Introduction
Liver cancer is a common and devastating malignancy
with high mortality worldwide (1).
High-throughput sequencing of liver cancer tissues has revealed
that liver cancer samples possess extensive intra-tumor
heterogeneity (2,3). Most types of cancers contain a small
population of highly tumorigenic and drug-resistant cells known as
cancer stem cells (CSCs) or tumor initiating cells (TICs) (4,5). These
cells have the capacity to self-renew, differentiate, and generate
tumors identical to the original one in primary and metastatic
sites (6). CSCs are thought to be
responsible for tumor initiation, progression, chemoresistance and
postsurgical recurrence of liver cancer patients (7). Therefore, therapies that specifically
target liver CSCs may be beneficial for the treatment of liver
cancer.
Diverse surface markers such as cluster of
differentiation 133 (CD133) (8,9), CD90
(10), epithelial cellular adhesion
molecule (EpCAM) (11), CD13
(12,13), cytokeratin 19 (CK19) (14) and oval cell marker (OV-6) (15), or side population cells that efflux
the DNA-binding dye Hoechst 33342 have been used to identify liver
CSCs (16,17). However, due to the low percentage of
CSCs existing in tumor patients and cell lines, and the fact that
cell surface antigens are disrupted by enzymatic digestion, the
isolation efficiency of CSCs is very low. Therefore, an easy-to-use
method of tumor sphere formation has been increasingly used to
enrich CSC-like cells in many cancers including liver cancer
(18), since tumor sphere-forming
cells from tumor patients and cell lines possess CSC-like features
and tumorigenic potential. In the present study, we used this
method to enrich liver CSCs and to identify differentially
expressed miRNAs between CSC-enriched tumor spheres and adherent
cells.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs (~22 nucleotides) that repress gene expression at the
post-transcriptional level by binding to the 3′UTR of target genes
(19,20). Several miRNAs have been implicated
in liver cancer proliferation, metastasis, chemoresistance
(21,22), and interactions between the tumor
microenvironment and liver cancer cells (23). For example, miRNA-125b, which is
downregulated in liver cancer cells, significantly suppresses
epithelial-mesenchymal transition (EMT) associated traits in liver
cancer cells targeting SMAD2 and SMAD4 (24). A strategy of restoring
tumor-repressing miRNA expression in liver cancer cells may be used
to inhibit the proliferation, metastasis and self-renewal of liver
CSCs.
In the present study, we enriched liver CSCs using
tumor spheres or two surface markers, CD13 and EpCAM. Then, we
screened for significantly downregulated miRNAs from The Cancer
Genome Atlas (TCGA) database in CSCs and adherent cells. Among
them, miR-486 was found to be significantly downregulated in tumor
spheres and liver cancer tissues. Overexpression of miR-486
suppressed tumor sphere expansion, and the invasion and
tumorigenicity of liver cancer cells. The miRecords and RNAhybrid
algorithms predicted that Sirt1 was a direct target of miR-486.
Luciferase reporter assays and immunohistochemistry (IHC) staining
indicated that miR-486 significantly reduced Sirt1 expression. We
also found that Sirt1 was responsible for maintaining the
self-renewal and tumorigenicity of liver CSCs. The results of the
present study suggest that increasing miR-486 levels in liver
cancer or liver CSCs may be a promising strategy for liver cancer
treatment.
Materials and methods
Patient specimens
Liver cancer tissues and corresponding non-tumor
tissues were obtained from the Beijing 302 Hospital. Sixteen HCC
samples (14 males and 2 females; range, 49–65 years) were obtained
from consecutive patients undergoing initial hepatectomy from
January 2015 to December 2017. All human specimens were collected
in accordance with the Declaration of Helsinki, and the protocols
involving clinical samples were approved by the Research Ethics
Committees of the Academy of Military Medical Sciences, Beijing,
China. Written informed consent was obtained from all patients.
Cell culture
Liver cancer cells including Huh7, Hep3B, Li-7 and
PLC/PRF/5, (CRL-8024™; also referred to as PLC) from the American
Type Culture Collection (ATCC; Manassas, VA, USA), the MHCC97H
(briefly, 97H), MHCC97L (briefly, 97L), HCCLM3 (briefly, LM3) cells
were obtained from the Liver Cancer Institute of Fudan University
(Shanghai, China) (25). The HepG2
cells were obtained from the National Platform for Experimental
Cell Resources (Beijing, China). These cells were maintained in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS), 100 U/ml penicillin and 100 ng/ml
streptomycin at 37°C under 5% CO2.
TCGA database
The TCGA database (https://tcga-data.nci.nih.gov/tcga/) was used to
analyze differentially expressed miRNAs between liver cancer and
adjacent non-tumor tissues.
Quantitative real-time PCR (qPCR)
Total RNAs from liver cancer specimens and cells
were extracted using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Reverse transcription of total RNA into
cDNA was performed with the miScript Reverse Transcription kit
(Qiagen Sciences, Inc., Gaithersburg, MD, USA), and qPCR was
performed using the miScript SYBR-Green PCR Master Mix on the ABI
Prism 7900 system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Briefly, a 20 µl RTqPCR system was performed for 40 cycles
according to the following conditions: An initial denaturation was
performed at 95°C for 3 min, followed by denaturation at 95°C for
15 sec, annealing at 58°C for 30 sec, and extension at 72°C for 7
min. Relative quantification of miRNA or mRNA expression was
calculated using the 2−∆∆Cq method (26). 18S was used as an mRNA internal
control. U6 RNA was used as a miRNA internal control. Each
experiment was conducted with at least three independent
replicates. Primer sequences are listed in Table I.
 | Table I.The sequences of qRT-PCR primers in
the study. |
Table I.
The sequences of qRT-PCR primers in
the study.
