Introduction
Tongue squamous cell carcinoma (TSCC) is one of the
most common types of head and neck cancer (HNC) (1,2);
>3,000 people worldwide are diagnosed with TSCC annually and its
accounts for 24% of all HNC cases (3). Although many patients are identified
at the earlier stages of the disease, the 5-year survival rate for
advanced-stage TSCC is only ~50%, due to high invasiveness and
metastasis (4). In addition, no
effective drugs are currently available for the treatment and
control of regional and distant metastases of TSCC. Therefore,
screening novel methods for the diagnosis, prognosis and treatment
of TSCC may result in a significant clinical advancement.
TSCC is caused by the interaction of numerous genes,
the increased expression of proto-oncogenes and inhibition of tumor
suppressor genes. Furthermore, cell signaling pathways, such as p53
(5), Ras (6), mitogen-activated protein kinase, Janus
kinase/signal transducer and activator of transcription (7), Wnt and hedgehog (8) signaling pathways serve important roles
in the development of TSCC. The Wnt protein family is a group of 19
secreted glycoproteins (9); Wnt
proteins transmit signals to cells through ≥15 receptors or
collaborative receptors, thus resulting in regulation of the
differentiation, apoptosis and proliferation of epithelial cells
(10,11). Postoperative metastasis leads to
unfavorable outcomes in patients with TSCC, and
epithelial-mesenchymal transition (EMT) is considered the
fundamental event of cancer metastasis and recurrence (12,13).
It is well known that EMT is regulated by a series of molecular
factors, including Wnt signaling-related genes (14). Wnt7a is a member of the Wnt family,
which has not yet been studied in TSCC. β-catenin is a core
transducer in the canonical Wnt signaling pathway and is an
EMT-associated marker (15).
Previous studies have indicated the important role of β-catenin in
EMT, thus bridging a crosstalk between the Wnt pathway and EMT
(16,17). In addition, the positive correlation
between Wnt7a and the Wnt/β-catenin pathway has been discussed
previously (18); therefore, it is
of great value to explore the role of Wnt7a in the EMT process
during TSCC progression. The present study focused on Wnt7a and the
Wnt pathway; however, research regarding the interaction between
Wnt7a and other pathways may be conducted in the future.
Wnt7a has been reported to be highly associated with
the progress of cell growth through regulating transforming growth
factor (TGF)-β receptor and activating the cancer-associated
fibroblast phenotype (19). Wnt7a
has been reported to be overexpressed in colorectal, pancreatic,
gastric, breast and ovarian carcinoma (20). Conversely, in non-small cell lung
carcinoma, Wnt7a may function as a tumor suppressor gene (21). In addition, the Wnt7a gene has been
reported to be downregulated by high-frequency hypermethylation in
pancreatic carcinoma (22). The
overexpression of Wnt7a has been demonstrated to enhance the
invasiveness and metastasis of ovarian cancer cells, possibly
through the Frizzled receptor (23). These results suggested that Wnt7a
may regulate the proliferation and adhesion of cancer cells;
however, its role in tumor cell proliferation and migration may be
dependent on tumor type. However, the clinical significance of
Wnt7a expression in TSCC has not yet been revealed. Therefore, it
is of great significance to determine the expression levels of
Wnt7a in TSCC tissues, and to assess its clinical implications.
Furthermore, the mechanism underlying the regulatory effects of
Wnt7a on cell proliferation, metastasis and EMT in TSCC was
investigated.
Materials and methods
Tissue samples
Two independent cohorts, comprising 109 patients
with TSCC who were diagnosed, treated and followed up at the
Stomatological Hospital, Southern Medical University (Guangzhou,
China) between November 2010 and May 2014, were recruited to the
present study. All patients provided informed consent for the use
of their tissues and clinical data. Specimens, including paired
cancerous and non-cancerous tissues, were obtained following
surgical resection. Samples were immediately frozen using liquid
nitrogen after resection and were stored at −80°C prior to RNA
extraction (cohort 1, n=48), or were fixed in 4% paraformaldehyde
at room temperature for 24–48 h and embedded in paraffin for
immunohistochemistry (IHC; cohort 2, n=61). All patients were
initially diagnosed with tongue cancer and did not receive any
other treatment prior to surgery. The present study was approved by
the Independent Ethics Committee of the Stomatological Hospital,
Southern Medical University (approval no. [2018]05).
Cell culture
The CAL-27, SCC-15 and SCC-9 human TSCC cell lines
were obtained from the American Type Culture Collection (Manassas,
VA, USA). HSC-4 was purchased from the Human Science Research
Resources Bank of the Japan Health Sciences Foundation (Tokyo,
Japan). Human oral keratinocytes were purchased from ScienCell
Research Laboratories, Inc. (San Diego, CA, USA). CAL-27 cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), HSC-4 cells were
cultured in Eagle's Minimum Essential Medium (Gibco; Thermo Fisher
Scientific, Inc.), and SCC-15 and SCC-9 cells were maintained in a
1:1 mixture of DMEM and Ham's F12 medium (Gibco; Thermo Fisher
Scientific, Inc.). For complete growth medium, fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) was added to the media
at a final concentration of 10%. The cells were maintained at 37°C
in a humidified atmosphere containing 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cell lines
using RNAiso Plus reagent (Takara Bio, Inc., Otsu, Japan), followed
by RT using PrimeScript First-Strand cDNA Synthesis kit (Takara
Bio, Inc.), according to the manufacturer's protocols. The mRNA
expression levels of Wnt7a were evaluated by qPCR using a
SYBR® Premix Ex Taq™ kit (cat. no. RR420A; Takara Bio,
Inc.) on the StepOnePlus Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The RT-qPCR primers used were as
follows: Wnt7a, forward 5′-CCAGCAAAAACCTTGAGCCC-3′, reverse
5′-CCTTGTTCAGGTAGGCAGCA-3′; and β-actin, forward
5′-CGTCACCAACTGGGACGACA-3′ and reverse 5′-CTTCTCGCGGTTGGCCTTGG-3′.
β-actin was employed as an endogenous control. Target and
endogenous control genes were amplified in triplicate and the
relative mRNA expression levels of Wnt7a for each sample were
calculated using the 2−ΔΔCq method (24).
IHC
Paraffin-embedded sections (4 µm) were cut and
mounted onto slides, after which, paraffin was removed with xylene
and the sections were rehydrated in an alcohol gradient.
Subsequently, the sections were treated with citrate buffer for
antigen retrieval in a microwave oven for 10 min at 95°C and were
then cooled to room temperature. The sections were then immersed in
3% H2O2 for 15–20 min at room temperature,
blocked with 5% bovine serum albumin (EMD Millipore, Billerica, MA,
USA) solution for 30 min at room temperature, and incubated with
the Wnt7a primary antibody (cat. no. 10605-1-AP; 1:200; ProteinTech
Group, Inc., Chicago, IL, USA) at 4°C overnight. The next day,
sections were incubated with HRP-conjugated goat anti-rabbit IgG
(cat. no. TA130023; OriGene Technologies, Inc., Beijing, China) for
15 min at room temperature to detect primary antibody. Finally, the
slides were incubated with DAB (cat. no. PV-6000; ZSGB-BIO; OriGene
Technologies, Inc.) and were observed under an upright light
microscope.
