. Slides were incubated in
Strept Avidin-Biotin Complex-fluorescein isothiocyanate (FITC;
1:100; Wuhan Boster Biological Technology, Ltd.) at 37°C for 30
min, washed three times and directly observed by fluorescent
microscopy, with positive regions indicated by yellow or green.
The intensity histoscore with the following
categories according to Donahue's description was used: 0,
negative, 1, weakly positive, 2, moderately positive and 3,
strongly positive. The final score between 0–1 was determined as
low expression and a score of 2–3 was determined as high
expression.
Transfection of miRNA and siRNA
Human antisense miR-200a mimics and mimic negative
control were purchased from Guangzhou Ribobio Co., Ltd.,
(Guangzhou, China): Human antisense miR-200a mimic:
5′-UAACACUGUCUGGUAACGAUGUAUCGUUACCAGACAGUGUUAUU-3′. Negative
control: 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′.
Human antisense miR-200a mimic and mimic negative
control were referred to as miR-200a and mimic con, respectively.
Complete medium without antibiotics was used to culture the cells
at least 24 h prior to transfection. The cells were washed with 1X
PBS (pH 7.4) and then transiently transfected with 100 nM miR-200a
mimic or mimic con using Lipofectamine™ 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
protocol.
A total of three siRNA duplexes targeting human HGF
were designed and synthesized by Guangzhou RiboBio Co., Ltd. The
effective siRNA target sequence was 5′-GCAGAGGGACAAAGGAAAA-3′.
Negative control siRNA: 5′-UUUTGATCAUTGATGAAA-3′.
For transfection, the cells were plated on an
antibiotic-free growth medium at 60% confluence ~24 h prior to
transfection. RNA oligonucleotides were transfected at a final
concentration of 100 nM, using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol.
Rescue experiment
At 24 h following siRNA transfection into H1299
cells, HGF was transfected into the cells. Western blot detection
of HGF expression was performed after 24 h.
Transwell migration and invasion
assays
A549 and H1299 cells were grown in DMEM containing
10% FBS and transfected with 50 nM miR-200a mimic or negative
control, or 50 nM si-HGF or negative control. Following 24 h, the
cells were harvested by trypsinization and washed once with PBS. To
measure cell migration, 8-mm pore size culture inserts (Transwell;
Costar; Corning, Inc.) were placed into the wells of 24-well
culture plates, separating the upper and the lower chambers. In the
lower chamber, 500 µl of DMEM containing 10% FBS was added. Then,
serum-free medium containing 5×104 cells was added to
the upper chamber for migration assays. Invasion assays were
performed similarly except Transwells were precoated with 16 µg of
Matrigel (Corning, Inc.). Cells were incubated at 37°C with 5%
CO2 for 24 and 48 h for migration and invasion,
respectively, and cell morphology was observed by staining with
0.1% crystal violet staining solution was used for 10 min at room
temperature, and observed under light microscope following washing.
Filters were washed thoroughly with 1X PBS. Each experiment was
performed at least three times.
Dual-Luciferase reporter assay
Dual-Luciferase reporter assay system was purchased
from Promega Corp. (Madison, WI, USA). HGF wild-type and mutant
type were generated by Shanghai GenePharma Co., Ltd., (Shanghai,
China). The pmirGLO Dual-Luciferase miRNA Target Expression Vector
was used (Promega Corp.).
HGF 3′-UTR wild type 5′-CTGTTGTTTTGTTTGTCAGTGTTA-3′
HGF and 3′-UTR mutant type 5′-CTGTTGTTTTGTTTGTGCATACAA-3′.
HGF wild-type and mutant (100 ng) were transfected
using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) into H1299 cells together with 100 nM miR-200a mimic into
cells. After 36 h, growth media were removed from cultured cells,
which were rinsed in 1X PBS. The rinsing solution was removed and
20 µl of 1X Passive Lysis Buffer was added into each culture vessel
of 96-well culture plates, which were agitated for 30 min at room
temperature. Then, 100 µl of LAR II was added and firefly
luciferase activity was measured using a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA). Then, 100 µl of Stop &
Glo reagent was added and Renilla luciferase activity was
measured. The cycle was repeated for all wells in the plate.
Renilla luciferase (hRluc-neo) was used as a control
reporter for normalization and selection.
Clonogenic assay
Cells were transfected with miR-200a mimic or mimic
negative control, together with siR-HGF or negative control, as
described above. A total of 24 h later, transfected cells were
trypsinized, counted and replated at a density of 300, 500, 800,
1,200 and 2,000 cells/6-cm dish for 0, 2, 4, 6 and 8 Gy dose
irradiation, separately. A total of 10 days later, colonies
resulting from the surviving cells were fixed with 4%
paraformaldehyde for 15 min at room temperature, stained with 0.1%
crystal violet for 20 min and counted. Colonies containing at least
50 cells were scored under a light microscope. Each assay was
performed in triplicate.
Flow cytometric analysis of
apoptosis
Flow cytometric analysis of apoptosis was performed
with Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of
Biotechnology; cat. no. C1063). A549 and H1299 cells transfected
with miR-200a mimic or negative control, siRNA HGF or negative
control performed as described above. After 24 h, cells were
treated with 4 Gy dose irradiation. A total of 48 h later, cells
were trypsinized and resuspended, counted, and 5–10×105
cells in 1X binding buffer were incubated with 5 µl of Annexin
V-FITC and 10 µl propidium iodide (PI) for 15 min in the dark.
Cells were analyzed using a flow cytometer and associated BD
FACSuite™ v1.0.6.5230. (FACSCalibur; BD Biosciences) and
FITC-Annexin V-positive and PI-negative cells were considered as
apoptotic and the experiments were carried out in triplicates.
Immunofluorescence assay
Cells at 70–80% confluence were placed on sterilized
coverslips directly or 24 h following transfection. After
attachment, cells were treated with irradiation (4.0 Gy). A total
of 4 h later, the cells were fixed in ice-cold acetone for 15 min,
washed with PBS and then stained with rabbit anti-γ-H2AX (dilution
1:200; cat. no. ab2893; Abcam) 1 h following blocking with 3%
bovine serum albumin (BSA; Beyotime Institute of Biotechnology) for
30 min at room temperature. After washing, the cells were treated
with goat anti-rabbit FITC conjugated secondary antibody (Beyotime
Institute of Biotechnology; cat. no. P0176; 1.5 µg/ml) for 1 h at
room temperature and then counterstained with DAPI (Beyotime
Institute of Biotechnology; cat. no. C1005) for 5 min at room
temperature. Images were captured with a fluorescent
microscope.
Statistical analysis
All the quantitative data in the present study are
expressed as the mean of at least three independent experiments ±
standard error (SE). Comparisons of the control groups and the
treated groups were analyzed using the Student's t-test of GraphPad
Prism 5 software (Graphpad Software, Inc., La Jolla, CA, USA) and a
one-way analysis of variance (ANOVA) with a Fisher's post-hoc test
using SPSS statistics 19 software (IBM Corp., Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significantly
difference.
