Introduction
The treatment of acute myeloid leukemia (AML) is
hampered by the development of resistance to chemotherapy.
Multidrug resistance (MDR) is a phenomenon by which cancer cells
become simultaneously resistant to several structurally and
functionally distinct molecules, rendering them ineffective.
Resistance to anticancer therapy can arise via different
contributing mechanisms, including metabolic inactivation of drugs,
alteration of drug targets, increased drug efflux, evasion of cell
death, increased repair of damaged DNA, epithelial-mesenchymal
transition, and epigenetic mechanisms including DNA methylation and
histone modification (1,2). Cancer cell heterogeneity and the tumor
microenvironment are also involved in the development of acquired
resistance to chemotherapeutics (3). Drug efflux is an evolutionarily
conserved mechanism observed in microbes, which is also exhibited
by cancer cells. The efflux of substrates is an important
protective mechanism in normal cell physiology, which prevents the
accumulation of toxins and is an integral property of the
blood-brain barrier (4).
Members of the ATP-binding cassette (ABC)
transporters serve a key role in mediating the extracellular
pumping of substrates (5). ABC
subfamily B member 1 (ABCB1) is the first member of the ABC
superfamily of transporters to be implicated in MDR (6). Structurally, it consists of two
homologous arms, each comprising six transmembrane domains
connected to an ATP-binding domain via flexible polypeptide
linkers. ABCB1 substrates include an array of hydrophobic
compounds, including anthracyclines, Vinca alkaloids,
epipodophyllotoxins, taxanes and several tyrosine kinase inhibitors
(7). The inhibition of
ABCB1-mediated drug efflux is a relevant therapeutic approach for
overcoming MDR (8). In this regard,
several generations of ABCB1 inhibitors have been developed but
have failed to produce the desired clinical response and are
associated with systemic toxicity. Therefore, there is an urgent
requirement for agents capable of modulating the activity of ABCB1
and producing therapeutically relevant inhibition without exerting
adverse effects. Small molecule inhibitors, such as alectinib,
bafetinib, trametinib and quizartinib, which target specific
molecules in oncogenic signaling, have been reported to modulate
ABC pumps and reverse resistance by acting as substrates/inhibitors
(9–12).
The phosphoinositide 3-kinase (PI3K)/AKT/mammalian
target of rapamycin (mTOR) signaling pathway constitutes the
central axis of intracellular growth signaling, which is commonly
deregulated in cancer. In total, 50–70% of patients with AML
exhibit abnormal/hyperactivated PI3K signaling (13). The therapeutic inhibition of mTOR
has resulted in anticancer effects in vitro and in
vivo in various cancer types, including AML. Several mTOR
inhibitors have been approved by the USA Food and Drug
Administration and are currently used as a single agent or in
combination (14–17). Furthermore, inhibitors of mTOR have
been demonstrated to overcome chemoresistance (18–20).
Classical mTOR inhibitors, including rapamycin and its analogs
(rapalogs), are only modestly effective as anticancer agents as
they only partially suppress the PI3K/AKT/mTOR signaling pathway,
leading to a compensatory overactivation of the pathways via a
negative feedback loop (21). The
mechanistic insufficiency of rapalogs in completely deactivating
PI3K signaling is overcome by a novel class of ATP-competitive mTOR
inhibitors that target the kinase domain of mTOR, thereby
effectively repressing mTOR complex (mTORC)1 and mTORC2 (22). WYE-354 is a synthetic mTOR kinase
inhibitor, which has demonstrated robust anticancer activity via
dual inhibition of the mTORC1 and mTORC2 complexes in several cell
lines (23–25). The present study evaluated WYE-354
as a potent chemosensitizing agent and assessed its role in
reversing MDR mediated by ABCB1. Furthermore, the present study
attempted to elucidate the mechanisms by which WYE-354 causes
chemosensitization and aimed to understand its interaction with the
ABCB1 protein using in silico methods.
Materials and methods
Cell culture and reagents
The Adriamycin (Adr)-resistant cell lines
K562/Adr200 and K562/Adr500 were generated by culturing K562 cells
(CLS Cell Lines Service GmbH, Eppelheim, Germany) in step-wise
incremental doses of Adr, ranging between 0.002 and 0.5 µM over a
period of 2 months at 37°C with 5% CO2 in a humidified
incubator. Resistant clones were selected upon plating of the cells
in methylcellulose semi-solid medium (MethoCult™ H4230; Stemcell
Technologies, Inc., Vancouver, BC, Canada). The resistant cells
were maintained without Adr for >2 weeks prior to
experimentation. The cells were cultured in RPMI medium
supplemented with 10% fetal bovine serum and ciprofloxacin (10
µg/ml) and were maintained at 37°C with 5% CO2 in a
humidified incubator. All cell culture reagents were purchased from
Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Adr, daunorubicin, idarubicin, etoposide and WYE-354
were purchased form Selleck Chemicals (Houston, TX, USA). Verapamil
and cisplatin were purchased from Merck KGaA (Darmstadt, Germany).
The CellTiter®-Blue Cell Viability assay was acquired
from Promega Corporation (Madison, WI, USA). The RNeasy Mini kit
was purchased from Qiagen GmbH (Hilden, Germany). The SuperScript
VILO cDNA Synthesis kit was acquired from Thermo Fisher Scientific,
Inc. and Power SYBR®-Green PCR Master Mix was from
Applied Biosystems; Thermo Fisher Scientific, Inc. The primers used
to amplify ABCB1 and GAPDH were purchased from Metabion
International AG (Planegg, Germany). Mammalian Cell Lysis kit was
obtained from Sigma-Aldrich; Merck KGaA. Protein concentration was
determined using the DC Protein Assay Reagents Package from Bio-Rad
Laboratories, Inc. (Hercules, CA, USA). NuPAGE® gels and
buffers, in addition to the Invitrogen WesternBreeze™
chemiluminescent kit for western blotting, were purchased from
Thermo Fisher Scientific, Inc. The antibody against ABCB1 (cat. no.
MA-126529) was obtained from Thermo Fisher Scientific, Inc.,
whereas anti-β-tubulin (cat. no. MAB8527), anti-human
phosphorylated (p)-p70S6K (T389) (cat. no. MAB8963) and anti-p70S6K
(cat. no. MAB8962) antibodies were purchased from R&D Systems,
Inc. (Minneapolis, MN, USA). The Pgp-Glo™ assay system was
purchased from Promega Corporation. The BD Pharmingen APC-Annexin V
kit from BD Biosciences (San Jose, CA, USA) was used for the
analysis of apoptosis.
Cell viability assay
The cells (104/well) were incubated with
a concentration gradient of chemotherapeutic drugs ranging from
0.01 to 100 µM, alone or in combination with WYE-354 (0.2 or 1 µM)
or Verapamil (5 µM) in 96-well plates for 48 h at 37°C.
Subsequently, 20 µl CellTiter®-Blue Cell Viability
reagent was added to each well and incubated for an additional 2 h
for the development of fluorescence. The fluorescence emission was
measured at 590 nm using the SpectraMax® i3× Multi-Mode
microplate reader (Molecular Devices, LLC, San Jose, CA, USA) and
plotted against drug concentrations to determine the mean
inhibitory concentration of the drug combination producing 50%
decrease in cell viability (IC50).