Two individual pathologists evaluated positive
staining of Wnt7a in TSCC tissues. Cytoplasmic brown staining in
tumor cells was considered positive and was scored based on the
following criteria: i) Staining intensity: 0, negative Wnt7a
staining; 1, weak staining; 2, moderate staining; and 3, strong
staining. ii) Percentage of stained tumor cells was categorized
into four classes, 0, negative; 1, ≤25%; 2, 26–50%; 3, 51–74%; and
4, ≥76%. Finally, Wnt7a protein expression was scored
semi-quantitatively by multiplying the aforementioned scores.
Gene silencing
When cells reached a confluence of ~70%, small
interfering (si)RNAs (Sangon Biotech Co., Ltd., Shanghai, China)
targeting Wnt7a mRNA (siWnt7a-1: 5′-AAAUUGUUAAAUAUUGCUGUG-3′;
siWnt7a-2: 5′-UUAAUAUUAUUUUUAUCAGAA-3′) were transfected at a
concentration of 50 nmol/l to induce transient knockdown of Wnt7a
expression using Lipofectamine® 3000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The cells were collected for
subsequent experiments after 48–72 h incubation. The negative
control (NC) sequence was as follows:
5′-UUCUCCGAACGUGUCACGUTT-3′.
For the construction of stable Wnt7a-knockdown cell
models, CAL-27 cells underwent lentivirus-mediated infection with
short hairpin (sh)RNA (Sangon Biotech Co., Ltd.) targeting Wnt7a
mRNA (5′-AAAUUGUUAAAUAUUGCUGUG-3′) or the NC sequence
(5′-UUCUCCGAACGUGUCACGUTT-3′) at a multiplicity of infection (MOI)
of 20, according to the manufacturer's protocol. After 8 h culture,
the lentivirus-containing culture medium was replaced with fresh
medium. The efficiency of lentivirus-mediated RNA interference was
determined by RT-qPCR and western blotting.
Cell proliferation assay
Cell proliferation was measured using the Cell
Counting Kit-8 (CCK-8) (Dojindo Molecular Laboratories, Inc.,
Kumamoto, Japan). CAL-27 and SCC-15 cells were collected,
resuspended and seeded onto 96-well plates, at a density of
3×103 cells/well. After adherence, cells were
transfected with Wnt7a siRNA or NC siRNA. After a further
incubation for 0, 24, 48, 72 and 96 h, 10 µl CCK-8 reagent was
added to each well, and absorbance was measured at 450 nm using a
microplate reader (SpectraMax Plus 384; Molecular Devices, LLC,
Sunnyvale, CA, USA) after 2 h incubation at 37°C.
Tumorigenicity assay
BALB/c male nude mice (specific pathogen-free grade;
weight, 14–16 g; age, 3–4 weeks) were purchased from the Laboratory
Animal Center of Southern Medical University. Lentiviral-mediated
stable Wnt7a-knockdown CAL-27 cells and NC cells were
subcutaneously injected into the flanks of nude mice (n=5 in each
group). Subsequently, the mice were maintained in a specific
pathogen-free grade lab, under the following conditions: Controlled
temperature, 23±2°C; humidity, 40–70%; 12-h light/dark cycle) at
the Laboratory Animal Center of Nanfang Hospital, Southern Medical
University (Guangzhou, China) with ad libitum access to food
and water for 4 weeks. The volume of xenograft tumors was monitored
every 3 days by measuring the length and width (Volume = length ×
width × width × π/6). The animal study was conducted in accordance
with the Institutional Animal Care and Use Committee (IACUC)
guidelines, and was approved by Experimental Animal Ethics
Committee of Southern Medical University (approval no.
L2016113).
Transwell migration and invasion
assays
For the migration assay, a total of 5×104
CAL-27 or SCC-15 cells transfected with Wnt7a siRNA or NC siRNA
were collected and resuspended in 200 µl serum-free medium. The
cells were added to the upper chamber of a Transwell insert
(Corning Incorporated, Corning, NY, USA) placed in a 24-well plate,
whereas 500 µl complete medium containing 10% FBS was added to the
lower chamber. After 24 h culture, cells on the upper surface of
the filter were removed and cells on the lower surface were stained
with 0.1% crystal violet for 20 min at room temperature. Cells that
migrated through the filters were counted and images were captured
in five random fields at a magnification of ×400 using an inverted
light microscope. For the invasion assay, the chambers were
pre-coated with Matrigel matrix (Corning Incorporated) diluted with
serum-free medium in a ratio of 1:8.
Western blotting
Cells were lysed in radioimmunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China)
supplemented with 1% protease inhibitors and phenylmethylsulfonyl
fluoride. Subsequently, after determining the concentration of
samples using a bicinchoninic acid protein assay kit (cat. no.
K3000; Shanghai Bocai Biotechnology Co., Ltd., Shanghai, China), 30
µg protein was separated by 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes (EMD Millipore). After blocking
non-specific antigens with 5% skimmed milk for 1 h at room
temperature, membranes were incubated with primary antibodies at
4°C overnight. Subsequently, the membranes were washed and
incubated with HRP-conjugated goat anti-rabbit secondary antibody
(cat. no. D110058; 1:10,000; BBI Life Sciences, Shanghai, China)
for 1 h at room temperature. Finally, signals were visualized using
the enhanced chemiluminescence detection system (Hangzhou Fude
Biological Technology Co., Ltd., Hangzhou, China). The primary
antibodies used were as follows: Anti-Wnt7a rabbit polyclonal
antibody (cat. no. 10605-1-AP; 1:1,000; ProteinTech Group, Inc.),
anti-E-cadherin rabbit polyclonal antibody (cat. no. 20874-1-AP;
1:1,000; ProteinTech Group, Inc.), anti-N-cadherin rabbit
polyclonal antibody (cat. no. 22018-1-AP; 1:1,000; ProteinTech
Group, Inc.), anti-Vimentin rabbit monoclonal antibody (cat. no.
5741T; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA),
anti-β-catenin rabbit monoclonal antibody (cat. no. 8480T; 1:1,000;
Cell Signaling Technology, Inc.), anti-Snail rabbit monoclonal
antibody (cat. no. 3879T; 1:1,000; Cell Signaling Technology, Inc.)
and anti-β-actin rabbit polyclonal antibody (20536-1-AP; 1:10,000;
ProteinTech Group, Inc.). β-actin was used as an endogenous protein
for normalization.
Statistical analysis
Statistical analysis was conducted using IBM SPSS
Statistics 20.0 (IBM Corporation, Armonk, NY, USA). Data from
triplicate experiments are presented as the means ± standard error
of the mean. The associations between Wnt7a mRNA/protein expression
and clinicopathological parameters were assessed using the
χ2 test. Student's t-test was used to analyze the
differences between two groups. One-way analysis of variance
followed by least significant difference test, or Kruskal-Wallis
test, was used to compare multiple groups. Survival data were
evaluated using the Kaplan-Meier method and log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Upregulation of Wnt7a in human TSCC
tissues
Wnt7a expression was detected in the two cohorts of
patients with TSCC, which comprised 109 pairs of TSCC specimens and
adjacent non-cancerous tissues, using RT-qPCR and IHC. The present
study aimed to determine the role of Wnt7a in TSCC. In cohort 1,
upregulation of Wnt7a mRNA was observed in TSCC tissues in 72.9%
(35/48) of patients (Fig. 1A). The
mRNA expression levels of Wnt7a were significantly increased in
TSCC tissues compared with in the corresponding adjacent normal
tissues (Fig, 1B). Furthermore, IHC
staining revealed that Wnt7a protein, which is primarily located in
the cytoplasm, was significantly overexpressed in TSCC tissues
compared with in matching adjacent normal tissues (Fig. 1C and D). These results suggested
that Wnt7a was upregulated in TSCC tissues and may exert a role in
carcinogenesis and its progression.