Results
HGF expression is significantly
associated with NSCLC
HGF serves a central role in the HGF/c-Met pathway.
To gain insight in to the association between HGF and cancer, the
expression level of HGF in lung cancer was investigated. IHC
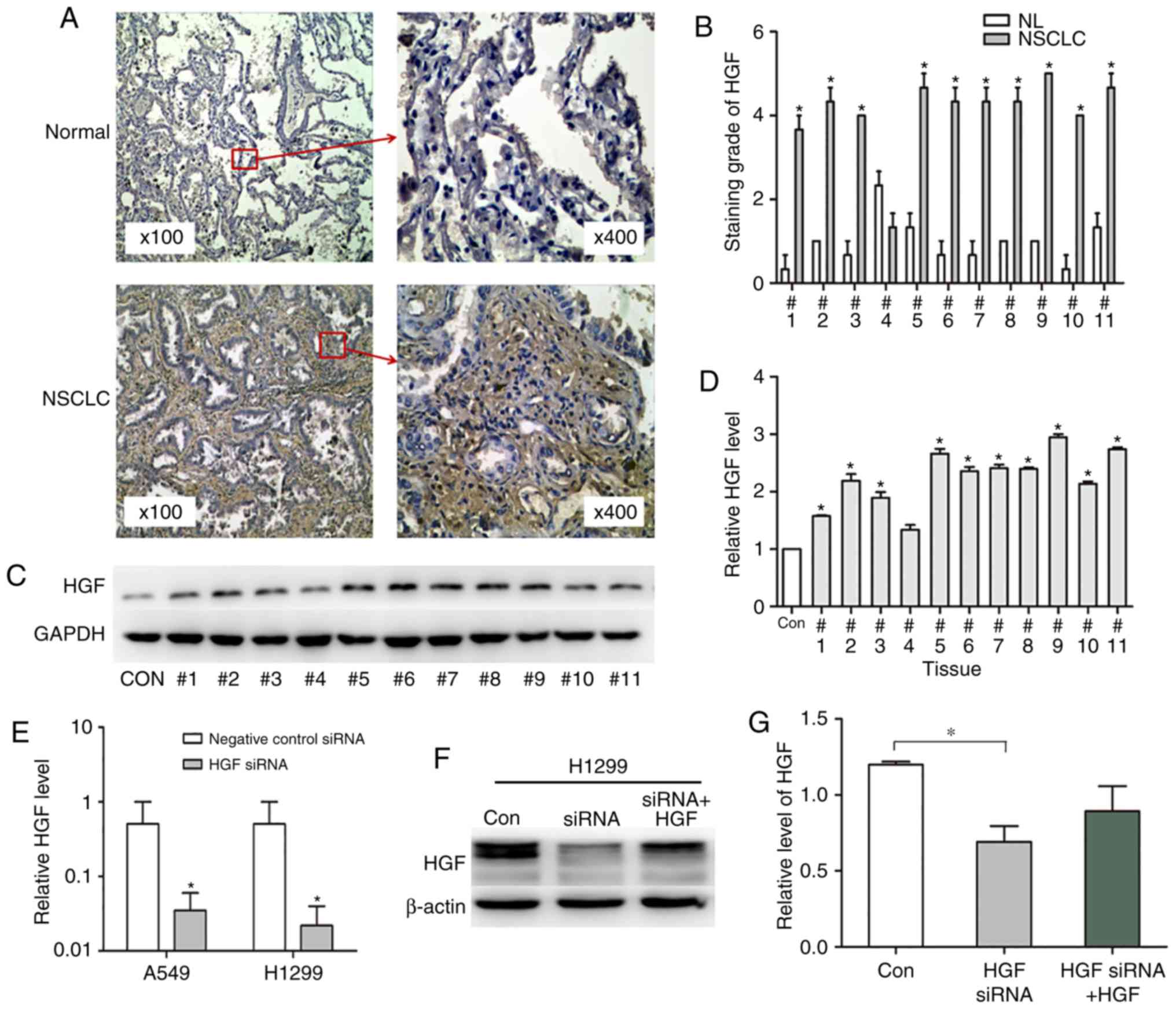
analysis of clinical lung cancer tissues (Fig. 1A) demonstrated that the HGF positive
ratio was 91% in lung cancer tissues, compared with 9% in normal
lung tissues (P<0.05; Fig. 1B),
indicating that the expression level of HGF was increased in NSCLC
compared within normal tissues. Next, the proteins from the above
samples were isolated and separated by gel electrophoresis to
perform grey value analysis (Fig.
1C). The relative protein expression level was significantly
increased in all the lung cancer tissues except 4 compared with
normal tissues (P<0.05; Fig.
1D). Western blot analysis confirmed that HGF expression was
increased in NSCLC tissues compared within normal lung tissues.
To assess the HGF expression level and activity
in vitro, HGF or mock siRNA were transfected and/or
co-cultured with HGF in A549 and H1299 NSCLC cells. The results of
RT-qPCR demonstrated that the HGF mRNA expression levels were
significantly reduced in A549 and H1299 cells (0.06- and 0.04-fold
in siRNA HGF and mock; P<0.05; Fig.
1E). Western blot analysis demonstrated similar results, as HGF
siRNA reduced the HGF expression level by 0.5-fold compared with
the control group (P=0.0086; Fig. 1E
and F). Treatment with HGF rescued the HGF siRNA effect
(Fig. 1F). As presented in the grey
value analysis, the HGF protein value decreased from 1 to 0.75 in
the HGF plus siRNA group compared with the siRNA alone group.
(P=0.14; Fig. 1G). These results
indicated that HGF can be upregulated or downregulated through HGF
siRNA or HGF treatment.