Adr accumulation assay
The cells (105) were counted, seeded in a
6-well plate and incubated with 10 µM Adr for 2 h at 37°C to allow
drug uptake to occur. Subsequently, the cells were washed twice
with 1X ice-cold PBS and immediately analyzed on a BD FACSAria III
(BD Biosciences). Adr florescence was measured using the 561-nm
excitation laser line and the emission was acquired using a
582/15-nm filter. A minimum of 10,000 events were acquired for
analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Primer-BLAST (National Center for Biotechnology
Information; National Institutes of Health, Bethesda, MD, USA) was
used to design primer oligonucleotide sequences for the ABCB1 gene.
The messenger RNA (mRNA) expression levels of ABCB1 were determined
using the following forward and reverse primer sequences,
respectively: 5′-TTGCTGCTTACATTCAGGTTTCA-3′ and
5′-AGCCTATCTCCGTCGCATT-3′. The primer pair used for GAPDH was
forward, 5′-TGAAGGTGCCATCATTCTTG-3′ and reverse,
5′-ATGAGCGACGTGGCTATTGT-3′. RNA (100 ng) was used to prepare cDNA
using the SuperScript® VILO™ cDNA synthesis kit as per
manufacturer's instructions. RT-qPCR was performed using 1 µl cDNA
in a 20 µl reaction mix containing 10 µl Power
SYBR®-Green PCR Master mix and 100 nM each of forward
and reverse primers. Amplifications were performed under following
conditions: 95°C for 20 sec, followed by 40 cycles of 95°C for 15
sec, 60°C for 60 sec and 72°C for 15 sec. Data was collected at the
end of the extension step (72°C). Melt curve analysis was performed
to ensure specificity of the amplified product. Relative gene
expression was calculated by the 2−ΔΔCq method (26).
Western blot analysis
Total protein was extracted with RIPA lysis buffer
containing protease and phosphatase inhibitor cocktails (cat. nos.
sc-24948 and sc-45044 respectively; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and quantified using the DC™ Protein assay
(Bio-Rad Laboratories, Inc.) according to the manufacturer's
protocol. A total of ~40 µg protein sample was prepared under
reducing conditions with the NuPAGE reducing agent (Thermo Fisher
Scientific, Inc.) at 37°C for 30 min and loaded onto 4–12% NuPAGE
gradient gels for electrophoresis, which was performed at 200 V for
2 h. The transfer onto a methanol-activated 0.45-micron PVDF
membrane (Thermo Fisher Scientific, Inc.) was conducted overnight
at 30 V and 4°C with a transfer buffer containing 0.1% SDS.
Subsequently, the membrane was washed, blocked for 60 min at room
temperature using buffers supplied with the WesternBreeze™
Chemiluminescent kit and probed with anti-ABCB1 (dilution 1:200),
anti-β-tubulin (dilution 1:1,000), anti-p-p70S6K (dilution 1:1,000)
or anti-p70S6K (dilution 1:4,000) antibodies overnight at 4°C. The
primary antibody was completely removed by washing and the membrane
was re-incubated with the secondary antibody from the
WesternBreeze™ Chemiluminescent kit for 60 min at room temperature.
The chemiluminescent agent from the WesternBreeze™ Chemiluminescent
kit was applied upon washing the membrane several times, and the
reaction was allowed to develop for 5 min prior to acquisition of
the results on a C-DiGit Blot Scanner (LI-COR Biosciences, Lincoln,
NE, USA). Western blot images were analyzed using Image Studio
Digits version 4.0 (LI-COR Biosciences).
Cell cycle analysis
The cells (3.5×105) were incubated with
the corresponding drugs at 37°C for 72 h. Subsequently, the cells
were collected and washed twice with ice-cold PBS (1X). The washed
cells were fixed on ice for 20 min using a fixation buffer
containing paraformaldehyde. Hoechst 33342 (10 µg/ml; Thermo Fisher
Scientific, Inc.) was used for staining. The cells were then
incubated in the dark for 30 min on ice. A total of 20,000 events
were acquired using a BD FACSAria III. FlowLogic version 7.2.1
software (Inivai Technologies, Victoria, Australia) was used to
obtain the percentages of cells in the G1, S and G2/M phases in the
singlet-gated population.
Apoptosis assay
The cells (1.5×105) were counted and
plated in a 12-well plate. Following 48 h of drug treatment, the
cells were harvested and washed twice with PBS (1X). APC-Annexin V
and 7-AAD (both from BD Biosciences) were added to the cell
suspension according to the manufacturer's protocol. The cells were
mixed gently on a vortex and incubated in the dark for 20 min at
room temperature. The labelled cells were analyzed by acquiring
10,000 events using a BD FACSAria III.
Rhodamine 6G (R6G) efflux assay
The cells (105) were counted and
suspended in 1 ml RPMI medium. R6G (0.5 µM) was added to the medium
alongside 1, 5 or 10 µM WYE-354, or 100 µM Verapamil as control
inhibitors and incubated for 60 min at 37°C. Upon dye uptake, the
cells were pelleted by centrifugation at 250 × g for 5 min at 4°C
in a cold centrifuge. The harvested cells were washed twice with 1X
ice-cold PBS and resuspended in 1 ml RPMI with the inhibitors
alone, and reincubated for an additional 60 min at 37°C to allow
the efflux of R6G. The cells were subsequently washed twice with
ice-cold PBS and analyzed immediately on a BD FACSAria III. A
minimum of 5,000 events were recorded for analysis.
ABCB1-ATPase assay
The ABCB1-ATPase assay was performed using the
Pgp-Glo™ assay system. Briefly, recombinant human ABCB1
(hABCB1)-containing membranes were incubated with different
concentrations of WYE-354 and 0.25 mM sodium orthovanadate in the
presence of 5 mM MgATP at 37°C for 40 min. The unconsumed ATP was
quantified by adding ATP detection buffer, and luminescence was
recorded after 20 min at room temperature using a
SpectraMax® i3× Multi-Mode microplate reader. The basal
and drug-stimulated ABCB1-ATPase activities were calculated
according to the manufacturer's protocol.
Docking analysis
The three-dimensional structure of the ligand was
generated from PubChem and Balloon (https://pubchem.ncbi.nlm.nih.gov/ and http://users.abo.fi/mivainio/balloon/index.php).
BLASTp and HHBlits (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins;
http://toolkit.tuebingen.mpg.de/#/tools/hhblits)
were used to interrogate the human ABCB1 (UniProt ID: P08381) amino
acid sequence against the SWISS-MODEL (https://swissmodel.expasy.org/) template library.
Based on the target-template alignment, hABCB1 mouse homology
models were established using Promod3. CLC Drug Discovery Workbench
4.0 (Qiagen GmbH) was used for docking. Based on the binding
scores, the top 100 binding conformations were selected and
analyzed.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism Software version 6.07 (GraphPad Software, Inc., La
Jolla, CA, USA). Student's t-test was used to compare paired data
points between each group. P<0.05 over a 95% confidence interval
was considered to indicate a statistically significant
difference.