| Gene/miRNA | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| miRNA universal
primer |
GATTGAATCGAGCACCAGTTAC |
|
| U6 |
CGCTTCGGCAGCACATATACTA | miRNA universal
primer |
| miR-486-5p |
TCCTGTACTGAGCTGCCCCGAG | miRNA universal
primer |
| miR-214 |
ACAGCAGGCACAGACAGGCAGT | miRNA universal
primer |
| miR-187 |
TCGTGTCTTGTGTTGCAGCCGG | miRNA universal
primer |
| miR-511-3p |
AATGTGTAGCAAAAGACAGA | miRNA universal
primer |
| miR-101 |
TACAGTACTGTGATAACTGAA | miRNA universal
primer |
| miR-99a |
AACCCGTAGATCCGATCTTGTG | miRNA universal
primer |
| miR10a |
TACCCTGTAGATCCGAATTTGTG | miRNA universal
primer |
| miR-337 |
CTCCTATATGATGCCTTTCTTC | miRNA universal
primer |
| miR-199a-3p |
ACAGTAGTCTGCACATTGGTTA | miRNA universal
primer |
| miR-483 |
TCACTCCTCTCCTCCCGTCTT; | miRNA universal
primer |
| miR-33b |
GTGCATTGCTGTTGCATTGC | miRNA universal
primer |
| miR-490 |
CAACCTGGAGGACTCCATGCTG | miRNA universal
primer |
| miR-451 |
AAACCGTTACCATTACTGAGTT | miRNA universal
primer |
| miR-138 |
AGCTGGTGTTGTGAATCAGGCCG | miRNA universal
primer |
| miR-223 |
TGTCAGTTTGTCAAATACCCCA | miRNA universal
primer |
| miR-135b |
TATGGCTTTTCATTCCTATGTGA | miRNA universal
primer |
| miR-195 |
TAGCAGCACAGAAATATTGGC | miRNA universal
primer |
| miR-375 |
TTTGTTCGTTCGGCTCGCGTGA | miRNA universal
primer |
| miR-139 |
TCTACAGTGCACGTGTCTCCAGT | miRNA universal
primer |
| miR-1258 |
AGTTAGGATTAGGTCGTGGAA | miRNA universal
primer |
| miR-145 |
GTCCAGTTTTCCCAGGAATCCCT | miRNA universal
primer |
| miR-199a-5p |
CCCAGTGTTCAGACTACCTGTTC | miRNA universal
primer |
| miR-383-5p |
AGATCAGAAGGTGATTGTGGCT | miRNA universal
primer |
| miR-411 |
TAGTAGACCGTATAGCGTACG | miRNA universal
primer |
| miR-592 |
TTGTGTCAATATGCGATGATGT | miRNA universal
primer |
| miR-144 |
TACAGTATAGATGATGTACT | miRNA universal
primer |
| miR-450 |
TTTTGCGATGTGTTCCTAATAT | miRNA universal
primer |
| miR-424 |
CAGCAGCAATTCATGTTTTGAA | miRNA universal
primer |
| miR-326 |
CCTCTGGGCCCTTCCTCCAG | miRNA universal
primer |
| miR-150 |
TCTCCCAACCCTTGTACCAGTG | miRNA universal
primer |
| miR-379 |
TGGTAGACTATGGAACGTAGG | miRNA universal
primer |
| miR-154 |
TAGGTTATCCGTGTTGCCTTCG | miRNA universal
primer |
| miR-497 |
CAGCAGCACACTGTGGTTTGT | miRNA universal
primer |
| miR-204 |
TTCCCTTTGTCATCCTATGCCT | miRNA universal
primer |
| miR-378 |
CTCCTGACTCCAGGTCCTGTGT | miRNA universal
primer |
| 18S |
AACCCGTTGAACCCCATT |
CCATCCAATCGGTAGTAGCG |
| Sox2 |
GCCGAGTGGAAACTTTTGTCG |
GGCAGCGTGTACTTATCCTTCT |
| OCT4 |
GGGAGATTGATAACTGGTGTGTT |
GTGTATATCCCAGGGTGATCCTC |
| CD13 |
TTCAACATCACGCTTATCCACC |
AGTCGAACTCACTGACAATGAAG |
| Sirt1 |
TAGCCTTGTCAGATAAGGAAGGA |
ACAGCTTCACAGTCAACTTTGT |
| TNC |
GCCCCTGATGTTAAGGAGCTG |
GGCCTCGAAGGTGACAGTT |
| ANXA13 |
GCTAAAGCGAGCAGTCCTCAG |
GTCCTGCCCGATAAGATTTCAA |
Flow cytometry and sorting
Fluorescence-activated cell sorting was performed to
enrich EpCAM+ and CD13+ liver cancer cells.
Briefly, Huh7 cells were trypsinized and washed twice with
phosphate-buffered saline (PBS), then incubated with APC-anti-human
EpCAM (dilution 1:200; cat. no. 324208; BioLegend, San Diego, CA,
USA) or APC-anti-human CD13 antibody (dilution 1:100; cat. no.
301706; BioLegend) for 30 min at 4°C. After incubation, Huh7 cells
were washed 3 times and then sorted on the FACSAria II instrument
(BD Biosciences, San Jose, CA, USA). Huh7-EpCAM+ (or
CD13+) and Huh7-EpCAM− (or CD13−)
cells were collected for qPCR analysis.
Tumor sphere culture
A total of 1,000 tumor cells were seeded in the
6-well ultra-low attachment plates (Corning Inc., Corning, NY, USA)
and cultured in DMEM/F-12 supplemented with B27, N2 (both from
Invitrogen; Thermo Fisher Scientific, Inc.), 10 ng/ml of epidermal
growth factor (EGF), 5 ng/ml of basic fibroblast growth factor
(bFGF), 100 U/ml penicillin and 100 ng/ml streptomycin at 37°C
under 5% CO2 for 7–10 days as we previously described
(27,28).
Oligonucleotides
Transient expression of the miR-486 mimics or
miR-486 inhibitors have been previously described (29). Scramble miRNA, lenti-miR-486 and
lenti-miR-486 inhibitor were purchased from Suzhou GenePharma Co.,
Ltd. (Suzhou, China). The sequence of hsa-miR-486 mimics was:
TCCTGTACTGAGCTGCCCCGAG, that of hsa-miR-486 inhibitor was:
ACCCCTATCACGATTAGCATTAA, and that of the miRNA inhibitor negative
control (NC) was: CAGTACTTTTGTGTAGTACAA.
Short hairpin RNA (shRNA) and
lentivirus package
To generate lentivirus plasmids for stable RNA
interference, short hairpins were designed using online software
(http://rnaidesigner.lifetechnologies.com/rnaiexpress/design.do)
as previously described (30).
Based on the Sirt1 sequence, the small interfering RNA (siRNAs)
Sirt1 sequence was 5′-GGUGCCGUGCUACUCAUAUTT-3′. The Sirt1-specific
short hairpin (shRNA) expression vector and the scrambled
ineffective shRNA cassette were cloned into the pSicoR-GFP plasmid.
Virus packaging was performed in 293T cells after co-transfection
of lentiviral expression plasmids with the packaging plasmids
(pRRE, pCMV-VSVG and pRSV-REV; Addgene, Inc., Cambridge, MA, USA)
using Lipofectamine 2000. The viral supernatant was used to infect
liver cancer cells, and stable GFP-expressing clones were selected
by fluorescence-activated cell sorting (FACS).
Luciferase reporter assay
The 3′UTR sequences of Sirt1 that contained the
predicted complimentary sites of miR-486 were cloned into the pGL3
basic reporter vector (Promega Corp., Madison, WI, USA). The
pGL3-Sirt1-3′UTR vector or pGL3 basic vector was co-transfected
with miR-486 mimics or NC into 293T cells, with the Renilla
luciferase vector used as an internal control. After 48 h, cells
were harvested, and the luciferase activity was detected using the
Dual-Luciferase assay kit (Promega). The ratio of
firefly/Renilla luciferase activities was calculated and
designated as the relative promoter activity.
Cell invasion assays
Cell invasion assays were performed in Transwell
chambers coated with Matrigel (8 µm Transwell inserts; BD
Biosciences). A total of 1×106 cells in serum-free DMEM
were seeded in the upper chamber, and DMEM with 5% FBS was added to
the bottom chamber as an attractant. After 48 h of incubation, the
penetrated cells on the filters were fixed in 20% methanol and
stained with crystal violet. Ten randomly selected fields
(magnification, ×100) in each well were counted under a light
microscope.