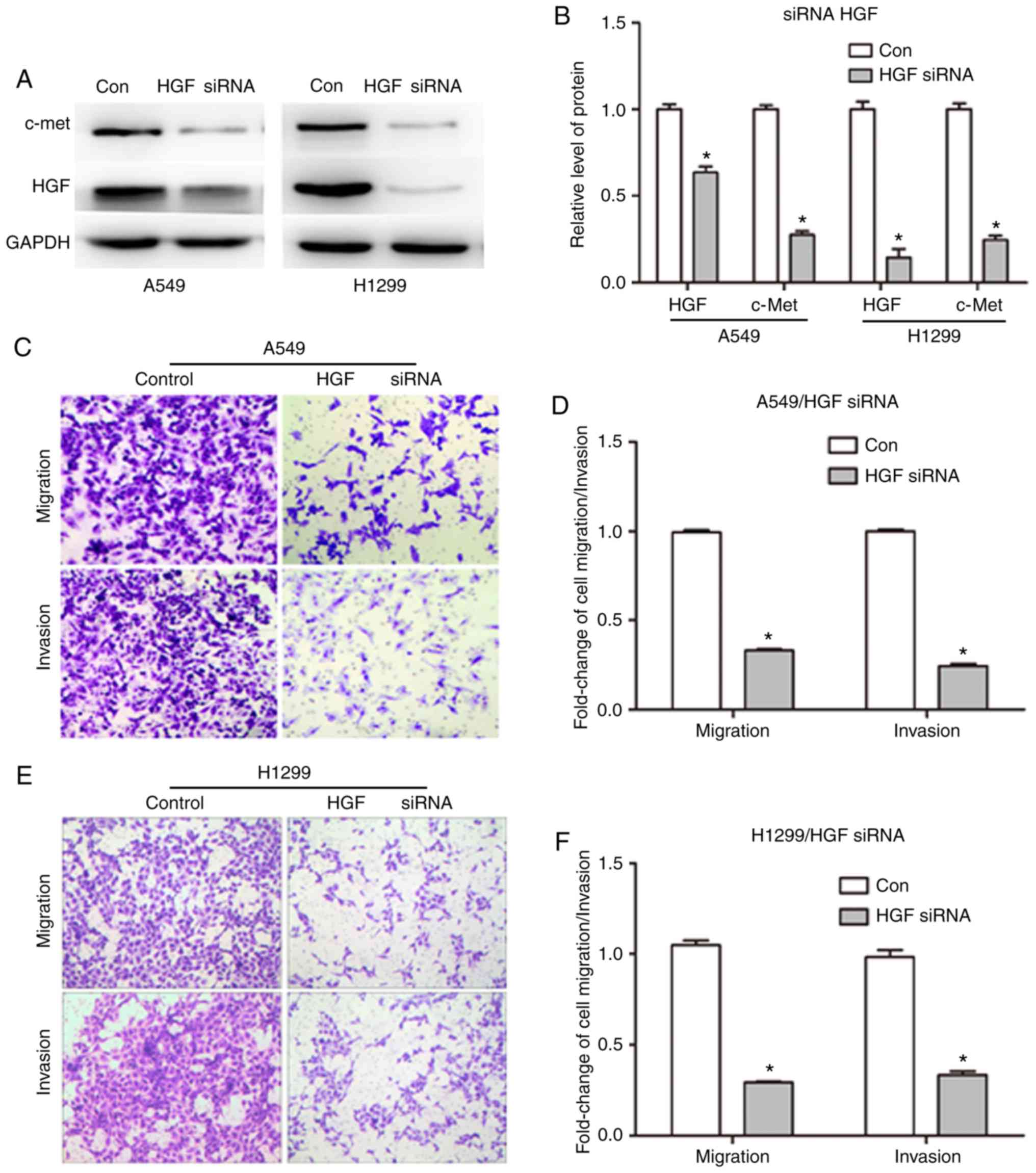
Downregulation of HGF expression
inhibits NSCLC cell migration and invasion
HGF was upregulated in NSCLC tissues compared with
normal tissues, suggesting that HGF upregulation affects lung
cancer malignancy. To clarify the role of HGF in tumor migration
and invasion, HGF was silenced in NSCLC cells using the siRNA
technique and the effect on cell migration and invasion was
assessed. A total of 48 h following transfection of siRNA into A549
and H1299 cells by Lipofectamine 2000, western blot analysis was
used to detect the protein expression of HGF and c-Met. As
presented in Fig. 2A, HGF siRNA
reduced the HGF expression. Grayscale value analysis demonstrated
that the expression level of HGF in A549 and H1299 cells was
decreased from 1 to 0.63±0.034 and 0.14±0.05 in the control group
and HGF siRNA groups, respectively (P<0.05; Fig. 2B). In addition, c-Met expression was
downregulated in response to HGF knockdown (Fig. 2A). The grey value of c-Met was
reduced to 0.28- and 0.25-fold in the siRNA group compared with the
con group (Fig. 2B), respectively.
The migration and invasion of NSCLC cells with silenced HGF were
examined by Transwell assay. The transmembrane cell rates of
migration and invasion were significantly decreased by 66.9 and
75.7% in A549 cells (Fig. 2C and
D), and by 70.7 and 66.7% in H1299 cells (Fig. 2E and F), respectively (P<0.05).
These results suggest that HGF is critical for the malignancy of
lung cancer through its effect on migration and invasion.
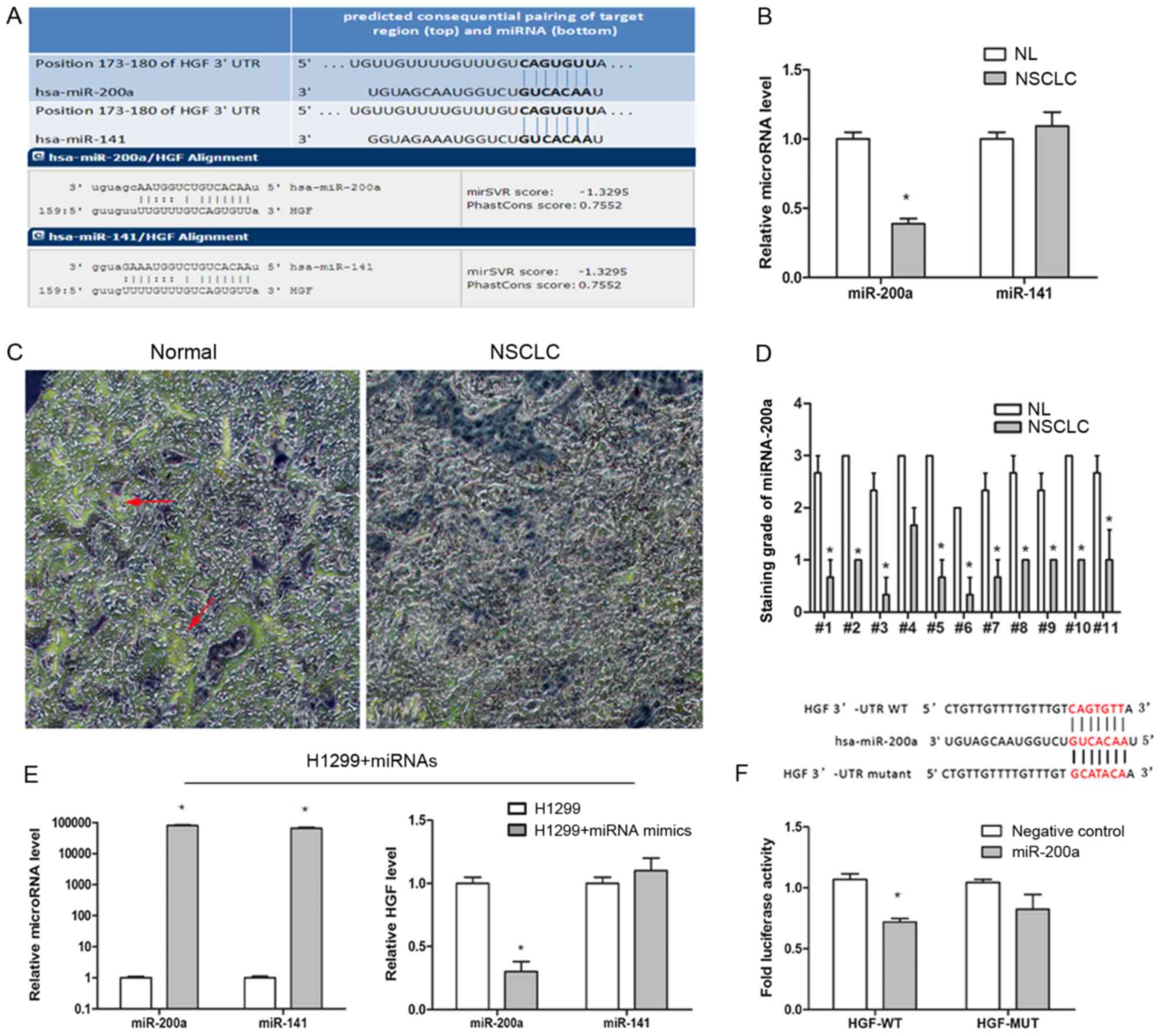
HGF is the target gene of
miR-200a
miRNAs serve important roles in the regulation of
the expression of various oncogenes by binding to complementary
3′-UTR mRNA sequences. First, the potential miRNAs involved in the
regulation of the HGF/c-Met signaling pathway in NSCLC cells were
investigated using targetscan.org and microrna.org
miRNA database websites. The intersection results demonstrated that
the conserved sequences of miR-200a and miR-141 have potential
binding sites in the HGF 3′-UTR mRNA region (Fig. 3A). Secondly, the expression level of
the two miRNAs in the NSCLC samples and normal lung tissues (n=11)
were investigated by RT-qPCR to determine whether miR-200a and
miR-141 were associated with NSCLC. The results demonstrated that
the miR-200a expression level was significantly decreased in NSCLC
samples compared with normal lung tissues (P<0.05; Fig. 3B). miR-141 expression levels did not