Results
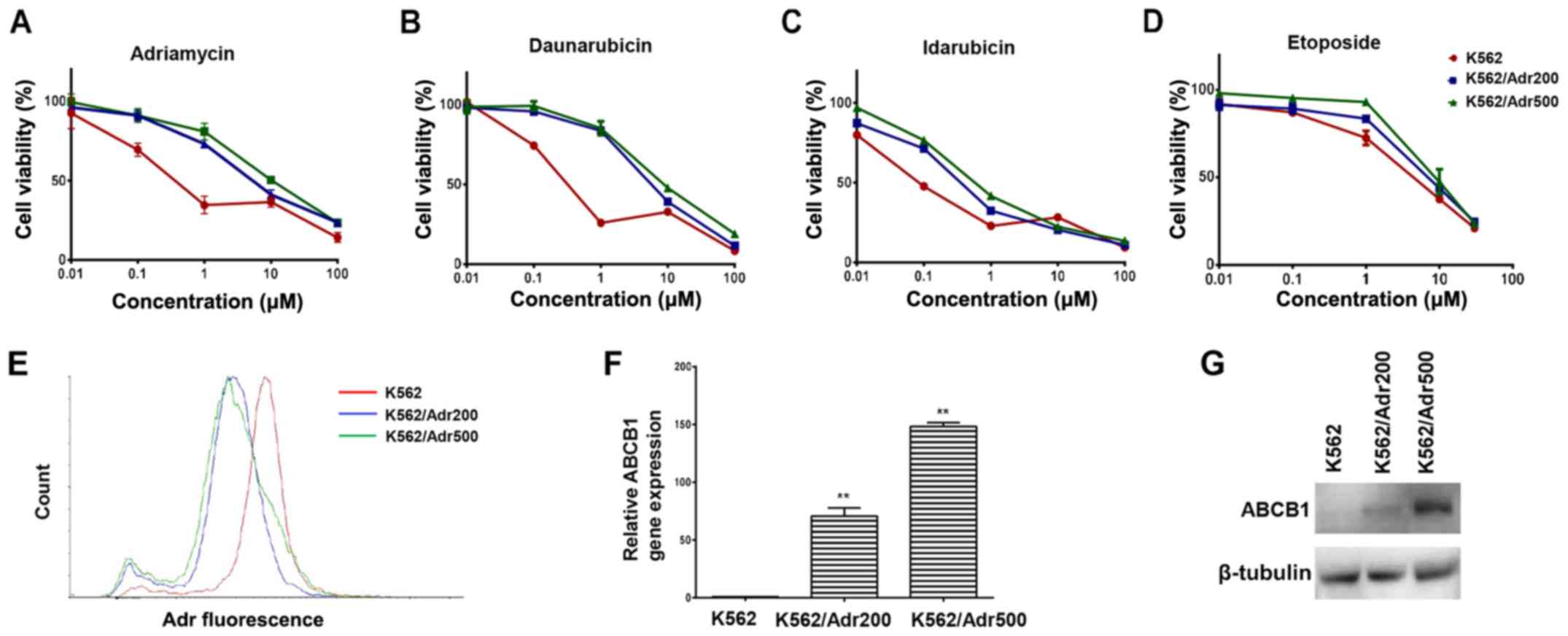
Adr-resistant cell lines exhibit MDR
and overexpress ABCB1
In order to screen agents capable of reversing
ABCB1-mediated resistance, the present study first developed
Adr-resistant K562 cells exhibiting varying levels of resistance.
The cell viability assay revealed 16-fold differences in
IC50 values in the K562/Adr200 cells and 30-fold
differences in IC50 values in the K562/Adr500 cells
compared with the parental cells (Fig.
1A). Additionally, the K562/Adr200 and K562/Adr500 cells
exhibited varying degrees of resistance to other chemotherapeutics
(daunorubicin, idarubicin and etoposide), demonstrating the
establishment of MDR cell lines (Fig.
1B-D and Table I). To clarify
the mechanism of MDR, the intracellular Adr concentration was
quantified by determining Adr fluorescence levels in the parental
and the two resistant cell lines using flow cytometry. As shown in
Fig. 1E, Adr fluorescence was
considerably reduced in the K562/Adr200 and K562/Adr500 cells
compared with the parental K562 cells. The RT-qPCR analysis
revealed increased mRNA levels of ABCB1 by ~70- and 148-fold in the
K562/Adr200 and K562/Adr500 cells, respectively (Fig. 1F and Table II). Increased protein levels of
ABCB1 were also confirmed by western blot analysis (Fig. 1G). The resistant cells did not show
increased expression of ABCG2, also known to efflux Adr (data not
shown). In addition, the cytotoxicity of cisplatin, which is not an
ABCB1 substrate, did not differ between parental and resistant
cells (Table I). Therefore, the
overexpression of ABCB1 was recognized as the underlying mechanism
for the observed MDR.
 | Table I.IC50 values of
chemotherapeutics. |
Table I.
IC50 values of
chemotherapeutics.
|
| IC50 µM
(RR) |
|---|
|
|
|
|---|
| Drug | K562 | K562/Adr200 | K562/Adr500 |
|---|
| Adriamycin | 0.153±0.011 | 2.506±0.304
(16.379)b | 4.659±0.786
(30.451)b |
| Daunorubicin | 0.167±0.011 | 0.972±0.523
(5.834) | 2.026±0.283
(12.163)b |
| Idarubicin | 0.070±0.004 | 0.174±0.126
(2.470) | 0.255±0.089
(3.627)a |
| Etoposide | 2.391±0.071 | 5.193±0.155
(2.172)b | 7.892±2.658
(3.301)b |
| Cisplatin | 12.821±0.86 | 11.942±1.71
(0.93) | 12.390±1.49
(0.97) |
 | Table II.Comparison of average Cq values for
ABCB1 and GAPDH using SYBR-Green reverse transcription-quantitative
polymerase chain reaction analysis. |
Table II.
Comparison of average Cq values for
ABCB1 and GAPDH using SYBR-Green reverse transcription-quantitative
polymerase chain reaction analysis.