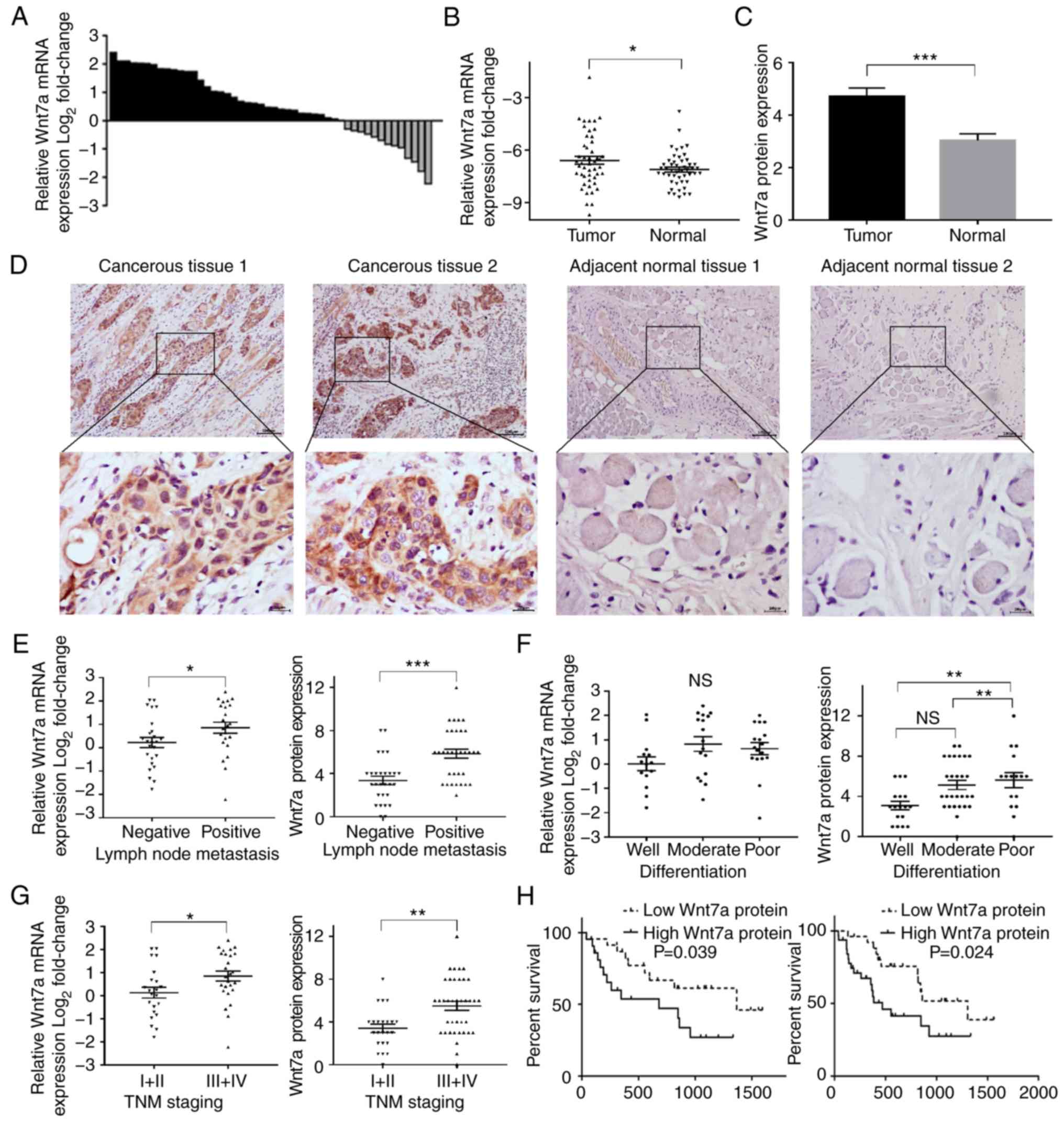 | Figure 1.Wnt7a mRNA and protein expression in
patients with TSCC, and the association between Wnt7a expression
and recurrence-free survival. (A) Alterations in Wnt7a expression
in TSCC tissues compared with in adjacent normal tissues. (B) Wnt7a
mRNA was upregulated in TSCC tissues compared with that in matching
non-cancerous tissues (Student's t-test). (C) Wnt7a protein was
upregulated in TSCC tissues compared with that in the matching
non-cancerous tissues (Student's t-test). (D) Immunohistochemical
staining exhibited Wnt7a protein expression in TSCC and adjacent
normal tissues (magnification: Upper panel, ×100, scale bars=100
µm; lower panel, ×400, scale bars=20 µm). (E) Wnt7a mRNA and
protein expression was higher in tissues obtained from patients
with TSCC with lymph node metastasis than those without lymph node
metastasis (Student's t-test). (F) Wnt7a protein expression was
higher in ESCC tissues with poor differentiation than those with
well or moderate differentiation (one-way analysis of variance and
least significant difference test); however, no significant
difference was observed in Wnt7a mRNA expression between groups
(Kruskal-Wallis test). (G) Wnt7a mRNA and protein expression were
higher in tissues obtained from patients with advanced TSCC than
those with early stage TSCC (Student's t-test). (H) Kaplan-Meier
analysis demonstrated that patients with higher Wnt7a mRNA and
protein expression had a shorter recurrence-free survival compared
with those with lower Wnt7a expression (log-rank test). *P<0.05,
**P<0.01, ***P<0.001. NS, not significant; TSCC, tongue
squamous cell carcinoma. |
Association between Wnt7a expression
and clinicopathological parameters in patients with TSCC
The association between Wnt7a expression and
clinicopathological parameters was analyzed to assess the effects
of Wnt7a on TSCC. In cohort 1, the patients were categorized into
low or high expression groups according to the median value. Wnt7a
mRNA expression was strongly associated with T classification
(P=0.039), lymph node metastasis (P=0.020), pathological
differentiation (P=0.037) and clinical stage (P=0.020; Table I). In cohort 2, TSCC tissues with a
staining index of >4 were defined as the high expression group,
whereas those with a staining index ≤4 were defined as the low
expression group. Results demonstrated that Wnt7a protein
expression was positively associated with T classification
(P=0.033), lymph node metastasis (P=0.001), pathological
differentiation (P<0.0001), and clinical stage (P=0.002;
Table II). Further analyses
revealed that Wnt7a protein expression was higher in tissues
obtained from patients with lymph node metastasis (Fig. 1E), poor differentiation (Fig. 1F) and advanced TSCC (Fig. 1G). Wnt7a mRNA expression was also
higher in tissues obtained from patients with lymph node metastasis
(Fig. 1E) and advanced TSCC
(Fig. 1G); however, it was not
significantly associated with differentiation (P=0.087; Fig. 1F). Furthermore, patients with higher
Wnt7a mRNA (median recurrence-free survival, 1,366 days vs. 681
days, P=0.039) and protein (median recurrence-free survival, 1,305
days vs. 469 days, P=0.024) expression had a shorter
recurrence-free survival (Fig.
1H).
 | Table I.Association between Wnt7a mRNA
expression and clinicopathological parameters in patients with
tongue squamous cell carcinoma (n=48). |
Table I.