differ significantly between NSCLC and normal lung tissues. To
further confirm the miR-200a distribution and expression in lung
cancer, miR-200a expression in lung cancer and normal tissues was
examined by ISH (Fig. 3C). The
score of miR-200a was significantly lower in NSCLC lung cancer
tissues than in normal tissues (0.85±0.37 and 2.64±0.35,
respectively; P<0.05; Fig. 3D).
Collectively, these results indicated that miR-200a expression was
decreased in NSCLC, suggesting that miR-200a serves a role in NSCLC
by targeting HGF. To confirm this hypothesis, miR-200a mimics and
the corresponding negative controls were transfected into H1299
cells for 48 h. The RT-qPCR results demonstrated that HGF
expression was significantly reduced following transfection of
miR-200a mimics compared with the control (0.3±0.08; P<0.05),
whereas miR-141 had no significant effect on HGF expression
(Fig. 3E). To confirm this result,
an HGF-3′-UTR Dual-Luciferase reporter vector and mock vector was
constructed and co-transfected them into H1299 cells with miR-200a
mimics/control for 48 h. As presented in Fig. 3F, the HGF-WT image intensity was
significant weaker in miR-200a mimics compared within the control
(P<0.05). Taken together, these data imply that reduced
expression of miR-200a and increased expression of HGF in NSCLC
tissues are due to the miR-200a-mediated negative regulation of HGF
expression (Fig. 3F).
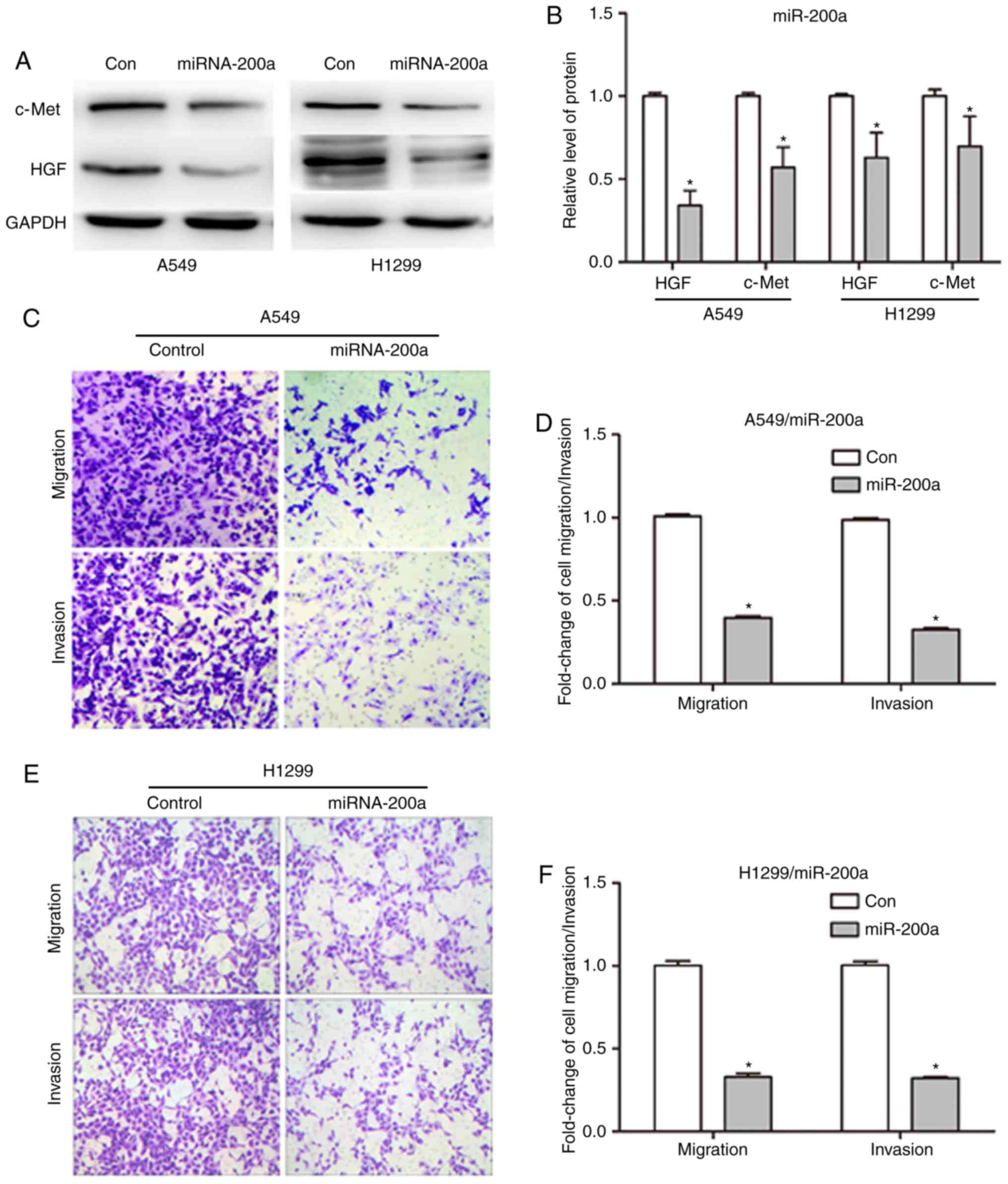
miRNA-200a reduces the migration and
invasion of NSCLC cells by negatively regulating HGF
expression
miR-200a is a negative regulator of HGF and is
significantly associated with HGF expression in NSCLC; however, how
miR-200a affects the development of NSCLC by targeting HGF remains
unclear. Migration and invasion are two major characteristics of
malignancies. Therefore, the expression of associated proteins was
assessed by western blot analysis of A549 and H1299 cells
transfected with miR-200a mimics and control. As presented in
Fig. 4A, following miR-200a
overexpression in cells by transfection for 48 h (Fig. 3E), the HGF protein band was weaker
in the miR-200a mimics group compared with the control (Fig. 4A). The HGF protein expression level
was significantly decreased to 0.34±0.09 and 0.63±0.15 in A549 and
H1299 cells (P<0.05; Fig. 4B) as
determined by protein band gray value analysis. A similar c-Met
protein expression trend was observed in the miR-200a mimics group
compared with the control group (Fig.