| Cell line | ABCB1 Cq value | GAPDH Cq value |
|---|
| K562 | 26.45±0.08 | 18.57±0.28 |
| K562/Adr200 | 20.25±0.06 | 18.53±0.20 |
| K562/Adr500 | 19.54±0.26 | 19.12±0.22 |
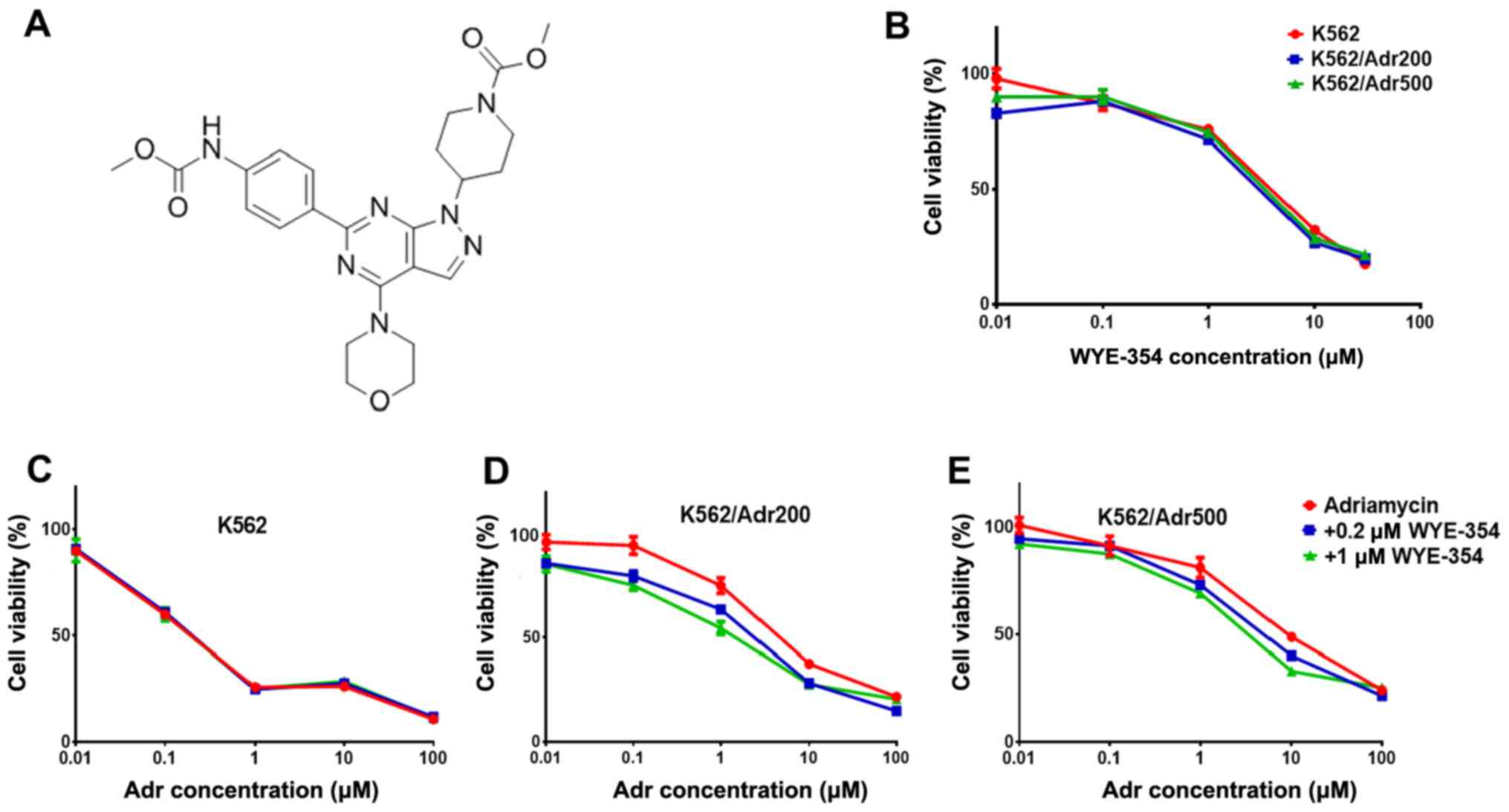
WYE-354 increases Adr cytotoxicity in
Adr-resistant cell lines
WYE-354 cytotoxicity (Fig. 2A) was first determined in the
parental K562 and resistant cell lines K562/Adr200 and K562/Adr500
using the CellTiter®-Blue Cell Viability assay. Based on
the IC50 of WYE-354 in the parental and resistant cell
lines (>3.2 µM; Fig. 2B), two
doses of WYE-354 (0.2 and 1 µM), producing <10 and <30% cell
death respectively, were selected for examining Adr sensitivity in
the K562/Adr200 and K562/Adr500 cells. As shown in Fig. 2C-E, 0.2 µM WYE-354 produced a
marginal decrease in Adr IC50 in the K562/Adr200 cells,
with a significant decrease in Adr IC50 observed for the
K562/Adr500 cells. The addition of 1 µM WYE-354 significantly
increased Adr cytotoxicity in both resistant cell lines by
decreasing the IC50 from 2.5±0.3 to 1.3±0.2 µM in the
K562/Adr200 cells and from 4.6±0.7 to 1.7±0.3 µM in the K562/Adr500
cells. Verapamil at 5 µM was used as the positive control. Of note,
Adr cytotoxicity in the parental K562 cells was unchanged by the
addition of 0.2 or 1 µM WYE-354, including the positive control
(Table III). These results
suggest that WYE-354 is a potent modulator of Adr resistance in
vitro.
 | Table III.Adr IC50 values in the
presence of indicated doses of WYE-354. |
Table III.
Adr IC50 values in the
presence of indicated doses of WYE-354.
|
| IC50 µM
(FR) |
|---|
|
|
|
|---|
| Drug | K562 | K562/Adr200 | K562/Adr500 |
|---|
| Adriamycin | 0.153±0.011
(1.000) | 2.506±0.304
(16.379) | 4.659±0.786
(30.451) |
| +0.2 µM
WYE-354 | 0.124±0.09
(0.810) | 2.230±0.661
(14.575) | 2.879±0.381
(18.816)a |
| +1 µM WYE-354 | 0.110±0.238
(0.718) | 1.358±0.279
(8.875)a | 1.778±0.391
(11.620)a |
| +5 µM
verapamil | 0.109±0.053
(0.712) | 1.683±0.21
(11.000)b | 2.160±0.23
(14.117)b |
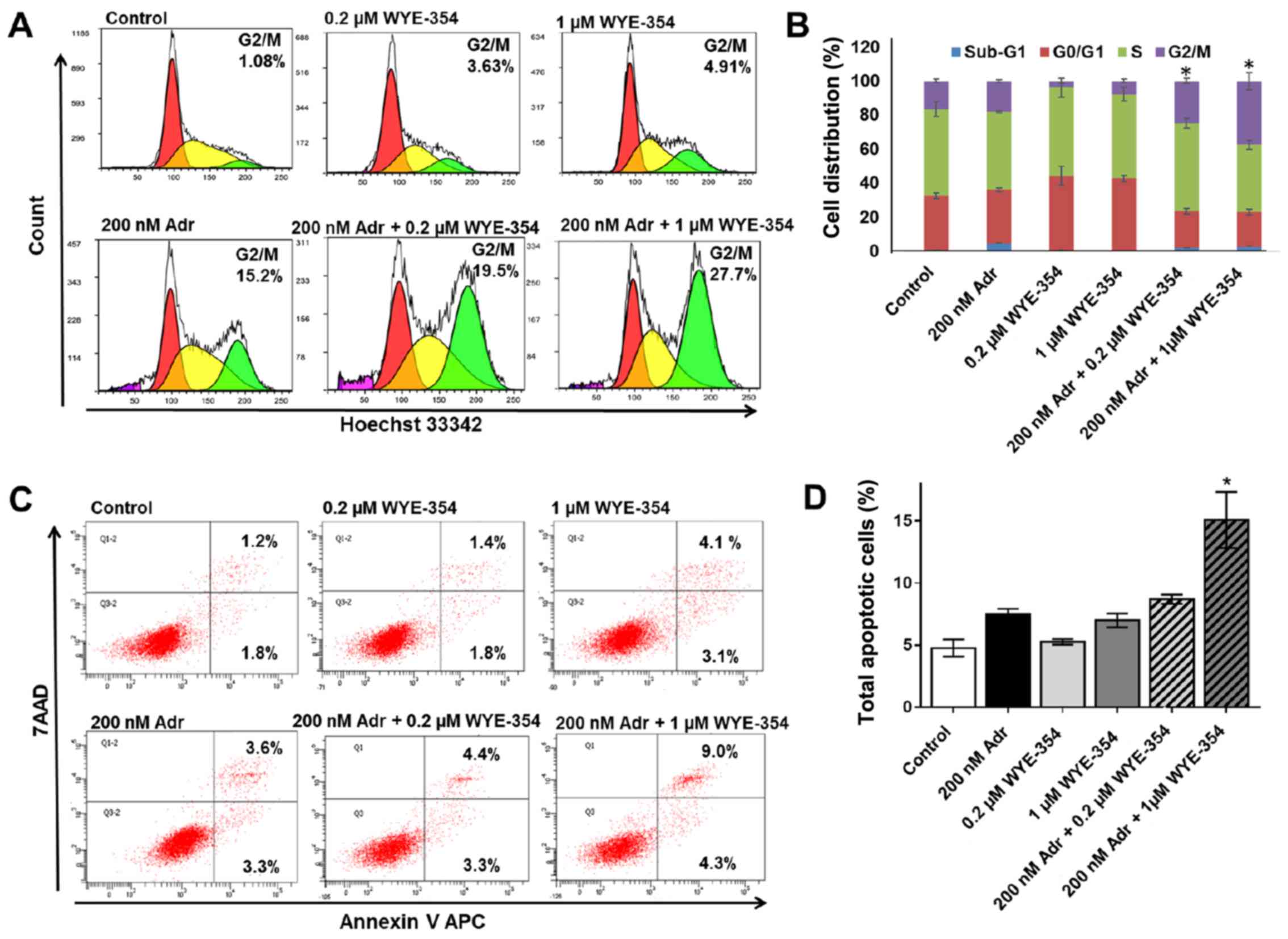
WYE-354 increases G2/M cell cycle
arrest and apoptosis in combination with Adr in Adr-resistant cell
lines
To elucidate the mechanism by which WYE-354 enhances
Adr cytotoxicity in Adr-resistant cell lines, cell cycle analysis
and apoptosis were assessed in the K562/Adr200 cells by flow
cytometry. A viable dose of 200 nM Adr, corresponding to 1/10 of
the Adr IC50 for K562/Adr200 cells, was selected for the
assays. As shown in Fig. 3A and B,
treatment with 0.2 and 1 µM WYE-354 did not produce any significant
changes in the cell cycle distribution of K562/Adr200 cells, which
was comparable to that in the untreated control, however, Adr at
200 nM produced marked G2/M cell cycle arrest. The addition of 1 µM
WYE-354 to 200 nM Adr caused a significant increase in G2/M cell
cycle arrest in the K562/Adr200 cells. Similarly, 1 µM WYE-354
significantly increased apoptosis in combination with 200 nM Adr
(Fig. 3C and D). No significant
difference in apoptosis was observed upon treatment of the
K562/Adr200 cells with WYE-354 or Adr alone.