Association between Wnt7a mRNA
expression and clinicopathological parameters in patients with
tongue squamous cell carcinoma (n=48).
|
| Wnt7a mRNA |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | Low (n=24)(%) | High (n=24)(%) | P-value |
|---|
| Gender |
|
| 0.365 |
|
Male | 14 (45.2) | 17 (54.8) |
|
|
Female | 10 (58.8) | 7 (41.2) |
|
| Age (years) |
|
| 0.755 |
|
<60 | 17 (51.5) | 16 (48.5) |
|
|
≥60 | 7 (46.7) | 8 (53.3) |
|
| T
classification |
|
| 0.039 |
|
T1/T2 | 17 (65.4) | 9 (34.6) |
|
|
T3/T4 | 7 (31.8) | 15 (68.2) |
|
| Lymph node
metastasis |
|
| 0.020 |
|
Positive | 7 (30.4) | 16 (69.6) |
|
|
Negative | 17 (68.0) | 8 (32.0) |
|
| Pathological
differentiation |
|
| 0.037 |
|
Well | 11 (78.6) | 3 (21.4) |
|
|
Moderate | 7 (41.2) | 10 (58.8) |
|
|
Poor | 6 (35.3) | 11 (64.7) |
|
| Clinical stage |
|
| 0.020 |
|
I+II | 15 (68.2) | 7 (31.8) |
|
|
III+IV | 9 (34.6) | 17 (65.4) |
|
 | Table II.Association between Wnt7a protein
expression and clinicopathological parameters in patients with TSCC
(n=61). |
Table II.
Association between Wnt7a protein
expression and clinicopathological parameters in patients with TSCC
(n=61).
|
| Wnt7a protein |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | Low (n=27)(%) | High (n=34)(%) | P-value |
|---|
| Gender |
|
| 0.288 |
|
Male | 13 (38.2) | 21 (61.8) |
|
|
Female | 14 (51.9) | 13 (48.1) |
|
| Age (years) |
|
| 0.134 |
|
<60 | 24 (49.0) | 25 (51.0) |
|
|
≥60 | 3 (25.0) | 9 (75.0) |
|
| T
classification |
|
| 0.033 |
|
T1/T2 | 20 (55.6) | 16 (44.4) |
|
|
T3/T4 | 7 (28.0) | 18 (72.0) |
|
| Lymph node
metastasis |
|
| 0.001 |
|
Positive | 8 (24.2) | 25 (75.8) |
|
|
Negative | 19 (67.9) | 9 (32.1) |
|
| Pathological
differentiation |
|
| <0.0001 |
|
Well | 15 (88.2) | 2 (11.8) |
|
|
Moderate | 8 (28.6) | 20 (71.4) |
|
|
Poor | 4 (25.0) | 12 (75.0) |
|
| Clinical stage |
|
| 0.002 |
|
I+II | 16 (69.6) | 7 (30.4) |
|
|
III+IV | 11 (28.9) | 27 (71.1) |
|
Construction of Wnt7a-knockdown TSCC
cell models
Since high Wnt7a expression was associated with
malignant phenotypes in patients with TSCC, the present study aimed
to investigate the effects of Wnt7a on TSCC carcinogenesis and
progression. BioGPS (http://biogps.org/#goto=welcome) was used to observe
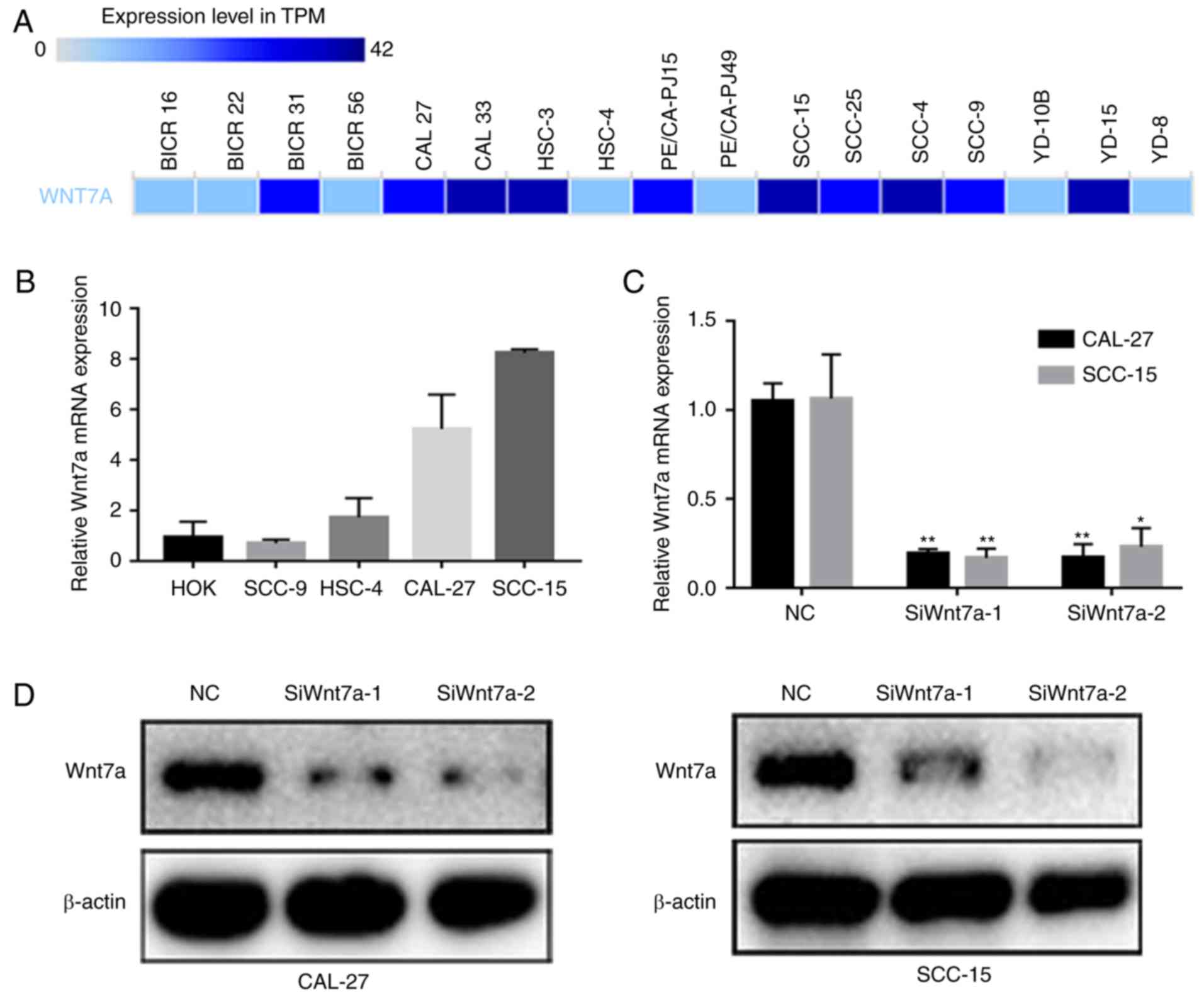
Wnt7a mRNA expression in TSCC cell lines. A total of 10/17 TSCC
cell lines recorded in the BioGPS exhibited elevated Wnt7a mRNA
expression levels (Fig. 2A). The
present study validated its expression in five cell lines using
RT-qPCR; the results revealed that CAL-27 and SCC-15 cells
exhibited the highest Wnt7a mRNA expression levels (Fig. 2B). To construct Wnt7a-knockdown TSCC
cell models, two specific siRNAs targeting Wnt7a were transfected
into CAL-27 and SCC-15 cell lines. Wnt7a siRNA transfection
effectively knocked down the mRNA and protein expression of Wnt7a
in CAL-27 and SCC-15 cells, as verified by RT-qPCR and western
blotting, respectively (Fig. 2C and
D).