Scratch healing assays
The effects of miR-486 on the migration of tumor
cells were determined by wound healing assays. A total of
5×105 cells/well were seeded and cultured in 6-well
plates to create a confluent monolayer. After the cells attached
overnight, a straight line was scraped with a 200-µl pipette tip
across the cell monolayer to create a ‘scratch’. After 48 h,
microscopic images of the ‘scratch closure’ were captured under a
light microscope at an original magnification of ×4.
In vivo tumorigenicity assays
Female nude mice (6–8 weeks old) were purchased from
Weitonglihua Company (Beijing WeitongLihua Experimental Animal
Technology Co., Ltd., Beijing, China) and raised under specific
pathogen-free conditions. Mice were allowed ad libitum
access to food and water and were maintained on a constant light
dark cycle with constant temperature and humidity. All animal
experiments in the present study were approved by the Ethics
Committee of the Academy of Military Medical Sciences. Huh7-miR-486
or HepG2-miR-486 and corresponding control cells (2×106)
were subcutaneously injected into the right and left side of mice,
respectively. Similarly, Hep3B-shSirt1 or HepG2-shSirt1 and control
cells were subcutaneously injected into nude mice. Approximately 5
weeks after injection, mice were anesthetized with 1% pentobarbital
sodium (75 mg/kg) through the intraperitoneal (IP) administration,
and sacrificed by cervical dislocation, and the tumors were weighed
as previously described (23).
Non-retrospective ethical approval was obtained for the animal
experiments conducted in the study. Tumor burden did not exceed the
recommended dimensions.
Statistical analysis
The data are presented as the means ± standard
deviation (SD) from at least 3 independent experiments.
Quantitative results were compared using GraphPad Prism version 5.0
software (GraphPad Software, Inc., La Jolla, CA, USA). A two-tailed
Student's paired t-test was used to test for significance between
two groups. Multiple groups were compared by a one- or two-way
analysis of variance with Tukey's post hoc correction. Statistical
significance was regarded as P<0.05 or P<0.01.
Results
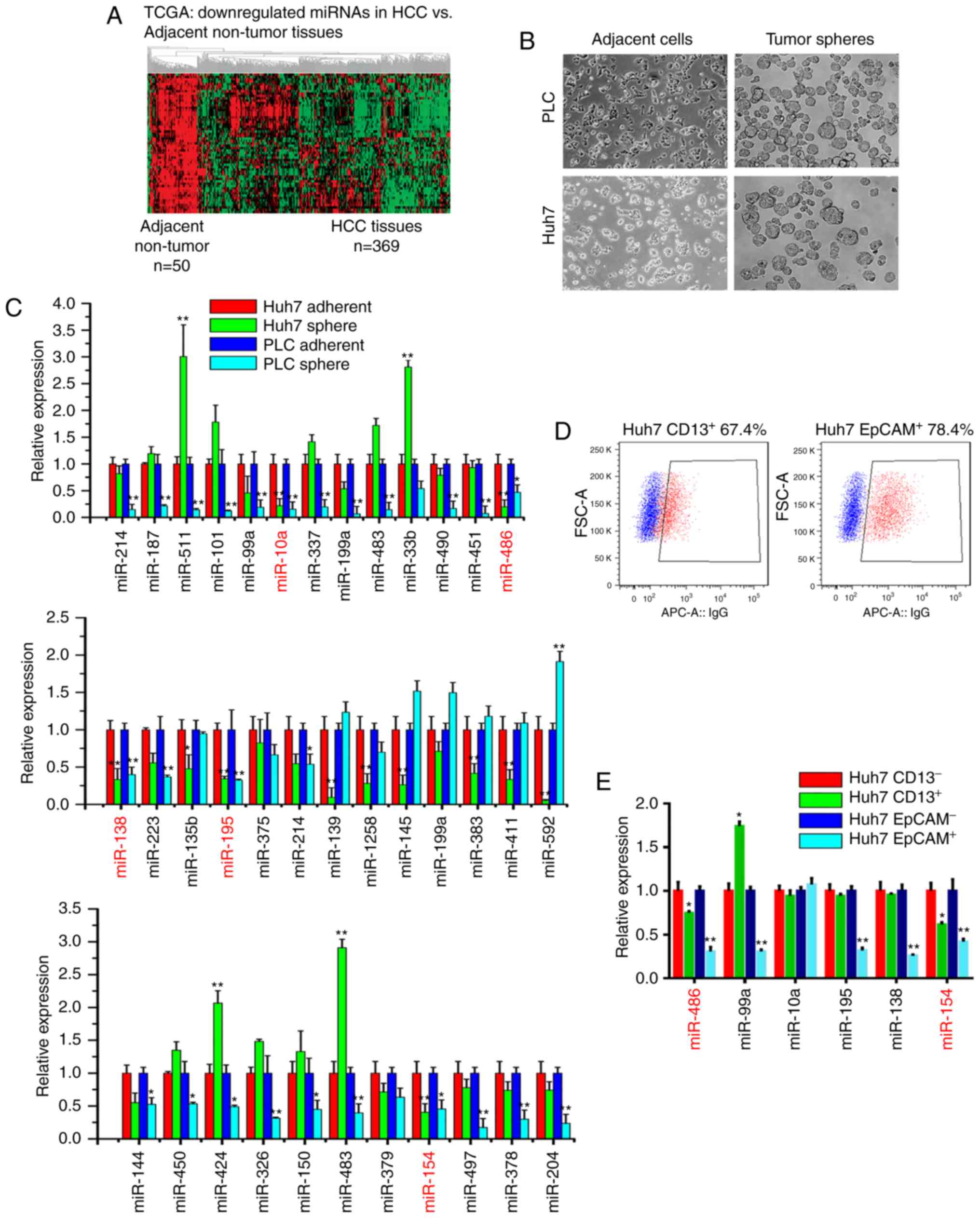
miR-486 is preferentially
downregulated in liver CSC-like cells and liver cancer tissues
To identify differentially expressed miRNAs between
CSC-like cells and adherent cells, we first used the TCGA miRNA
sequence database to analyze the altered miRNAs between liver
cancer specimens (n=369) and adjacent non-tumor tissues (n=50). We
mainly focused on the downregulated miRNAs in liver cancer tissues,
and identified a set of 59 miRNAs that were significantly
downregulated in liver cancer specimens compared with adjacent
non-tumor tissues, including miR-486, miR-99a, miR-195 and miR-154
(Fig. 1A). Then, we enriched liver
CSC-like cells using tumor sphere models (Fig. 1B), and qPCR assays were further
utilized to compare the differentially expressed miRNAs between
tumor spheres and adherent cells. As shown in Fig. 1C, the expression levels of miR-99a,
miR-10a, miR-486, miR-195, miR-154 and miR-138 in liver tumor
spheres were significantly decreased compared with adherent cells.
Moreover, liver CSCs were enriched with CSC surface markers
including EpCAM and CD13. Thus, we performed fluorescence-activated
cell sorting (FACS) to separate Huh7 cells into CD13−
and CD13+, and EpCAM− and EpCAM+
subsets, respectively (Fig. 1D).