4A and B).
Since the upregulation of miR-200a reduced HGF and
c-Met expression, the migration and invasion of A549 and H1299
cells was examined next following miR-200a overexpression by
Transwell experiments. As presented in Fig. 4C, the number of migrated and invaded
cells was decreased in the miR-200a mimics A549 group compared with
the control group (Fig. 4C). The
migration and invasion rates were significantly reduced to
39.8±1.64 and 32.6±1.70%, respectively in the miR-200a transfected
A549 group compared with the control (P<0.05 and P<0.05;
Fig. 4D). Next, whether the
reduction of migration and invasion was unique to A549 cells was
verified. The results were similar in the H1299 cell line. Crystal
violet staining demonstrated in Fig.
4C indicated that the number of migrated and invaded cells was
decreased in the miR-200a mimics H1299 group (Fig. 4E). The migration and invasion rates
were significantly reduced to 32.8±4.0 and 32.2±0.98% in the
miR-200a H1299 group (P<0.05 and P<0.05; Fig. 4F). Collectively, these results
indicated that miR-200a serves an important role in regulating
NSCLC migration and invasion by negatively regulating HGF
expression.
High miR-200a and low HGF expression
promote DNA double strand breaks (DSBs) and apoptosis, and inhibits
colony formation in NSCLC cells in response to irradiation
Chemoradiation therapy is one of the main treatment
methods for advanced NSCLC. Histone H2AX on serine 139
phosphorylation (γ-H2AX) was selected as the DNA DSB marker to
assess the effect of the miR-200a/HGF pathway on DNA damage and
repair following exposure of A549 and H1299 cells to X-ray
radiation. Following transfection of miR-200a mimics and control,
A549 and H1299 cells were cultured for 48 h and exposed to 4 Gy
X-rays. Cells were fixed after 4 h and subjected to
immunofluorescence assays with the anti-γ-H2AX antibody (Fig. 5A). Compared with the control IR
group, the miR-200a mimics group formed more γ-H2AX foci (Fig. 5A). Compared with the control group,
the γ-H2AX foci number in the miR-200a mimics group was
significantly 1.51- and 1.79-fold increased (P<0.05; Fig. 5B). To confirm that the increase in
γ-H2AX foci was caused by the downregulation of HGF expression, HGF
siRNA or mock was directly transfected into A549 and H1299 cells
and immunofluorescence assays were performed. The results of
transfection of miR-200a mimics and siRNA HGF were similar, which
resulted in a greater number of γ-H2AX foci compared with the
control group (Fig. 5A). The γ-H2AX
foci number was significantly 1.46- and 1.82-fold increased in the
A549 and H1299 siRNA HGF groups compared within the control group
(P<0.05; Fig. 5B). This
suggested that miR-200a downregulated HGF expression and inhibited
the DNA DSB repair pathway, resulting in a higher number of DSBs.
Next, whether the miR-200a upregulation-induced DSBs increased the
cell death rate was tested. An Annexin-PI cell apoptosis kit was
used to assess the apoptosis rate in miR-200a-transfected A549 and
H1299 cells following exposure to 4 Gy irradiation and recovery for
48 h. Compared with the mock group, the miR-200a mimics group
exhibited a significantly higher apoptosis rate (Fig. 5C) by 1.8- and 1.7-fold (P<0.05;
Fig. 5D). The same assay was
performed in HGF siRNA transfected cells. As presented in Fig. 5C and D, the apoptosis rate was 1.44-
and 1.67-fold increased in the siRNA HGF group compared with the
mock group. In addition, transfection of miR-200a or HGF siRNA or
control into A549 and H1299 cells was used to investigate
radiosensitivity by comparing colony formation rate at radiation
doses of 0, 2, 4, 6 and 8 Gy. The survival curves demonstrated that
HGF siRNA or miR-200a mimics transfected A549 and H1299 groups were
more radiosensitive than the corresponding control groups (Fig. 5E and F). These results indicated
that miR-200a serves a key role in regulating NSCLC cell
radiosensitivity by targeting HGF in response to irradiation.