Knockdown of Wnt7a suppresses
proliferation of TSCC cells in vitro and in vivo
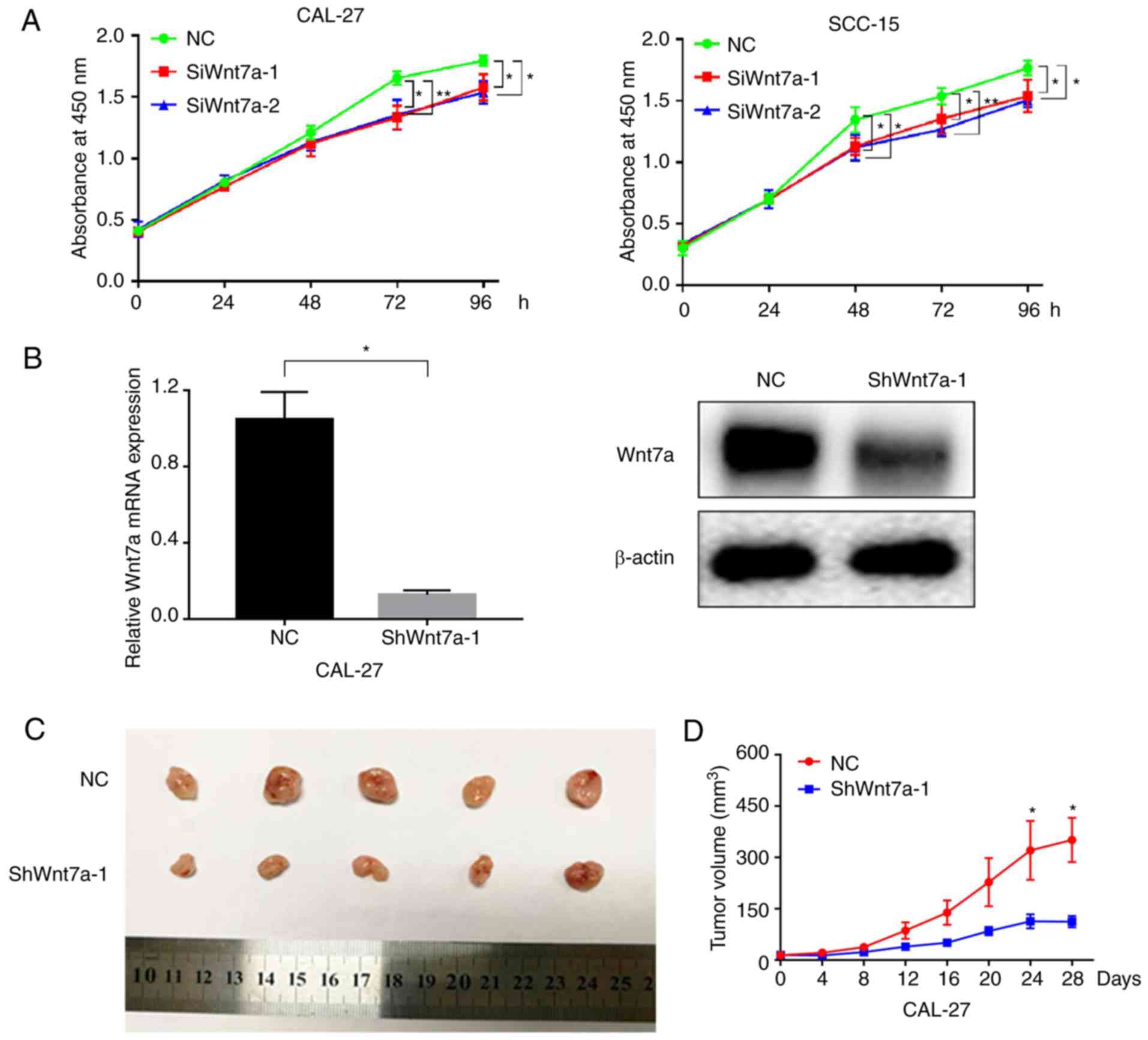
To detect the effects of Wnt7a on cell proliferation
in TSCC cell lines, a CCK-8 assay was performed. In addition, a
tumorigenicity assay was performed in nude mice following
implantation of Wnt7a-knockdown cells. As shown in Fig. 3A, cells transfected with siRNAs
against Wnt7a exhibited reduced growth compared with in the NC
group, thus indicating that the knockdown of Wnt7a may
significantly suppress the proliferative ability in CAL-27 and
SCC-15 cells. In addition, xenograft models were successfully
constructed in BALB/c nude mice, using stable Wnt7a-knockdown
CAL-27 cells (Fig. 3B). The results
revealed that tumor volumes in the shRNA-Wnt7a group were markedly
smaller than in the NC group (Fig.
3C). The tumor growth curves also revealed that knockdown of
Wnt7a substantially inhibited tumor growth (Fig. 3D). These results indicated that
Wnt7a knockdown suppressed the proliferation of TSCC cells.
Knockdown of Wnt7a suppresses
migration and invasion of TSCC cells in vitro
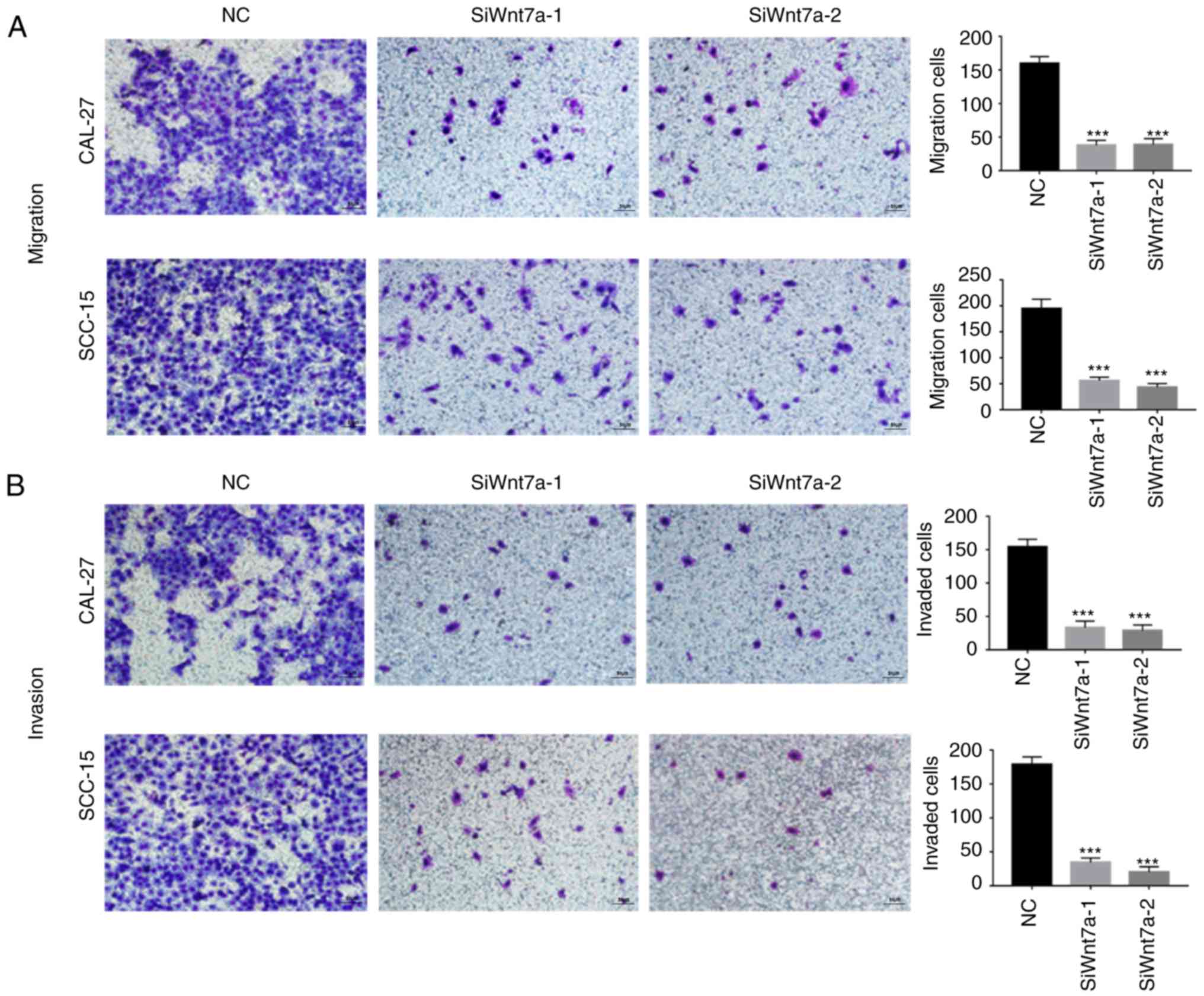
Since Wnt7a expression was positively associated
with lymph node metastasis, the present study further assessed the
effects of Wnt7a on migration and invasion. Following Wnt7a
knockdown, the numbers of cells that migrated (P<0.001; Fig. 4A) and invaded (P<0.001; Fig. 4B) through the membrane were
significantly decreased in the CAL-27 and SCC-15 cell lines. These
results revealed a functional role for Wnt7a in TSCC
metastasis.