According to the qPCR results, miR-486 was significantly
downregulated in EpCAM+ and CD13+
subpopulations (Fig. 1E). Moreover,
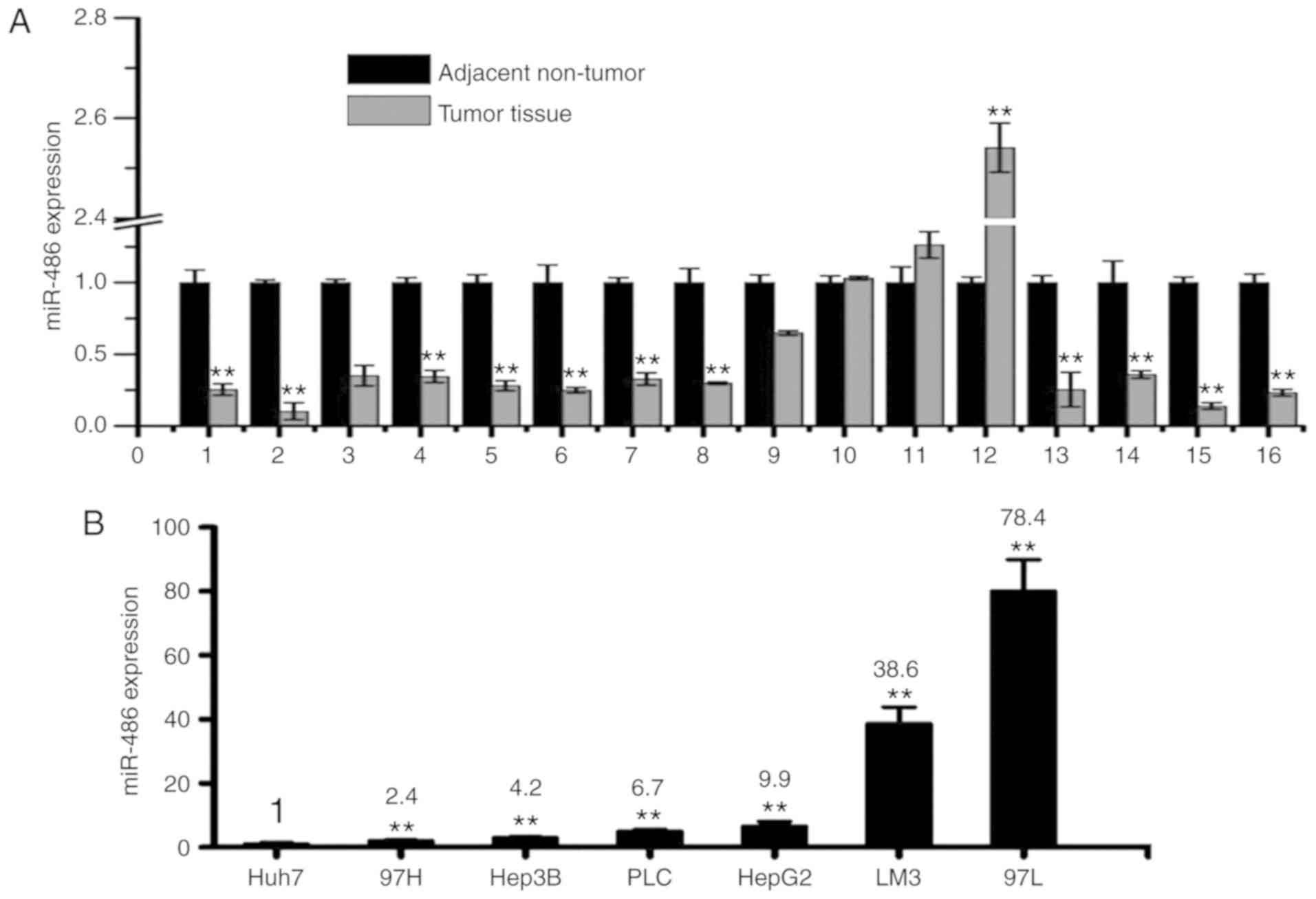
miR-486 levels were significantly decreased in tumor samples
compared with corresponding non-tumor tissues (Fig. 2A) and most tumor cell lines
(Fig. 2B). Among the 10 most
frequently used liver cancer cell lines, miR-486 was lowly
expressed in Huh7 (highly proliferative) (23,29)
and 97H cells (highly invasive) (25,31).
In our laboratory, only 3 liver cancer cell lines including Huh7,
PLC/PRF/5 and Li-7 cells could form tumor spheres easily in
vitro (data not shown). The tumor sphere-forming cells from
tumor patients and cell lines possessed more CSC-like features and
tumorigenic potential. In addition, the Huh7 cells could easily
form tumors in vivo than PLC/PRF/5 and Li-7 cells (data not
shown). These results indicated that miR-486 was significantly
downregulated in liver CSCs and liver cancer tissues.