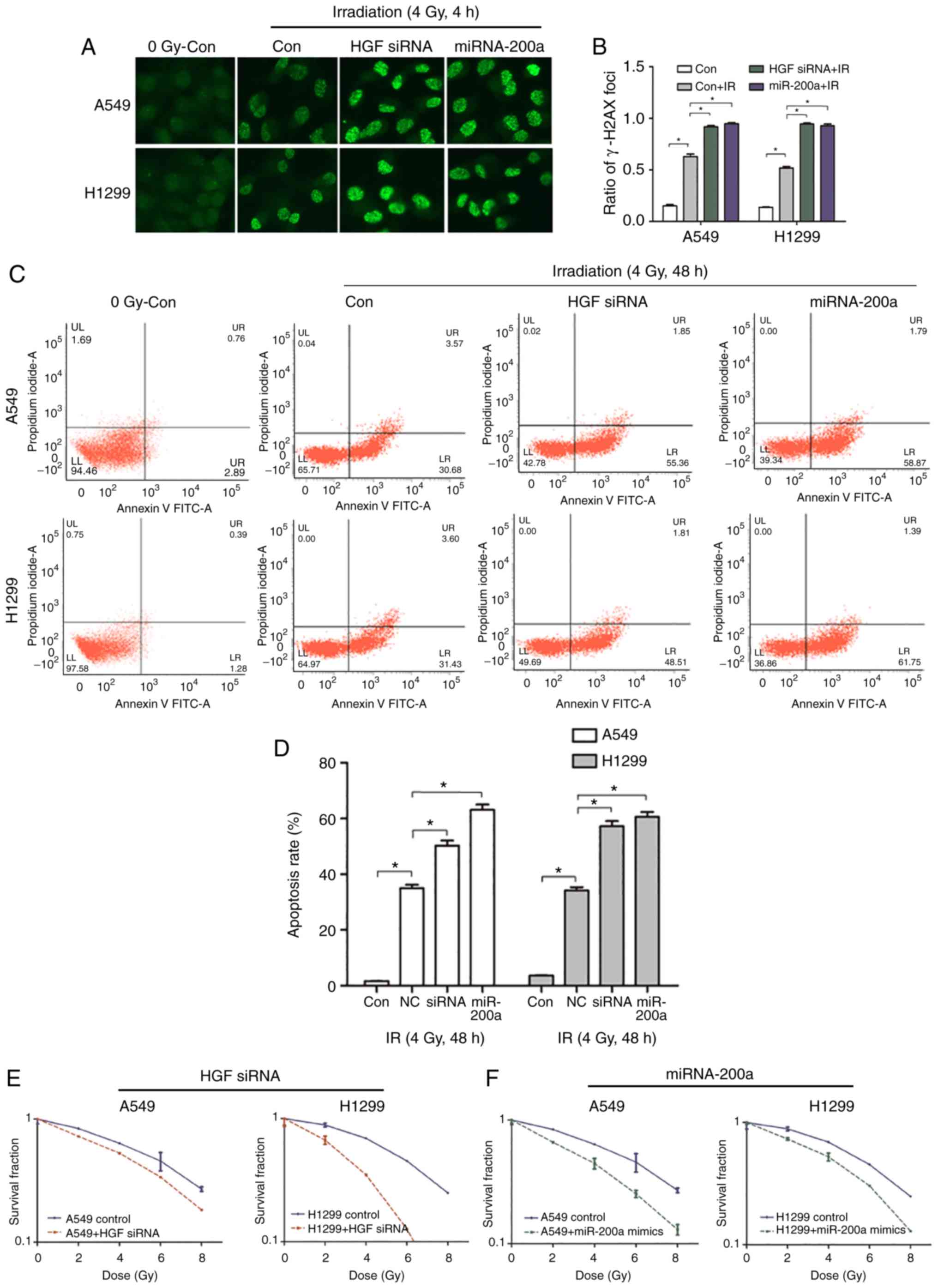 | Figure 5.High miR-200a and low HGF expression
promotes apoptosis, DNA double strand breaks, and inhibits the
cloning rate of NSCLC cells following irradiation. (A and B)
immunofluorescence detection of γ-H2AX foci in miR-200a transfected
or siRNA HGF-transfected A549 and H1299 cells treated with or
without irradiation. Magnification, ×100. *P<0.05 con+IR vs.
con. At 4 h following irradiation, the rates of γ-H2AX foci in
miR-200a mimics. *P<0.05 miR-200a+IR vs. con+IR or HGF siRNA.
*P<0.05, HGF siRNA+IR vs. con+IR. Transfected cells were higher
than in negative cells. (C and D) Flow cytometric analysis results
demonstrated that the rates of apoptosis of miR-200a mimics.
*P<0.05 miR-200a vs. con+IR or HGF siRNA, *P<0.05 HGF siRNA
vs. con+IR transfected cells were significantly increased compared
with negative cells at 48 h following irradiation. The colony
formation experiment results demonstrated that the radiosensitivity
of A549 and H1299 cells was enhanced following transfection of (E)
siRNA (*P<0.05 HGF siRNA vs. con+IR) or (F) miR-200a mimics.
*P<0.05 miR-200a vs. con +IR. HGF, hepatocyte growth factor; si,
small interfering; NSCLC, non-small cell lung cancer; Con, control;
miR, microRNA; IR, ionizing radiation. |
Discussion
Lung cancer remains the leading cause of
cancer-associated mortality (2).
Approximately 70% of NSCLC patients present with locally advanced
(clinical III) or metastatic disease at the time of diagnosis.
Identifying the regulatory molecules and the underlying mechanism
is important for improving our understanding of NSCLC and
developing novel treatments. In the present study, it was
demonstrated that HGF, a key c-Met growth pathway regulator, was
associated with NSCLC compared with normal clinical samples. HGF is
an autocrine and paracrine secretory factor that serves critical
roles in cell dissociation, migration, proliferation and
differentiation by targeting the c-Met receptor. The results of the
present study identified HGF as a potential marker for NSCLC. HGF
serves important roles through its secretion into the tumor
microenvironment and the activation of c-Met and downstream
effectors. HGF and its receptor c-Met serve important roles in
regulating cell proliferation. Overexpression of HGF has been
reported in a variety of types of human cancers, including NSCLC,
renal, breast, ovarian cancer and pleural mesothelioma (5). Its high level of expression represents
an unfavorable prognostic factor (6,7).
Therefore, focusing on the HGF status and regulation in tumors is
important to further develop novel therapeutic strategies. In the
present study, evidence was provided that the HGF expression level
was correlated with NSCLC tumor malignancy (10,28).
Concurrent radio-chemotherapy is the indicated
treatment for patients with NSCLC. Whether HGF expression affects
cancer radioresistance and its potential applications in
radiotherapy remain unclear. Ionizing radiation (IR) could activate
HGF and c-Met expression and signaling cascades, which serve
critical roles in promoting proliferation, invasion and resistance
to apoptosis (29,30). Analysis of HGF and Met expression in
205 pre-menopausal and 184 post-menopausal patients randomized to
receive chemo- or radiotherapy demonstrated that higher HGF or
metexpression in breast cancer patients was associated with a
better response to radiotherapy (31). Saigusa et al (32) demonstrated that inhibition of
radiation-induced HGF upregulation and blockade of
autocrine/paracrine HGF/c-Met signaling are potential novel
strategies for controlling distant recurrence in rectal cancer
patients following preoperative chemoradiotherapy. These findings
indicate that HGF is important for cancer radiotherapy research and
application. HGF and its receptor c-Met are overexpressed in NSCLC,
and their overexpression is associated with poor prognosis in
patients with NSCLC (33–35). The present study also demonstrated
that HGF expression was increased in NSCLC samples compared within
normal lung tissues. Several studies demonstrated that activation
of the HGF/c-Met signaling pathway not only promotes invasiveness
and distant metastasis in cancer, but also increases the
radioresistance of cancer (36,37).
The present study revealed that the silencing of HGF in A549 and
H1299 lung cancer cells inhibited their migration and invasion and
enhanced their radiosensitivity. Furthermore, the inhibition of HGF
expression promoted apoptosis and DNA DSBs in lung cancer cells
following irradiation. The results indicated that the HGF/c-Met
signaling pathway may be an effective target for increasing the
radiosensitivity of lung cancer.
In recent years, an increasing number of studies
have demonstrated that several inhibitors can specifically target
HGF/c-Met in various types of cancer (38,39).