Knockdown of Wnt7a suppresses EMT in
TSCC cells
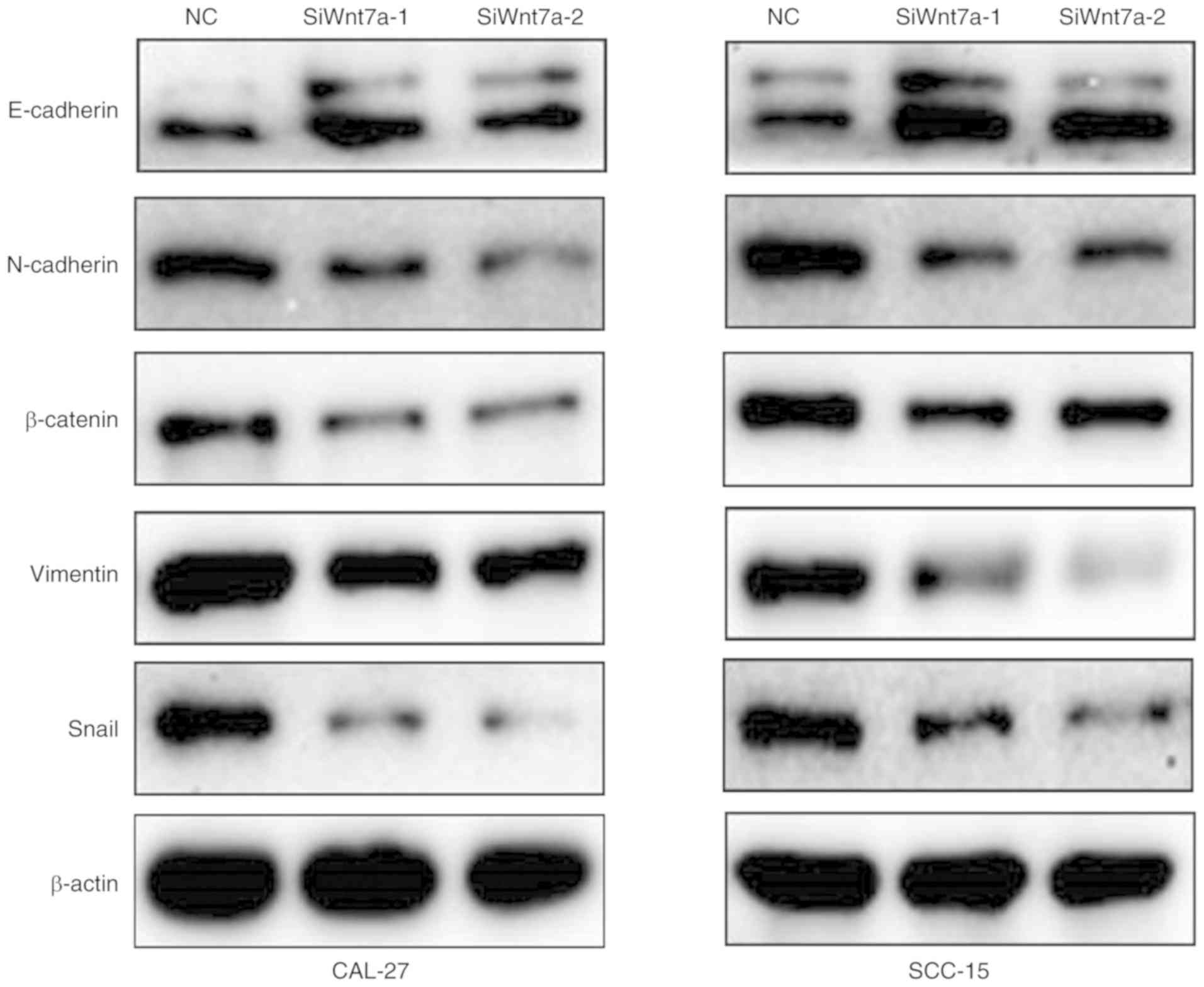
Wnt7a modulates EMT and extracellular matrix
degradation by targeting the canonical Wnt/β-catenin signaling in
bladder cancer (25). In the
present study, western blotting was performed to investigate the
expression of EMT markers in Wnt7a-knockdown TSCC cell lines.
Silencing Wnt7a in CAL-27 and SCC-15 cells upregulated the
epithelial marker, E-cadherin, whereas the expression levels of the
mesenchymal markers, N-cadherin, β-catenin and Vimentin, were
inhibited, as was the EMT-associated transcriptional factor Snail
(Fig. 5). These results indicated
that Wnt7a may promote migration and invasion in TSCC by activating
EMT activity.