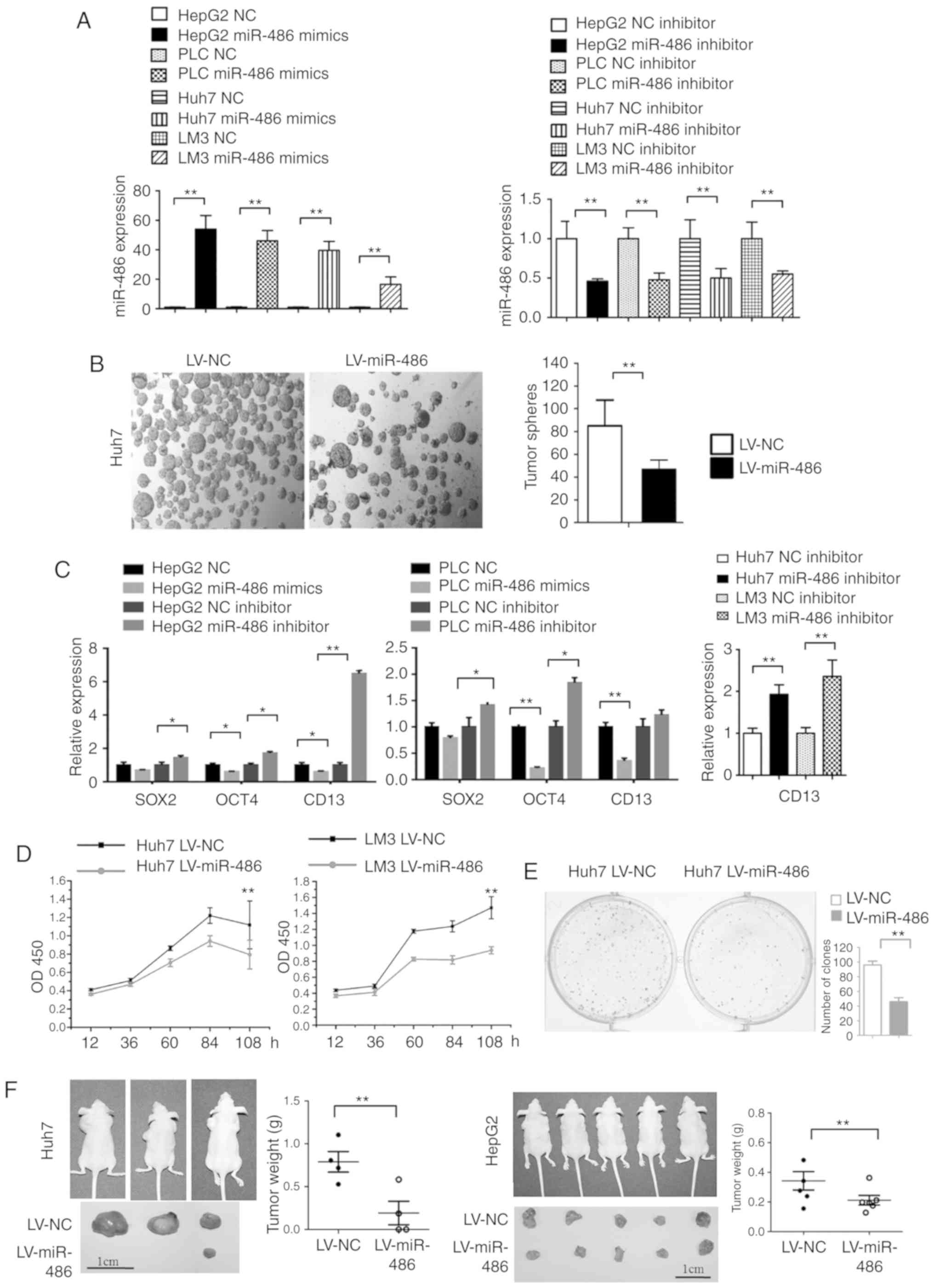
miR-486 significantly suppresses the
tumor stemness and invasion of liver cancer cells
First, we confirmed the relative expression of
miR-486 in miR-486 overexpression and inhibitor expression in liver
cancer cells (Fig. 3A). Compared to
the NC group, the number of tumor spheres was decreased in Huh7
cells transfected with lentivirus miR-486 (LV-miR-486) (Fig. 3B). miR-486 overexpression suppressed
the expression of stemness-related gene CD13. Conversely, miR-486
inhibitor promoted the expression of CD13 in liver cancer cells
(Fig. 3C). The Cell Counting Kit-8
(CCK-8) assays indicated that miR-486 overexpression significantly
suppressed the proliferation of Huh7 and LM3 cells in vitro
(Fig. 3D). Moreover, the
clone-forming capacity in miR-486 overexpression Huh7 cells was
significantly decreased compared with NC group (Fig. 3E). Then, Huh7-miR-486 and Huh7-NC
cells were subcutaneously injected into nude mice. Consequently,
the tumor sizes and weights in the Huh7-miR-486 group were
significantly decreased compared to the Huh7-NC group (P<0.01;
Fig. 3F, left panel). Similar
results were observed in mice injected with HepG2-miR-486 cells
(P<0.01) (Fig. 3F, right panel).
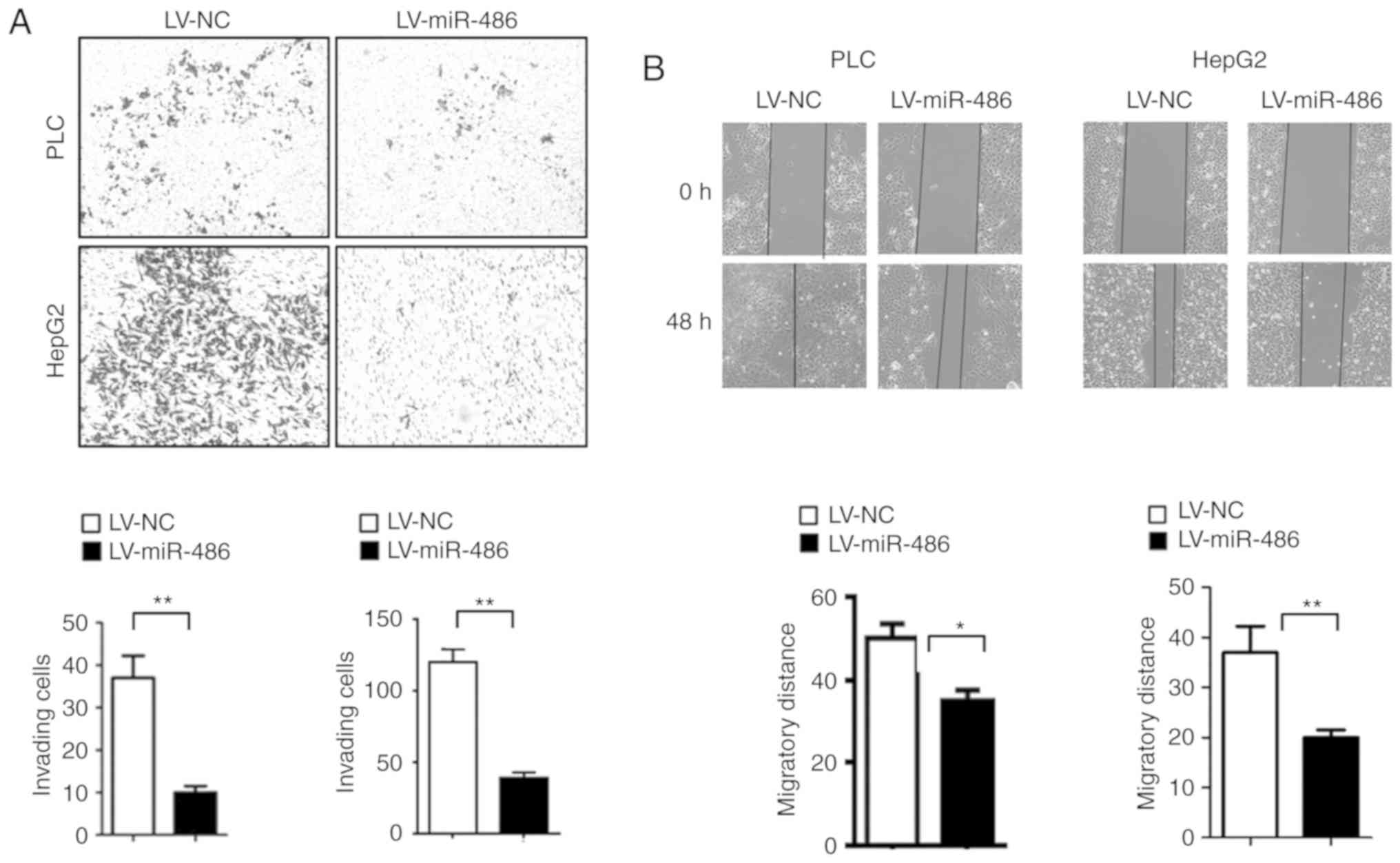
The cell invasion and scratch healing assays revealed that miR-486
mimics significantly decreased the number of invading cells
(Fig. 4A) and the migration
distance of liver cancer cells (Fig.