Several c-Met inhibitors are currently under clinical studies
(40,41). In addition, HGF analogues, anti-HGF
humanized antibodies and c-Met induced receptor antibodies directed
against the c-Met extracellular domain are being used to prevent
HGF-specific binding and activation of c-Met (42,43).
Inhibition of HGF/c-Met signaling can also be achieved by using
selective c-Met small molecule tyrosine kinase inhibitors (TKIs) or
nonselective receptor TKIs. However, these methods have their own
limitations. For example, TKIs primarily inhibit tyrosine kinase
sites, not just only c-Met tyrosine kinases (44). Therefore, additional treatment
methods need to be identified.
In the last decade, the biological actions of miRNAs
have attracted much attention. A number of studies demonstrated
that miRNAs serve an important role in tumor pathogenesis and
provided novel insights into the biology of radiotherapy (45–47).
In the present study, miRNAs associated with NSCLC that function in
negatively regulating HGF expression was identified. miR-200a and
miR-141 were associated with NSCLC. Furthermore, miR-200a
negatively regulated HGF expression in the present study's
experiments. miR-200a belongs to the miR-200 family and
miR-200a-dependent signaling is associated with improved survival
rates of patients (48). In the
present study, it was demonstrated that transfection of miR-200a
mimics inhibited the migration and invasion of A549 and H1299 cells
and increased radiosensitivity compared with the corresponding
control cells. Similar to the results of HGF downregulation,
following radiation, the apoptosis and DNA DSBs of these NSCLC
cells were increased by miR-200a overexpression.
Although the potential for miR-200a in clinical
applications was identified, how to achieve the same expression
in vivo requires further investigation. Certain studies
demonstrated that xenografts can be used to upregulate specific
miRNAs for radiosensitization in vivo. In clinically
relevant studies, the delivery of synthetic miRNAs as tumor
suppressors was tested to develop alternative therapies (49,50).
For example, the use of liposomal nanoparticles containing miR-200c
mimics was tested in animal lung cancer models and this method can
increase tumor radiosensitivity by regulating cellular oxidative
stress (51). Therefore, the
application of miR-200a to clinical treatment may be achieved in
the future.
In conclusion, the present study demonstrated that
HGF is one of the target genes of miR-200a and miR-200a can inhibit
migration and invasion, increase the apoptosis rate and γ-H2AX
foci, and enhance the radiosensitivity of NSCLC cells by inhibiting
the HGF/c-Met signaling pathway. miR-200a could therefore be used
as a potential radiosensitization molecule in clinical
practice.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Jiangsu Provincial Natural Science Foundation Project (grant no.
BK20141185), the Shanghai Natural Science Foundation Project (grant
no. 17ZR1406100) and the Fudan University Shanghai Cancer Center
Foundation Project (grant no. YJRC1601).
Availability of data and materials
The datasets generated during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
MD completed all trial procedures, data analysis and
was a major contributor in writing the manuscript. JW was involved
in the collection and processing of human lung cancer specimens. HC
screened the microRNAs and organized the patient-related pathology
data. SW guided the histopathological study of
immunohistochemistry, the in situ hybridization and all the
methods and ideas of cell experiments. LC provided solutions and
detailed operations for ionizing radiation, provided the
accelerator use rights, control and calculation of radiation dose.
YX participated in the completion of the PCR and the in situ
hybridization test procedures, and participated in the cultivation
of cells. XL and FS designed the overall idea of the experiment and
provided theoretical guidance throughout the process, and
participated in the revision of the manuscript and the processing
of data. All authors read and approved the manuscript and agree to
be accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Soochow University
(Suzhou, China) and written informed consent was provided by all
patients.
Patient consent for publication
Written informed consent was provided by all
patients.
Competing interests
The authors declare that there is no conflict of
interest regarding the publication of this paper.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blumenschein GR Jr, Mills GB and
Gonzalez-Angulo AM: Targeting the hepatocyte growth factor-cMET
axis in cancer therapy. J Clin Oncol. 30:3287–3296. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim YJ, Go H, Wu HG, Jeon YK, Park SW and
Lee SH: Immunohistochemical study identifying prognostic
biomolecular markers in nasopharyngeal carcinoma treated by
radiotherapy. Head Neck. 33:1458–1466. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raghav KP, Wang W, Liu S, Chavez-MacGregor
M, Meng X, Hortobagyi GN, Mills GB, Meric-Bernstam F, Blumenschein
GR Jr and Gonzalez-Angulo AM: cMET and phospho-cMET protein levels
in breast cancers and survival outcomes. Clin Cancer Res.
18:2269–2277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Preusser M, Streubel B, Berghoff AS,
Hainfellner JA, von Deimling A, Widhalm G, Dieckmann K, Wöhrer A,
Hackl M, Zielinski C and Birner P: Amplification and overexpression
of CMET is a common event in brain metastases of non-small cell
lung cancer. Histopathology. 65:684–692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhardwaj V, Cascone T, Cortez MA, Amini A,
Evans J, Komaki RU, Heymach JV and Welsh JW: Modulation of c-Met
signaling and cellular sensitivity to radiation: Potential
implications for therapy. Cancer. 119:1768–1775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Firtina Karagonlar Z, Koc D, Iscan E,
Erdal E and Atabey N: Elevated hepatocyte growth factor expression
as an autocrine c-Met activation mechanism in acquired resistance
to sorafenib in hepatocellular carcinoma cells. Cancer Sci.
107:407–416. 2007. View Article : Google Scholar
|
|
11
|
Xie LQ, Bian LJ, Li Z, Li Y and Liang YJ:
Co-elevated expression of hepatocyte growth factor and
Interleukin-8 contributes to poor prognosis of patients with
primary nasopharyngeal carcinoma. Oncol Rep. 23:141–150.
2010.PubMed/NCBI
|
|
12
|
Sipos F, Galamb O, Herszényi L, Molnár B,
Solymosi N, Zágoni T, Berczi L and Tulassay Z: Elevated
insulin-like growth factor 1 receptor, hepatocyte growth factor
receptor and telomerase protein expression in mild ulcerative
colitis. Scand J Gastroenterol. 43:289–298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okazaki M, Yoshimura K, Uchida G and Harii
K: Elevated expression of hepatocyte and keratinocyte growth factor
in cultured buccal-mucosa-derived fibroblasts compared with
normal-skin-derived fibroblasts. J Dermatol Sci. 30:108–115. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Li M, Zhang G and Pang Z:
MicroRNA-10b overexpression promotes non-small cell lung cancer
cell proliferation and invasion. Eur J Med Res. 18:412013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F,
Bao G, Kong H, Ge C, Zhang F, et al: miRNA-200c inhibits invasion
and metastasis of human non-small cell lung cancer by directly
targeting ubiquitin specific peptidase 25. Mol Cancer. 13:1662014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang R, Chen DQ, Huang JY, Zhang K, Feng
B, Pan BZ, Chen J, De W and Chen LB: Acquisition of radioresistance
in docetaxel-resistant human lung adenocarcinoma cells is linked
with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling.