Discussion
In most cases, diagnosis of TSCC is made when the
disease has already progressed to the advanced stage. Invasion and
metastasis are fundamental characteristics of advanced TSCC, and
are the principal reasons behind the unfavorable outcomes for
patients with TSCC. Numerous studies have suggested that Wnt
signaling is associated with the metastasis and progression of TSCC
(26–30); however, to the best of our
knowledge, the specific ligand partners associated with Wnt
signaling in this disease have not been identified. The present
study revealed that Wnt7a was upregulated in TSCC tissues compared
with in the corresponding adjacent non-cancerous tissues from two
patient cohorts. Wnt7a expression was also revealed to be closely
associated with tumor growth, lymph node metastasis and tumor
staging. These results were concordant with those of a previous
study that conducted gene expression profiling on advanced TSCC
tissues, further supporting the idea that Wnt7a is frequently
overexpressed in TSCC in the favorable prognostic predicted group
(31). However, an opposite result
was revealed in the present survival analysis; elevated Wnt7a
expression was detected in patients with TSCC and an unfavorable
prognosis. The results of the in vitro study revealed the
pathological and tumorigenic roles of Wnt7a in regulating invasion
and metastasis of TSCC. Knockdown of Wnt7a suppressed
proliferation, migration and invasion, and reversed the EMT
phenotype in TSCC cells. Therefore, the present study suggested
that Wnt7a may act as an oncogene in TSCC and should be considered
a candidate prognostic marker for patients with TSCC.
Local lymph node and distant organ metastases are
believed to be critical features of TSCC, which contribute toward
poor prognosis and patient mortality. Several biological behaviors
are involved in this process, including EMT (32–35).
EMT is an event whereby cancer cells lose their epithelial
characteristics to acquire a mesenchymal phenotype, thereby
enhancing their ability to migrate and invade, which is closely
associated with cancer metastasis. Furthermore, EMT in cancer is
associated with abnormal activation of several signaling pathways,
including TGF-β, Wnt, epidermal growth factor and fibroblast growth
factor (34,36). Wnt/β-catenin signaling has a major
impact on EMT; however, the specific Wnt ligand and the underlying
downstream mechanisms are not completely understood (37,38).
The present study aimed to elucidate the role of
Wnt7a in TSCC progression and EMT based on several reasons.
Overexpression of Wnt7a has been reported in numerous malignant
tumors, including endometrial cancer (39,40),
ovarian cancer (18,41,23),
colorectal cancer and pancreatic cancer (20); however, to the best of our
knowledge, its role in oral cancer has not been examined. Wnt7a is
mainly expressed in epithelial cells and has the capacity to
control cell growth and maintain regular functioning of female
reproductive organs (42–44). A previous study revealed that
overexpression of Wnt7a in an ovarian cancer cell line results in
the enhancement of its migratory and invasive capacities (45). In addition, it has been reported
that Wnt7a, interacting with the Wnt/β-catenin pathway, accelerates
the growth and progression of ovarian cancer (18). Therefore, it may be hypothesized
that Wnt7a is mechanically involved in the reprogramming of
EMT.
Using RNA interference-mediated depletion of Wnt7a
to investigate the biological function of Wnt7a in TSCC, it was
revealed that Wnt7a was required for the proliferation, migration
and invasion of CAL-27 and SCC-15 TSCC cells. Subsequently, Wnt7a
knockdown in TSCC cells was demonstrated to lead to an acquisition
of an epithelial phenotype, by enhancing the expression of
E-cadherin, and a loss of the mesenchymal phenotype, by suppressing
the expression of N-cadherin, Vimentin and the EMT-associated
transcription factor, Snail. In addition, silencing Wnt7a
suppressed the expression of β-catenin, thus indicating that Wnt7a
regulated EMT by partially antagonizing the canonical Wnt/β-catenin
pathway. Therefore, it may be hypothesized that Wnt7a expression
contributes to cancer aggressiveness by enhancing cell
proliferation and metastasis, and accelerating the EMT process of
TSCC.
In conclusion, the present results indicated that
elevated expression of Wnt7a frequently occurred in TSCC tissues,
and could be used to characterize tumor progression and predict its
earlier recurrence. The present study confirmed that Wnt7a was
involved in regulating cell proliferation, migration and invasion
of TSCC cells, and established an important functional role for
Wnt7a in TSCC pathogenesis, as revealed by its engagement in the
EMT machinery.
However, the present study has several limitations.
Firstly, the conclusions were based on the examination of two cell
lines in which Wnt7a was silenced, and may not reflect the
oncogenic role of Wnt7a in TSCC. A rescue experiment needs to be
performed in the future to confirm that the re-expression of Wnt7a
can restore tumor growth and metastasis of TSCC cells. Secondly,
since this study was conducted under a retrospective design,
further prospective studies with eligibility criteria applicable to
clinical trials are required to confirm the findings that Wnt7a
could accurately predict outcomes for patients with TSCC.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Science and Technology Project of Guangzhou Province (201802020018)
and the Medical Science and Technology Research Project of
Guangdong (B2017103).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XQ and JZ were involved in the experimental
conception and design. XQ and BJ performed the experiments. HC
conducted the data analysis. XS contributed to the reagents,
materials and tools. SX and XZ contributed to the conception of the
study and wrote the manuscript. All authors read and approved the
final manuscript. All authors contributed toward data analysis,
drafting and critically revising the paper, and agree to be
accountable for all aspects of the work.
Ethics approval and consent to
participate
All patients provided informed consent for the use
of their tissues and clinical data. The present study was approved
by the Independent Ethics Committee of the Stomatological Hospital,
Southern Medical University (approval no. [2018]05). The animal
study was conducted in accordance with the Institutional Animal
Care and Use Committee guidelines, and was approved by the
Experimental Animal Ethics Committee of Southern Medical University
(approval no. L2016113).
Patient consent for publication
All patients recruited provided consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Magrath I and Litvak J: Cancer in
developing countries: Opportunity and challenge. J Natl Cancer
Inst. 85:862–874. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuen AP, Lam KY, Chan AC, Wei WI, Lam LK,
Ho WK and Ho CM: Clinicopathological analysis of elective neck
dissection for N0 neck of early oral tongue carcinoma. Am J Surg.
177:90–92. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Layland MK, Sessions DG and Lenox J: The
influence of lymph node metastasis in the treatment of squamous
cell carcinoma of the oral cavity, oropharynx, larynx, and
hypopharynx: N0 versus N+. Laryngoscope. 115:629–639. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bilancio A, Bontempo P, Di Donato M, Conte
M, Giovannelli P, Altucci L, Migliaccio A and Castoria G: Bisphenol
A induces cell cycle arrest in primary and prostate cancer cells
through EGFR/ERK/p53 signaling pathway activation. Oncotarget.
8:115620–115631. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tripathi K and Garg M: Mechanistic
regulation of epithelial-to-mesenchymal transition through RAS
signaling pathway and therapeutic implications in human cancer. J
Cell Commun Signal. 12:513–527. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Z, Wang J, Pan Z and Zhang Y:
miR-143-3p regulates cell proliferation and apoptosis by targeting
IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway
in rheumatoid arthritis. Exp Ther Med. 15:3781–3790.
2018.PubMed/NCBI
|
|
8
|
Song K, Zheng G and Zhao Y: Liver kinase
B1 suppresses growth of lung cancer cells through sonic hedgehog
signaling pathway. Cell Biol Int. 42:994–1005. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fracalossi AC, Silva MS, Oshima CT and
Ribeiro DA: Wnt/beta-catenin signalling pathway following rat
tongue carcinogenesis induced by 4-nitroquinoline 1-oxide. Exp Mol
Pathol. 88:176–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bondos S: Variations on a theme: Hox and
Wnt combinatorial regulation during animal development. Sci Stke
2006. pe382006.
|
|
11
|
Cadigan KM and Nusse R: Wnt signaling: A
common theme in animal development. Genes Dev. 11:3286–3305. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiao J, Zhao X, Liang Y, Tang D and Pan C:
FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC)
metastasis through epithelial-mesenchymal transition (EMT). Biochem
Biophys Res Commun. 466:327–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang H, Sun JD, Yan LJ and Zhao XP:
PDGF-D/PDGFRbeta promotes tongue squamous carcinoma cell (TSCC)
progression via activating p38/AKT/ERK/EMT signal pathway. Biochem
Biophys Res Commun. 478:845–851. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Wu B, Cai H, Li D, Ma Y, Zhu X, Lv
Z, Fan Y and Zhang X: Tiam1 promotes thyroid carcinoma metastasis
by modulating EMT via Wnt/β-catenin signaling. Exp Cell Res.