4B). Collectively, these results demonstrated that miR-486 has
tumor suppressive functions in CSC traits.
Sirt1 is a direct target of miR-486 in
liver CSC cells
miRNAs usually bind to the complementary sites in
the 3′UTRs of target mRNAs and trigger RNA degradation or
translation repression (32). Both
TargetScan and miRanda database predicted that Annexin A13
(ANXA13), Sirt1 and tenascin-C (TNC) could serve as potential
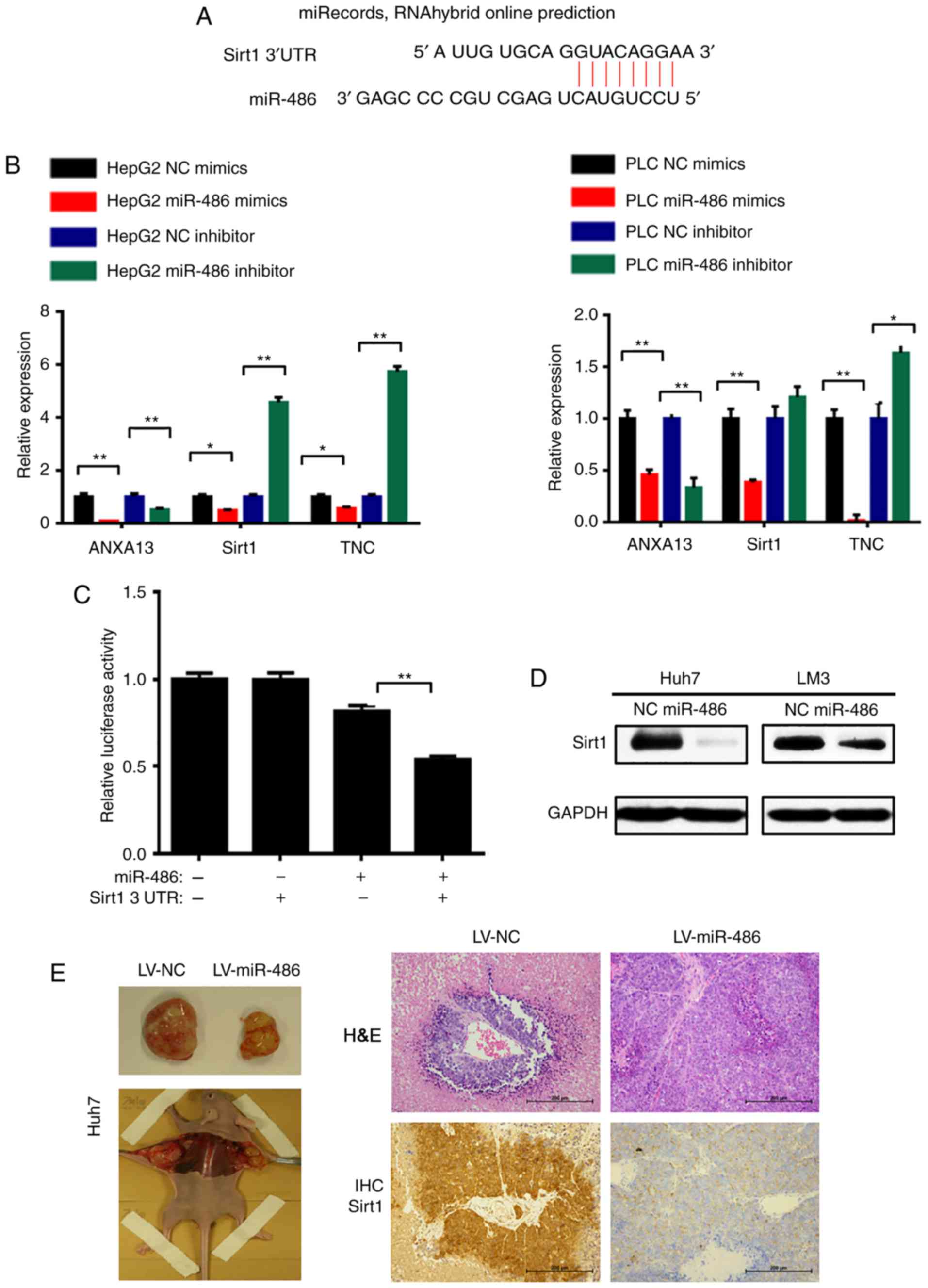
target genes of miR-486 (Fig. 5A).
The results of the qPCR analysis confirmed that miR-486 suppressed
the expression of Sirt1 and TNC in HepG2 and PLC cells. Conversely,
Sirt1 and TNC mRNA levels were increased with transfection of a
miR-486 inhibitor (Fig. 5B). Our
preliminary results indicated that knockdown of TNC did not inhibit
the growth of tumor spheres in vitro (data not shown). Thus,
in the present study, we mainly focused on Sirt1 as the target gene
of miR-486. To further elucidate the interaction between miR-486
and Sirt1 3′UTR, a luciferase assay was subsequently performed in
293T cells. The pGL3-Sirt1-3′UTR vector or pGL3 basic vector was
co-transfected with miR-486 mimics or NC into cells, and the
luciferase activity was detected using the Dual-Luciferase Assay
System. Co-transfection with miR-486 mimics significantly decreased
the firefly luciferase activity of Sirt1-3′UTR reporter but not
that of the pGL3 reporter (P<0.01; Fig. 5C). Next, we also observed
downregulation of Sirt1 in the miR-486 overexpression group by
western blot analysis (Fig. 5D).
Moreover, the IHC staining indicated that Sirt1 expression was
downregulated in miR-486 overexpression cell-derived tumor tissues
(Fig. 5E). In summary, these
findings indicated that Sirt1 is a direct target of miR-486.