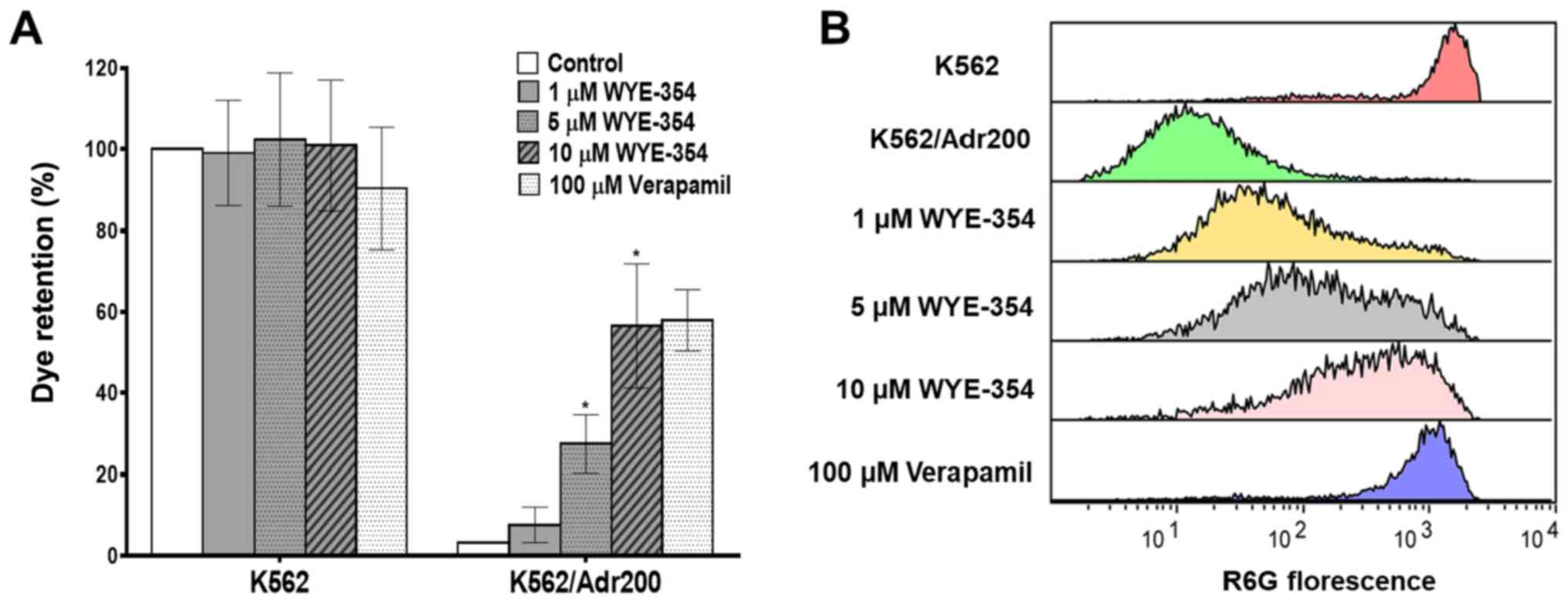
WYE-354 inhibits R6G efflux in
Adr-resistant cells
The effect of WYE-354 on the functional export of
R6G, a known substrate of the ABCB1 protein pump, was evaluated by
flow cytometry in K562 and K562/Adr200 cells in the presence of
increasing concentrations of WYE-354. As shown in Fig. 4A and B, WYE-354 dose-dependently
increased R6G retention in the K562/Adr200 cells. The addition of
1, 5 and 10 µM WYE-354 increased R6G fluorescence in the
K562/Adr200 cells from 3.26 to 7.64, 27.51 and 56.54%,
respectively, relative to the K562 controls. Verapamil, a
competitive ABCB1 inhibitor, was used as the positive control. No
significant differences in R6G fluorescence emission was observed
in the parental K562 cells following the addition of WYE-354
(Table IV). These findings clearly
indicate that WYE-354 exerts an inhibitory effect on the
ABCB1-mediated R6G efflux.
 | Table IV.R6G mean florescence intensity. |
Table IV.
R6G mean florescence intensity.