362:532–540. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui D, Zhao Y and Xu J: Activated
CXCL5-CXCR2 axis promotes the migration, invasion and EMT of
papillary thyroid carcinoma cells via modulation of beta-catenin
pathway. Biochimie. 148:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ghahhari NM and Babashah S: Interplay
between microRNAs and WNT/beta-catenin signalling pathway regulates
epithelial-mesenchymal transition in cancer. Eur J Cancer.
51:1638–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nie X, Liu Y, Chen WD and Wang YD:
Interplay of miRNAs and canonical wnt signaling pathway in
hepatocellular carcinoma. Front Pharmacol. 9:6572018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshioka S, King ML, Ran S, Okuda H,
MacLean JA II, McAsey ME, Sugino N, Brard L, Watabe K and Hayashi
K: WNT7A regulates tumor growth and progression in ovarian cancer
through the WNT/β-catenin pathway. Mol Cancer Res. 10:469–482.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Avgustinova A, Iravani M, Robertson D,
Fearns A, Gao Q, Klingbeil P, Hanby AM, Speirs V, Sahai E, Calvo F
and Isacke CM: Tumour cell-derived Wnt7a recruits and activates
fibroblasts to promote tumour aggressiveness. Nat Commun.
7:103052016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kirikoshi H and Katoh M: Expression of
WNT7A in human normal tissues and cancer, and regulation of
WNT7A, WNT7B in human cancer. Int J Oncol. 21:895–900.
2002.PubMed/NCBI
|
|
21
|
Bikkavilli RK, Avasarala S, Van Scoyk M,
Arcaroli J, Brzezinski C, Zhang W, Edwards MG, Rathinam MK, Zhou T,
Tauler J, et al: Wnt7a is a novel inducer of β-catenin-independent
tumor-suppressive cellular senescence in lung cancer. Oncogene.
34:54062015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sato N, Fukushima N, Maitra A,
Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH and Goggins M:
Discovery of novel targets for aberrant methylation in pancreatic
carcinoma using high-throughput microarrays. Cancer Res.
63:3735–3742. 2003.PubMed/NCBI
|
|
23
|
Zhang XL, Peng CJ, Peng J, Jiang LY, Ning
XM and Zheng JH: Prognostic role of Wnt7a expression in ovarian
carcinoma patients. Neoplasma. 57:545–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang X, Zhu H, Gao Z, Li J, Zhuang J,
Dong Y, Shen B, Li M, Zhou H, Guo H, et al: Wnt7a activates
canonical Wnt signaling, promotes bladder cancer cell invasion, and
is suppressed by miR-370-3p. J Biol Chem. 293:6693–6706. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paluszczak J, Kiwerska K and
Mielcarek-Kuchta D: Frequent methylation of DAB2, a Wnt
pathway antagonist, in oral and oropharyngeal squamous cell
carcinomas. Pathol Res Pract. 214:314–317. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hema KN, Smitha T, Sheethal HS and
Mirnalini SA: Epigenetics in oral squamous cell carcinoma. J Oral
Maxillofac Pathol. 21:252–259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Farooqi AA, Shu CW, Huang HW, Wang HR,
Chang YT, Fayyaz S, Yuan SF, Tang JY and Chang HW: TRAIL, Wnt,
Sonic Hedgehog, TGFβ, and miRNA signalings are potential targets
for oral cancer therapy. Int J Mol Sci. 18(pii): E15232017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gonzalez-Moles MA, Ruiz-Avila I,
Gil-Montoya JA, Plaza-Campillo J and Scully C: β-catenin in oral
cancer: An update on current knowledge. Oral Oncol. 50:818–824.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noguti J, DE Moura CF, Hossaka TA, Franco
M, Oshima CT, Dedivitis RA and Ribeiro DA: The role of canonical
WNT signaling pathway in oral carcinogenesis: A comprehensive
review. Anticancer Res. 32:873–878. 2012.PubMed/NCBI
|
|
31
|
Enokida T, Fujii S, Takahashi M, Higuchi
Y, Nomura S, Wakasugi T, Yamazaki T, Hayashi R, Ohtsu A and Tahara
M: Gene expression profiling to predict recurrence of advanced
squamous cell carcinoma of the tongue: Discovery and external
validation. Oncotarget. 8:61786–61799. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shetty AV and Wong DJ: Systemic treatment
for squamous cell carcinoma of the head and neck. Otolaryngol Clin
North Am. 50:775–782. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Almangush A, Heikkinen I, Makitie AA,
Coletta RD, Läärä E, Leivo I and Salo T: Prognostic biomarkers for
oral tongue squamous cell carcinoma: A systematic review and
meta-analysis. Br J Cancer. 117:856–866. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev CanceR. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vig N, Mackenzie IC and Biddle A:
Phenotypic plasticity and epithelial-to-mesenchymal transition in
the behaviour and therapeutic response of oral squamous cell
carcinoma. J Oral Pathol Med. 44:649–655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vincan E and Barker N: The upstream
components of the Wnt signalling pathway in the dynamic EMT and MET
associated with colorectal cancer progression. Clin Exp Metastasis.
25:657–663. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Y, Meng F, Xu Y, Yang S, Xiao M, Chen
X and Lou G: Overexpression of Wnt7a is associated with tumor
progression and unfavorable prognosis in endometrial cancer. Int J
Gynecol Cancer. 23:304–311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng C, Zhang X, Wang Y, Li L, Wang Q and
Zheng J: Expression and prognostic significance of wnt7a in human
endometrial carcinoma. Obstet Gynecol Int 2012. 1349622012.
|
|
41
|
King ML, Lindberg ME, Stodden GR, Okuda H,
Ebers SD, Johnson A, Montag A, Lengyel E, MacLean Ii JA and Hayashi
K: WNT7A/β-catenin signaling induces FGF1 and influences
sensitivity to niclosamide in ovarian cancer. Oncogene.
34:3452–3462. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tulac S, Nayak NR, Kao LC, Van Waes M,
Huang J, Lobo S, Germeyer A, Lessey BA, Taylor RN, Suchanek E, et
al: Identification, characterization, and regulation of the
canonical Wnt signaling pathway in human endometrium. J Clin
Endocrinol Metab. 88:3860–3866. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Miller C and Sassoon DA: Wnt-7a maintains
appropriate uterine patterning during the development of the mouse
female reproductive tract. Development. 125:3201–3211.
1998.PubMed/NCBI
|
|
44
|
Hayashi K, Erikson DW, Tilford SA, Bany
BM, Maclean JA II, Rucker EB III, Johnson GA and Spencer TE: Wnt
genes in the mouse uterus: Potential regulation of implantation.
Biol Reprod. 80:989–1000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Merritt MA, Parsons PG, Newton TR, Martyn
AC, Webb PM, Green AC, Papadimos DJ and Boyle GM: Expression
profiling identifies genes involved in neoplastic transformation of
serous ovarian cancer. BMC Cancer. 9:3782009. View Article : Google Scholar : PubMed/NCBI
|