Sirt1 is increased in liver CSCs and
is responsible for maintaining CSC properties
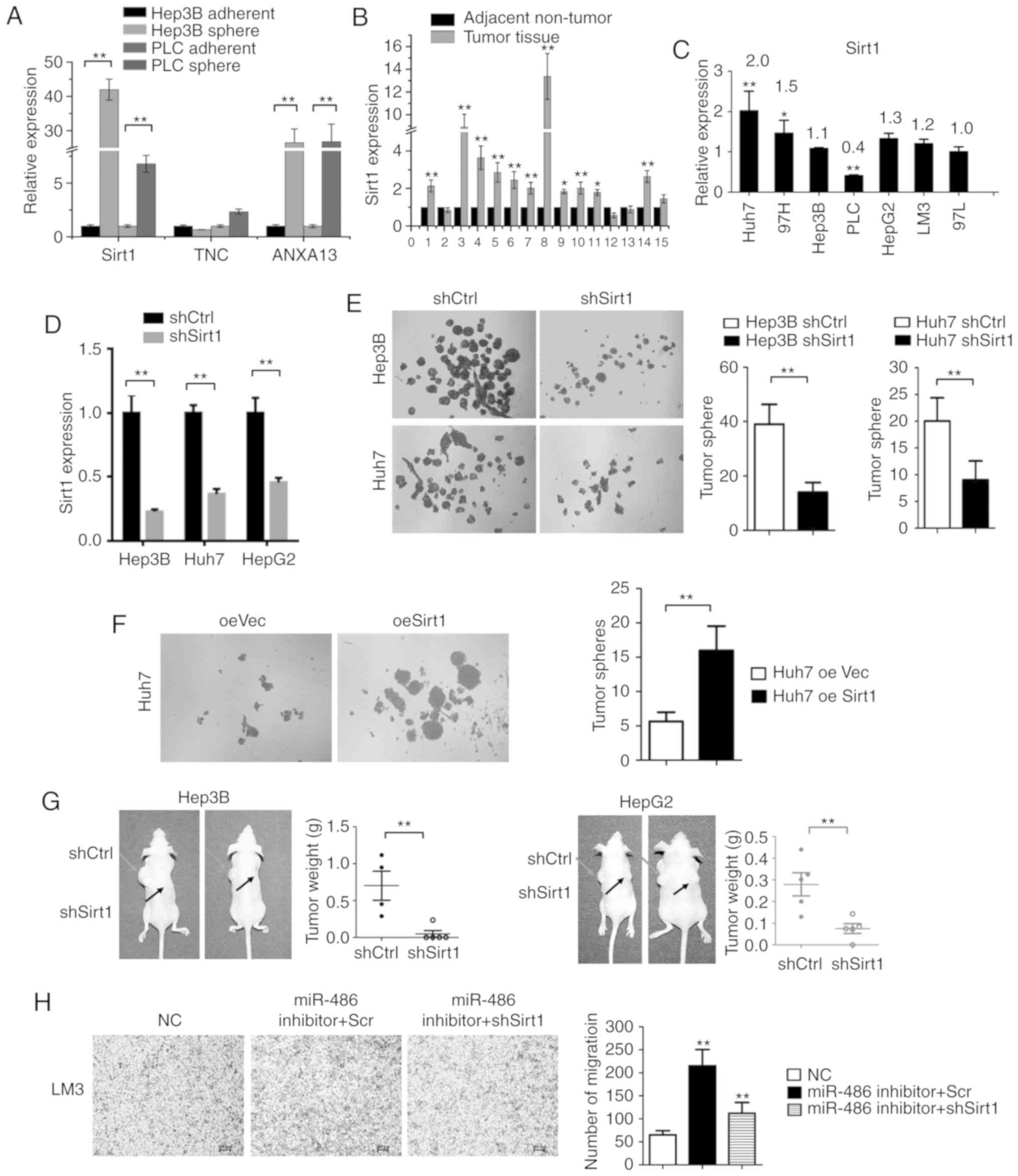
Sirt1 mRNA levels were assessed in tumor spheres and
adherent cells. The expression levels of Sirt1 were significantly
higher in tumor spheres compared with adherent cells (P<0.01;
Fig. 6A). This was consistent with
our results revealing that miR-486 expression was decreased in
tumor spheres and that Sirt1 expression was inversely associated
with miR-486 levels. Additionally, increased Sirt1 expression was
observed in liver cancer tissues compared with adjacent non-tumor
tissues (Fig. 6B). Then, the
expression of Sirt1 in liver cancer cell lines (Fig. 6C) was analyzed. To understand the
role of Sirt1 in the maintenance of liver CSC characteristics, we
transfected liver cancer cells with Sirt1-shRNA lentivirus, and
confirmed that its expression was suppressed by qPCR analyses
(Fig. 6D). To determine the
biological function of Sirt1 in maintaining CSC-like
characteristics, tumor sphere assays were performed. As revealed in
Fig. 6E, knockdown of Sirt1
markedly inhibited tumor sphere formation. Conversely,
overexpression of Sirt1 (oeSirt1) was capable of promoting tumor
sphere formation (Fig. 6F). To
further investigate the biological functions of Sirt1 in liver
cancer progression in vivo, HepG2-shSirt1 and HepG2 control
cells were subcutaneously injected into nude mice to monitor tumor
growth. Tumor weights were reduced in the HepG2-shSirt1 cells as
compared with the control group (Fig.
6G). Similar results were obtained with Hep3B-shSirt1 cells
(Fig. 6G). Moreover, Sirt1
knockdown reduced the invading capacity of the miR-486
inhibitor-transfected LM3 cells (Fig.
6H). Collectively, these data indicated that Sirt1 was
responsible for the maintenance of self-renewal of liver CSCs and
tumorigenesis in vivo.
Discussion
CSCs or TICs are believed to be the main cause of
drug resistance and relapse in liver cancer patients (10). However, few effective interventions
have been found for the elimination of CSCs. Liver CSCs have been
enriched through a variety of surface markers such as EpCAM, CD13,
CD24, CD90 and CD133. Subgroups of CD13+ and
EpCAM+ play important roles in liver cancer propagation
and relapse (12). miRNAs can
affect nearly 30% of human genes by binding to the 3′UTR of target
mRNAs. Recent studies have reported that several miRNAs participate
in carcinogenesis (33). In the
present study, we utilized easy-to-use tumor spheres to enrich
liver CSCs and to identify differentially expressed miRNAs between
tumor spheres and adherent cells. We mainly investigated the
differentially expressed miRNAs that were significantly
downregulated in the TCGA database and validated the expression of
these miRNAs in tumor spheres and liver CSCs (CD13+ or
EpCAM+).
Among these miRNAs, we found that miR-486 was
significantly downregulated in tumor spheres, CD13+ or
EpCAM+ liver CSCs. Previous studies have indicated that
miR-486 can function as a tumor suppressor in lung (34) and gastric cancer (35). However, it was unknown whether
miR-486 regulates the self-renewal properties of liver CSCs. In the
present study, it was revealed that miR-486 expression was
downregulated in CD13+ and EpCAM+ liver CSCs
compared with the control group. In addition, miR-486 significantly
suppressed liver CSC properties, tumorigenesis, chemoresistance and
the invasion process of liver cancer cells.
To further clarify the mechanism of miR-486
inhibition of cancer stemness, the TargetScan database and qPCR
assays to screen the potential target genes of miR-486 were used.
Consequently, it was revealed that Sirt1 was significantly
suppressed by miR-486. Moreover, the expression levels of Sirt1 in
tumor spheres were markedly higher compared with adherent cells.
The luciferase assay revealed that miR-486 significantly decreased
the firefly luciferase activity of the Sirt1 3′UTR reporter.
Collectively, these data revealed that Sirt1 was a direct target of
miR-486. Sirt1 expression has been revealed to be associated with
poor prognosis and development of liver cancer in patients
(36). The present findings
revealed that knockdown of Sirt1 inhibited tumor sphere formation.
Conversely, overexpression of Sirt1 significantly promoted tumor
sphere formation. Moreover, the present study demonstrated that
Sirt1 was required for liver cancer tumorigenesis in vivo.
Collectively, we found that Sirt1 was responsible for the
maintenance of the self-renewal of liver CSCs. Our data was
consistent with a study by Liu et al (36), which revealed that Sirt1 promoted
tumorigenesis by mediating the activation of SOX2. It was revealed
in the present results that the expression of both Sirt1 and SOX2
expression was suppressed by miR-486.