|
| R6G mean
florescence intensity |
|---|
|
|
|
|---|
| Treatment | K562 | K562/Adr200 |
|---|
| Control | 39.336±3910 | 1.663.3±286.61 |
| 1 µM WYE-354 | 40.561±401.5 |
4.142.5±1.216.16a |
| 5 µM WYE-354 | 41.636±499.5 |
7.963.25±664.47a |
| 10 µM WYE-354 |
41.581±1,939.33 |
25.690.25±1.817a |
| 100 µM
verapamil | 48.137±450.6 |
27.540.36±609.1 |
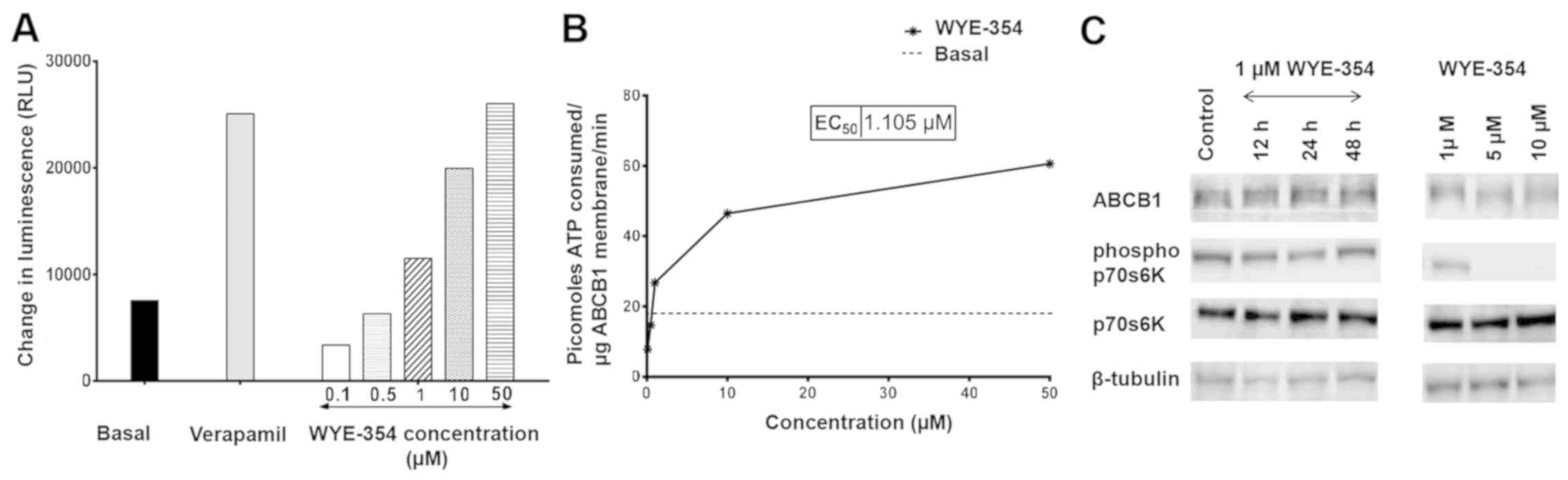
WYE-354 inhibits the activity of
ABCB1-ATPase and does not regulate the protein expression of
ABCB1
To ascertain the molecular interaction of WYE-354
with the ABCB1 protein, an ABCB1-ATPase assay was performed. ABCB1
is an active transporter and the rate of ATP hydrolysis is a direct
indicator of its activity. As shown in Fig. 5A, WYE-354 dose-dependently increased
luminescence in cell fractions containing recombinant hABCB1
protein, suggesting that WYE-354 is a potent substrate of the ABCB1
pump. Verapamil, an ABCB1 substrate and a competitive inhibitor,
was used as the positive control. Notably, WYE-354 exhibited
sub-basal ABCB1 ATPase activity, corresponding to the quantity of
ATP consumed, at concentrations <1 µM (Fig. 5B). This indicates that WYE-354 has
an inhibitory effect on the ATPase activity of ABCB1 at low
concentrations, which subsequently weakened as the WYE-354
concentration increased. A maximum increase by 3.45-fold in the
basal ATPase activity of ABCB1 was observed at a concentration of
50 µM, which was comparable to the increase produced by verapamil
at 500 µM. Furthermore, western blotting revealed no significant
differences in protein levels of ABCB1 in the K562/Adr500 cells
following treatment with 1 µM WYE-354 for 12, 24 and 48 h
time-points (Fig. 5C). At 1 µM,
WYE-354 did not produce significant suppression of mTOR signaling,
which was evident by the levels of p-p70S6K. Significant
downregulation of p-p70S6K was observed only at high concentrations
(5 and 10 µM), however, the protein expression of ABCB1 did not
differ substantially from that exhibited by the untreated control,
even at these concentrations (Table
V). This indicates that WYE-354 did not regulate the expression
of ABCB1 in the experiments.
 | Table V.Relative ABCB1 and p-p70S6K
expression detected by western blots. |
Table V.
Relative ABCB1 and p-p70S6K
expression detected by western blots.
|
| Normalized relative
intensities as compared to controla |
|---|
|
|
|
|---|
| Treatment | ABCB1 | p-p70S6K |
|---|
| Control | 1 | 1 |
| 1 µM WYE-354 12
h | 0.82±0.09 | 1.0±0.10 |
| 1 µM WYE-354 24
h | 0.95±0.12 | 0.75±0.14 |
| 1 µM WYE-354 48
h | 1.22±0.13 | 1.07±0.29 |
| 1 µM WYE-354 (24
h) | 1.22±0.50 | 0.76±0.11 |
| 5 µM WYE-354 (24
h) | 1.02±0.26 |
0.03±0.03b |
| 10 µM WYE-354 (24
h) | 1.37±0.29 |
0.04±0.04b |
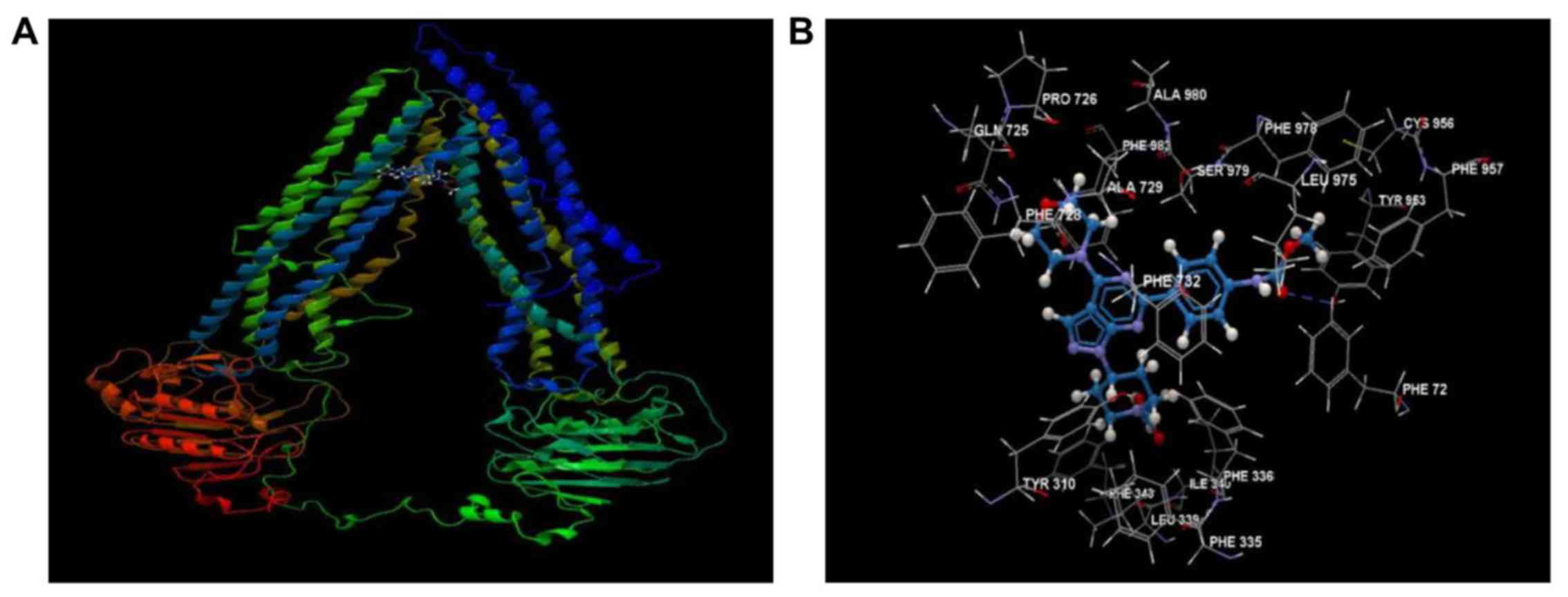
WYE-354 shows high affinity for the
hABCB1 drug-binding pocket
The mechanistic interactions of WYE-354 at the
drug-binding domain of the ABCB1 transmembrane protein were
predicted by docking using a hABCB1 mouse homology model (Fig. 6A). As shown in Fig. 6B, WYE-354 exhibited high affinity
for the drug-binding domain of hABCB1, interacting with several
amino acid residues, primarily via hydrophobic interactions,
forming a prominent hydrogen bond with Tyr953. The overall binding
and steric interaction scores, predicted using CLC Drug Discovery,
for WYE-354 were −85.194 and −83.273, respectively, indicating that
WYE-354 is a potent substrate of ABCB1.