Oncotarget. 5:6113–6129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Montoya V, Fan H, Bryar PJ, Weinstein JL,
Mets MB, Feng G, Martin J, Martin A, Jiang H and Laurie NA: Novel
miRNA-31 and miRNA-200a-mediated regulation of retinoblastoma
proliferation. PLoS One. 10:e01383662015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang JJ, Tao H, Hu W, Liu LP, Shi KH, Deng
ZY and Li J: MicroRNA-200a controls Nrf2 activation by target Keap1
in hepatic stellate cell proliferation and fibrosis. Cell Signal.
26:2381–2389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu YY, Chen MB, Cheng L, Zhang ZQ, Yu ZQ,
Jiang Q, Chen G and Cao C: microRNA-200a downregulation in human
glioma leads to Gαi1 over-expression, Akt activation, and cell
proliferation. Oncogene. 37:2890–2902. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Blum D and LaBarge S: Reproducibility
Project: Cancer Biology: Registered report: Tumour
micro-environment elicits innate resistance to RAF inhibitors
through HGF secretion. Elife. 3:e040342014.doi:10.7554/eLife.04034.
View Article : Google Scholar :
|
|
29
|
Rivera M, Sukhdeo K and Yu J: Ionizing
radiation in glioblastoma initiating cells. Front Oncol. 3:742013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Bacco F, Luraghi P, Medico E, Reato G,
Girolami F, Perera T, Gabriele P, Comoglio PM and Boccaccio C:
Induction of MET by ionizing radiation and its role in
radioresistance and invasive growth of cancer. J Natl Cancer Inst.
103:645–661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Veenstra C, Pérez-Tenorio G, Stelling A,
Karlsson E, Mirwani SM, Nordensköljd B, Fornander T and Stål O: Met
and its ligand HGF are associated with clinical outcome in breast
cancer. Oncotarget. 7:37145–37159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saigusa S, Toiyama Y, Tanaka K, Yokoe T,
Fujikawa H, Matsushita K, Okugawa Y, Inoue Y, Uchida K, Mohri Y and
Kusunoki M: Inhibition of HGF/cMET expression prevents distant
recurrence of rectal cancer after preoperative chemoradiotherapy.
Int J Oncol. 40:583–591. 2012.PubMed/NCBI
|
|
33
|
Hosoda H, Izumi H, Tukada Y, Takagiwa J,
Chiaki T, Yano M and Arai H: Plasma hepatocyte growth factor
elevation may be associated with early metastatic disease in
primary lung cancer patients. Ann Thorac Cardiovasc Surg. 18:1–7.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Navab R, Liu J, Seiden-Long I, Shih W, Li
M, Bandarchi B, Chen Y, Lau D, Zu YF, Cescon D, et al:
Co-overexpression of Met and hepatocyte growth factor promotes
systemic metastasis in NCI-H460 non-small cell lung carcinoma
cells. Neoplasia. 11:1292–1300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Y, Du M, Wang J, Xing P, Zhang Y, Li
F and Lu X: MiRNA-200a expression is inverse correlation with
hepatocyte growth factor expression in stromal fibroblasts and its
high expression predicts a good prognosis in patients with
non-small cell lung cancer. Oncotarget. 7:48432–48442.
2016.PubMed/NCBI
|
|
36
|
Jankowski K, Kucia M, Wysoczynski M, Reca
R, Zhao D, Trzyna E, Trent J, Peiper S, Zembala M, Ratajczak J, et
al: Both hepatocyte growth factor (HGF) and stromal-derived
factor-1 regulate the metastatic behavior of human rhabdomyosarcoma
cells, but only HGF enhances their resistance to radiochemotherapy.
Cancer Res. 63:7926–7935. 2003.PubMed/NCBI
|
|
37
|
Sylvester PW: Targeting met mediated
epithelial-mesenchymal transition in the treatment of breast
cancer. Clin Transl Med. 3:302014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kou J, Musich PR, Staal B, Kang L, Qin Y,
Yao ZQ, Zhang B, Wu W, Tam A, Huang A, et al: Differential
responses of MET activations to MET kinase inhibitor and
neutralizing antibody. J Transl Med. 16:2532018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cruickshanks N, Zhang Y, Hine S, Gibert M,
Yuan F, Oxford M, Grello CM, Pahuski M, Dube C, Guessous F, et al:
Discovery and therapeutic exploitation of mechanisms of resistance
to MET inhibitors in glioblastoma. Clin Cancer Res. September
10–2018.(Epub ahead of print). doi: 10.1158/1078-0432.CCR-18-0926.
PubMed/NCBI
|
|
40
|
Aliebrahimi S, Montasser Kouhsari S, Ostad
SN, Arab SS and Karami L: Identification of phytochemicals
targeting c-Met kinase domain using consensus docking and molecular
dynamics simulation studies. Cell Biochem Biophys. 76:135–145.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu X, Kou J, Xiao Z, Tian F, Hu J, Zheng
P and Zhu W: Design, synthesis and biological evaluation of
6,7-disubstituted-4-phenoxyquinoline derivatives bearing
pyridazinone moiety as c-Met inhibitors. Molecules. 23:E15432018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Harshman LC and Choueiri TK: Targeting the
hepatocyte growth factor/c-Met signaling pathway in renal cell
carcinoma. Cancer J. 19:316–323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Peters S and Adjei AA: MET: A promising
anticancer therapeutic target. Nat Rev Clin Oncol. 9:314–326. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Utsugi T: New challenges and inspired
answers for anticancer drug discovery and development. Jpn J Clin
Oncol. 43:945–953. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Redis RS, Berindan-Neagoe I, Pop VI and
Calin GA: Non-coding RNAs as theranostics in human cancers. J Cell
Biochem. 113:1451–1459. 2012.PubMed/NCBI
|
|
46
|
Cherni I and Weiss GJ: miRNAs in lung
cancer: Large roles for small players. Future Oncol. 7:1045–1055.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du L and Pertsemlidis A: microRNA
regulation of cell viability and drug sensitivity in lung cancer.
Expert Opin Biol Ther. 12:1221–1239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang X and Li GH: MicroRNA-16 functions as
a tumor-suppressor gene in oral squamous cell carcinoma by
targeting AKT3 and BCL2L2. J Cell Physiol. 233:9447–9457. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun GL, Li Z, Wang WZ, Chen Z, Zhang L, Li
Q, Wei S, Li BW, Xu JH, Chen L, et al: miR-324-3p promotes gastric
cancer development by activating Smad4-mediated Wnt/beta-catenin
signaling pathway. J Gastroenterol. 53:725–739. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cortez MA, Valdecanas D, Zhang X, Zhan Y,
Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al:
Therapeutic delivery of miR-200c enhances radiosensitivity in lung
cancer. Mol Ther. 22:1494–1503. 2014. View Article : Google Scholar : PubMed/NCBI
|