In summary, the present study demonstrated that
miR-486, which was significantly downregulated in liver CSCs,
suppressed liver CSCs by targeting Sirt1. Thus, the miRNA-486/Sirt1
axis may provide new insights into the mechanism of liver CSCs and
may be a useful therapeutic target for liver cancer.
Acknowledgements
We thank Zeng Fan (Institute of Health Service and
Transfusion Medicine, Beijing, China) for technical support. We
thank LetPub (www.letpub.com) for providing
linguistic assistance during the preparation of this
manuscript.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81472341, 81772617,
81773137 and 81502581), the Beijing Municipal Education Commission
(no. KM201710005031) and the Guangzhou Health Care and Cooperative
Innovation Major Project (no. 201803040005).
Availability of data and materials
The data sets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XY conceived and designed the study, performed the
experiments, analyzed the data and wrote the manuscript; XL and QC
performed the experiments and wrote the manuscript; ZW provided the
clinical specimens and analyzed the data. GJ, HY, LW, CG, QZ, JX,
LH and XN performed the experiments and analyzed the data; HH
constructed the plasmids and analyzed the data; WY and XP initiated
the study, organized, designed and wrote the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The Research Ethics Committees of the Academy of
Military Medical Sciences, Beijing, China. Written informed consent
was obtained from all patients. All animal experiments in the
present study were approved by the Ethics Committee of the Academy
of Military Medical Sciences.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Visvader JE: Cells of origin in cancer.
Nature. 469:314–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Easwaran H, Tsai HC and Baylin SB: Cancer
epigenetics: Tumor heterogeneity, plasticity of stem-like states,
and drug resistance. Mol Cell. 54:716–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kreso A and Dick JE: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Muthukrishnan SD, Alvarado AG and Kornblum
HI: Building bonds: Cancer stem cells depend on their progeny to
drive tumor progression. Cell Stem Cell. 22:473–474. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu C, Liu L, Chen X, Cheng J, Zhang H,
Shen J, Shan J, Xu Y, Yang Z, Lai M and Qian C: Sox9 regulates
self-renewal and tumorigenicity by promoting symmetrical cell
division of cancer stem cells in hepatocellular carcinoma.
Hepatology. 64:117–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Sun H, Zhao F, Lu P, Ge C, Li H,
Hou H, Yan M, Chen T, Jiang G, et al: BMP4 administration induces
differentiation of CD133+ hepatic cancer stem cells,
blocking their contributions to hepatocellular carcinoma. Cancer
Res. 72:4276–4285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang KH, Ma S, Lee TK, Chan YP, Kwan PS,
Tong CM, Ng IO, Man K, To KF, Lai PB, et al: CD133+
liver tumor-initiating cells promote tumor angiogenesis, growth,
and self-renewal through neurotensin/interleukin-8/CXCL1 signaling.
Hepatology. 55:807–820. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of
CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mani SK, Zhang H, Diab A, Pascuzzi PE,
Lefrançois L, Fares N, Bancel B, Merle P and Andrisani O:
EpCAM-regulated intramembrane proteolysis induces a cancer stem
cell-like gene signature in hepatitis B virus-infected hepatocytes.
J Hepatol. 65:888–898. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haraguchi N, Ishii H, Mimori K, Tanaka F,
Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H, Barnard
GF, et al: CD13 is a therapeutic target in human liver cancer stem
cells. J Clin Invest. 120:3326–3339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Christ B, Stock P and Dollinger MM: CD13:
Waving the flag for a novel cancer stem cell target. Hepatology.
53:1388–1390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawai T, Yasuchika K, Ishii T, Katayama H,
Yoshitoshi EY, Ogiso S, Kita S, Yasuda K, Fukumitsu K, Mizumoto M,
et al: Keratin 19, a cancer stem cell marker in human
hepatocellular carcinoma. Clin Cancer Res. 21:3081–3091. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang W, Wang C, Lin Y, Liu Q, Yu LX, Tang
L, Yan HX, Fu J, Chen Y, Zhang HL, et al: OV6+
tumor-initiating cells contribute to tumor progression and invasion
in human hepatocellular carcinoma. J Hepatol. 57:613–620. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamashita T and Wang XW: Cancer stem cells
in the development of liver cancer. J Clin Invest. 123:1911–1918.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chiba T, Kita K, Zheng YW, Yokosuka O,
Saisho H, Iwama A, Nakauchi H and Taniguchi H: Side population
purified from hepatocellular carcinoma cells harbors cancer stem
cell-like properties. Hepatology. 44:240–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN,
Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, et al: Hepatocellular
carcinoma-associated mesenchymal stem cells promote hepatocarcinoma
progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology.
57:2274–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chai S, Ng KY, Tong M, Lau EY, Lee TK,
Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, et al: Octamer
4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in
liver cancer stem cells. Hepatology. 64:2062–2076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Tang ZY, Ye SL, Liu YK, Chen J, Xue
Q, Chen J, Gao DM and Bao WH: Establishment of cell clones with
different metastatic potential from the metastatic hepatocellular
carcinoma cell line MHCC97. World J Gastroenterol. 7:630–636. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan XL, Fu CJ, Chen L, Qin JH, Zeng Q,
Yuan HF, Nan X, Chen HX, Zhou JN, Lin YL, et al: Mesenchymal stem
cells from primary breast cancer tissue promote cancer
proliferation and enhance mammosphere formation partially via
EGF/EGFR/Akt pathway. Breast Cancer Res Treat. 132:153–164. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miao X, Yan X, Qu D, Li D, Tao FF and Sun
Z: Red emissive sulfur, nitrogen codoped carbon dots and their
application in ion detection and theraonostics. ACS Appl Mater
Interfaces. 9:18549–18556. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST,
Nan X, Cao N, Fu CJ, Yan X, Jia YL, et al: MicroRNA-125b attenuates
epithelial-mesenchymal transitions and targets stem-like liver
cancer cells through small mothers against decapentaplegic 2 and 4.
Hepatology. 62:801–815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y,
Yan F, Zhu P, Wu J, Huang G, et al: Mesenchymal stem cells promote
hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and
miR-34a. Cancer Res. 77:6704–6716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Tian B, Yang J, Zhao L, Wu X, Ye SL,
Liu YK and Tang ZY: Stepwise metastatic human hepatocellular
carcinoma cell model system with multiple metastatic potentials
established through consecutive in vivo selection and studies on
metastatic characteristics. J Cancer Res Clin Oncol. 130:460–468.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down- regulated in human hepatocellular carcinoma.
Cancer Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan
IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, et al: Genomic loss
of miR-486 regulates tumor progression and the OLFM4 antiapoptotic
factor in gastric cancer. Clin Cancer Res. 17:2657–2667. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Liu C, Zhang Q, Shen J, Zhang H,
Shan J, Duan G, Guo D, Chen X, Cheng J, et al: SIRT1-mediated
transcriptional regulation of SOX2 is important for self-renewal of
liver cancer stem cells. Hepatology. 64:814–827. 2016. View Article : Google Scholar : PubMed/NCBI
|