Discussion
Despite advances in cancer research, chemotherapy
remains the first-line treatment for AML, the efficacy of which is
limited by the development of MDR. The overexpression of
drug-efflux pumps has been implicated in the failure of AML
chemotherapy, causing disease refraction or relapse (27). Therefore, substances that are
capable of restoring chemosensitivity are continuously being
investigated to maintain cytotoxicity and the clinical response of
chemotherapeutics. The role of ABCB1 in mediating MDR in cancer
cells has been extensively documented in different types of cancer.
The overexpression of ABCB1 is associated with poor clinical
response to chemotherapy in patients with AML (28). Therefore, the development or
identification of safe and effective ABCB1 inhibitors is of
particular interest. Besides specific inhibitors designed and
developed to target ABCB1, a number of small molecule inhibitors
targeting cellular signaling pathways have been demonstrated to
interact with ABCB1 as substrates or inhibitors. Various ABCB1
substrates may act as competitive inhibitors of ABCB1 efflux
activity (8,29).
Aberrant PI3K/AKT/mTOR signaling is common in AML
and is implicated in drug resistance (30,31).
First-generation mTOR inhibitors, including rapamycin and its
analogs, have been previously reported to reverse resistance in
different types of cancer (32,33).
Recently, BEZ235, a dual PI3K/mTOR inhibitor, demonstrated potent
activity as a resistance-modifying agent in AML cell lines, where
Adr-resistant K562 and vincristine-resistant HL60 cells were used
to represent MDR in AML (34). The
present study is the first, to the best of our knowledge, reporting
the effects of the mTOR kinase inhibitor WYE-354 on restoring Adr
chemosensitivity in MDR K562 cells in vitro. The present
study evaluated the cytotoxicity of WYE-354 in combination with Adr
in K562/Adr200 and K562/Adr500 cells. WYE-354 significantly
sensitized the two MDR cell lines to chemotherapy by decreasing Adr
IC50. Importantly, Adr cytotoxicity remained unchanged
in the parental cell line, suggesting that the mechanism may be
associated with the acquisition of drug resistance. The fact that
the overexpression of ABCB1 was identified as the mechanism
underlying Adr resistance in K562/Adr200 and K562/Adr500 cells
suggested that WYE-354 may be capable of interacting with the ABCB1
transporter, thus modulating its activity. Furthermore, the
combination of WYE-354 and Adr caused increased apoptosis and G2/M
cell cycle arrest, contributing to the increased chemosensitization
of Adr-resistant cells.
Adr is a well-known substrate of ABCB1 and
overexpression of the pump results in its increased clearance, thus
preventing its intracellular accumulation and reducing its
cytotoxicity as a consequence. The ABCB1 transporter activity was
assessed based on the efflux of R6G, a florescent substrate of
ABCB1. Detection of the natural florescence of rhodamine provides a
robust screening platform for examining ABCB1 efflux and, unlike
anthracyclines, quenching of florescence is not observed with
rhodamine dyes (35). WYE-354
potently inhibited the efflux of R6G in Adr-resistant cells. This
was consistent with the results from the cell viability assay,
indicating that the observed chemosensitization towards Adr was in
fact due to the inhibition of ABCB1-mediated Adr efflux, thus
leading to an increased intracellular accumulation of Adr.
Furthermore, WYE-354 dose-dependently increased the ATPase activity
of ABCB1, indicating that it is a potent substrate of ABCB1.
Notably, WYE-354 had a mild inhibitory effect on the ATPase
activity of ABCB1 at low concentrations (≤1 µM). This also
partially explains why the observed IC50 of WYE-354,
which was >3.2 µM, did not differ markedly between the parental
and resistant cell lines, as the substrate effect is expected to
become significant only at high concentrations. The phosphorylation
status of downstream targets directly reflects the potency of an
inhibitor. The present study revealed that the concentration of
WYE-354 required to re-sensitize resistant cell lines did not have
an effect on the phosphorylation status of p70S6K, an effector
molecule of the mTOR signaling pathway, even after ≤48 h of
incubation. Furthermore, there was minimal effect on the protein
expression of ABCB1 at high concentrations of WYE-354 (10 µM). The
molecular docking experiments revealed robust interactions of
WYE-354 with the hABCB1 mouse homology model. The above findings
suggest that the re-sensitization produced by WYE-354 is associated
with the competitive inhibition of Adr efflux, as a result of
WYE-354 being a substrate of ABCB1. These findings highlight
WYE-354 as a potential resistance reversal agent, due to its
interaction with the ABCB1 efflux pump, at the functional and
molecular level.
In conclusion, the present study described a novel
therapeutic application of the mTOR kinase inhibitor WYE-354 as a
potent modulator of ABCB1-mediated drug resistance. In addition,
the interaction between WYE-354 and ABCB1 may significantly improve
current understanding of drug availability and provide insights
into possible drug interactions in combination therapies. The use
of WYE-354 as a resistance-modulating agent can provide added
benefit due to its intrinsic anticancer activity. Further
investigations using patient-derived primary cell lines and animal
models are warranted to validate the clinical applicability of
using WYE-354 to potentially attenuate chemoresistance in
vivo.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Plan
for Science, Technology and Innovation (MAARIFAH), King Abdulaziz
City for Science and Technology (grant no. 09-BIO693-03).
Availability of data and materials
The datasets and certain material used and/or
analyzed during the present study are available from the
corresponding author on reasonable request.
Authors' contributions
FA and AGC conceived and designed the experiments.
SMI, AMI, SB and FA performed the experiments. Docking was
performed and analyzed by PNP. SMI, FA and SK analyzed the data.
SMI, and FA wrote the manuscript. JAK, AMA and MHAQ contributed
with reagents/materials/analysis tools and critically revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaffer BC, Gillet JP, Patel C, Baer MR,
Bates SE and Gottesman MM: Drug resistance: Still a daunting
challenge to the successful treatment of AML. Drug Resist Updat.
15:62–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin B, Ouillette P, Li Y, Keller J, Lam
C, Roulston D, Li C, Shedden K and Malek SN: Clonal evolution and
devolution after chemotherapy in adult acute myelogenous leukemia.
Blood. 121:369–377. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Löscher W and Potschka H: Role of drug
efflux transporters in the brain for drug disposition and treatment
of brain diseases. Prog Neurobiol. 76:22–76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohammad IS, He W and Yin L: Understanding
of human ATP binding cassette superfamily and novel multidrug
resistance modulators to overcome MDR. Biomed Pharmacother.
100:335–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dean M, Rzhetsky A and Allikmets R: The
human ATP-binding cassette (ABC) transporter superfamily. Genome
Res. 11:1156–1166. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kathawala RJ, Gupta P, Ashby CR Jr and
Chen ZS: The modulation of ABC transporter-mediated multidrug
resistance in cancer: A review of the past decade. Drug Resist
Updat. 18:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Binkhathlan Z and Lavasanifar A:
P-glycoprotein inhibition as a therapeutic approach for overcoming
multidrug resistance in cancer: Current status and future
perspectives. Curr Cancer Drug Targets. 13:326–346. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang K, Chen Y, To KK, Wang F, Li D, Chen
L and Fu L: Alectinib (CH5424802) antagonizes ABCB1- and
ABCG2-mediated multidrug resistance in vitro, in vivo and ex vivo.
Exp Mol Med. 49:e3032017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang YK, Zhang GN, Wang YJ, Patel BA,
Talele TT, Yang DH and Chen ZS: Bafetinib (INNO-406) reverses
multidrug resistance by inhibiting the efflux function of ABCB1 and
ABCG2 transporters. Sci Rep. 6:256942016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiu JG, Zhang YJ, Li Y, Zhao JM, Zhang WJ,
Jiang QW, Mei XL, Xue YQ, Qin WM, Yang Y, et al: Trametinib
modulates cancer multidrug resistance by targeting ABCB1
transporter. Oncotarget. 6:15494–15509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Kumar P, Anreddy N, Zhang YK, Wang
YJ, Chen Y, Talele TT, Gupta K, Trombetta LD and Chen ZS:
Quizartinib (AC220) reverses ABCG2-mediated multidrug resistance:
In vitro and in vivo studies. Oncotarget. 8:93785–93799.
2017.PubMed/NCBI
|
|
13
|
Dos Santos C, Récher C, Demur C and
Payrastre B: The PI3K/Akt/mTOR pathway: A new therapeutic target in
the treatment of acute myeloid leukemia. Bull Cancer. 93:445–447.
2006.(In French). PubMed/NCBI
|
|
14
|
Dinner S and Platanias LC: Targeting the
mTOR Pathway in Leukemia. J Cell Biochem. 117:1745–1752. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Motzer RJ, Escudier B, Oudard S, Hutson
TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA,
Hollaender N, et al RECORD-1 Study Group, : Phase 3 trial of
everolimus for metastatic renal cell carcinoma: Final results and
analysis of prognostic factors. Cancer. 116:4256–4265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hudes GR, Berkenblit A, Feingold J, Atkins
MB, Rini BI and Dutcher J: Clinical trial experience with
temsirolimus in patients with advanced renal cell carcinoma. Semin
Oncol. 36 (Suppl 3):S26–S36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park S, Chapuis N, Saint Marcoux F, Recher
C, Prebet T, Chevallier P, Cahn JY, Leguay T, Bories P, Witz F, et
al GOELAMS (Groupe Ouest Est d'Etude des Leucémies aiguës et Autres
Maladies du Sang), : A phase Ib GOELAMS study of the mTOR inhibitor
RAD001 in association with chemotherapy for AML patients in first
relapse. Leukemia. 27:1479–1486. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haritunians T, Mori A, O'Kelly J, Luong
QT, Giles FJ and Koeffler HP: Antiproliferative activity of RAD001
(everolimus) as a single agent and combined with other agents in
mantle cell lymphoma. Leukemia. 21:333–339. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grünwald V, DeGraffenried L, Russel D,
Friedrichs WE, Ray RB and Hidalgo M: Inhibitors of mTOR reverse
doxorubicin resistance conferred by PTEN status in prostate cancer
cells. Cancer Res. 62:6141–6145. 2002.PubMed/NCBI
|
|
20
|
Wang Z, Huang Y and Zhang J: Molecularly
targeting the PI3K-Akt-mTOR pathway can sensitize cancer cells to
radiotherapy and chemotherapy. Cell Mol Biol Lett. 19:233–242.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harrington LS, Findlay GM, Gray A,
Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S,
Shepherd PR, et al: The TSC1-2 tumor suppressor controls
insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol.
166:213–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun SY: mTOR kinase inhibitors as
potential cancer therapeutic drugs. Cancer Lett. 340:1–8. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu K, Toral-Barza L, Shi C, Zhang WG,
Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, et al:
Biochemical, cellular, and in vivo activity of novel
ATP-competitive and selective inhibitors of the mammalian target of
rapamycin. Cancer Res. 69:6232–6240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Zhu YR, Wang S and Zhao S:
Autophagy inhibition sensitizes WYE-354-induced anti-colon cancer
activity in vitro and in vivo. Tumour Biol. 37:11743–11752. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weber H, Leal P, Stein S, Kunkel H, García
P, Bizama C, Espinoza JA, Riquelme I, Nervi B, Araya JC, et al:
Rapamycin and WYE-354 suppress human gallbladder cancer xenografts
in mice. Oncotarget. 6:31877–31888. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salvia AM, Cuviello F, Coluzzi S,
Nuccorini R, Attolico I, Pascale SP, Bisaccia F, Pizzuti M and
Ostuni A: Expression of some ATP-binding cassette transporters in
acute myeloid leukemia. Hematol Rep. 9:74062017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu B, Li LJ, Gong X, Zhang W, Zhang H and
Zhao L: Co-expression of ATP binding cassette transporters is
associated with poor prognosis in acute myeloid leukemia. Oncol
Lett. 15:6671–6677. 2018.PubMed/NCBI
|
|
29
|
Beretta GL, Cassinelli G, Pennati M, Zuco
V and Gatt L: Overcoming ABC transporter-mediated multidrug
resistance: The dual role of tyrosine kinase inhibitors as
multitargeting agents. Eur J Med Chem. 142:271–289. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martelli AM, Evangelisti C, Chiarini F and
McCubrey JA: The phosphatidylinositol 3-kinase/Akt/mTOR signaling
network as a therapeutic target in acute myelogenous leukemia
patients. Oncotarget. 1:89–103. 2010.PubMed/NCBI
|
|
31
|
Tabe Y, Tafuri A, Sekihara K, Yang H and
Konopleva M: Inhibition of mTOR kinase as a therapeutic target for
acute myeloid leukemia. Expert Opin Ther Targets. 21:705–714. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ying L, Zu-An Z, Qing-Hua L, Qing-Yan K,
Lei L, Tao C and Yong-Ping W: RAD001 can reverse drug resistance of
SGC7901/DDP cells. Tumour Biol. 35:9171–9177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma Q, Chang Z, Wang W and Wang B:
Rapamycin-mediated mTOR inhibition reverses drug resistance to
adriamycin in colon cancer cells. Hepatogastroenterology.
62:880–886. 2015.PubMed/NCBI
|
|
34
|
Deng L, Jiang L, Lin XH, Tseng KF, Liu Y,
Zhang X, Dong RH, Lu ZG and Wang XJ: The PI3K/mTOR dual inhibitor
BEZ235 suppresses proliferation and migration and reverses
multidrug resistance in acute myeloid leukemia. Acta Pharmacol Sin.
38:382–391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JS, Paull K, Alvarez M, Hose C, Monks
A, Grever M, Fojo AT and Bates SE: Rhodamine efflux patterns
predict P-glycoprotein substrates in the National Cancer Institute
drug screen. Mol Pharmacol. 46:627–638. 1994.PubMed/NCBI
|