Introduction
Cancer stemness, which describes the stem-cell
functions of cancer cells (1), may
have profound implications for tumor aggressiveness and clinical
outcome (2,3). The majority of current cancer
therapies have been reported to induce cancer stemness, thereby
failing due to metastasis and relapse (4–7). In
the clinic, strategies that target cancer stemness may thus serve
as a novel approach to develop the next generation of cancer
therapeutics to suppress cancer relapse and metastasis (8,9).
Numerous signaling pathways have been reported to be
functionally associated with the induction of cancer stemness in
various cancer types, including WNT signaling (10,11),
NOTCH signaling (12), Hippo
signaling (13), hypoxia signaling
(14), DNA damage repair (15) and RAS signaling (16). However, previous conventional
molecular profiling studies have largely relied on bulk-tissue
measurements. Considering that clinical cancer tissues are
extremely heterogeneous, even within an individual patient
(17), characterizing the features
of cancer stemness in single cancer cells (i.e., intratumoral
cancer stemness) will likely provide more accurate and vital
information for identifying the potential pre-existing
treatment-resistant cancer cells in the cancer tissue. Furthermore,
the inhibition of these cancer stemness-associated signaling
pathways will ultimately improve the detection, diagnosis and
treatment of cancer.
To characterize the features of intratumoral cancer
stemness in two types of esophageal cancer, namely esophageal
squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC)
at a single-cell level, single-cell RNA-sequencing (RNA-Seq) data
of ESCC and EAC from the Sequence Read Archive (SRA) database was
utilized in the present study. Once the single cancer cells were
successfully separated from non-cancer cells in the cancer tissues
of patients with EAC and ESCC, cancer stemness analysis was applied
to identify high-and low-stemness cancer cells in each patient
(18). By comparing these cancer
cells within individual patients, the present study intended to
identify the signaling pathways and signature genes that were
responsible for the cancer stemness of EAC and ESCC. Furthermore,
the expression level of these significant genes was validated and
their association with prognosis was assessed using The Cancer
Genome Atlas (TCGA) cohort and our ESCC cohort.
Materials and methods
Single-cell transcriptome data for
bioinformatics analyses
All single-cell transcriptome data were obtained
from the SRA (https://www.ncbi.nlm.nih.gov/sra) under the accession
no. SRP119465. In these datasets, 5–10 M reads per single cell were
generated with Illumina Hiseq4000 (PE150).
RNA-Seq data preprocessing, alignment,
normalization and quantification
Fastqc and the R/Bioconductor package ‘ShortRead
1.40.0’ were used to perform quality control of all sequenced data
(19). Data were trimmed using
Trimmomatic 0.35 to remove and filter low quality and adapter
contaminated reads (20). The human
genome NCBI GRCh38 and its corresponding transcriptome gene
annotation, which can be downloaded from iGenome, was used for read
alignment. The Tophat2.1.1 (http://ccb.jhu.edu/software/tophat/) alignment tool
was used for alignment with default parameter settings. The
fragments per kilobase of transcript per million mapped reads
(FPKM) data from Cufflinks output were used for ‘Monocle 2.4.0’
gene differential expression methods (21). A quantile normalization method was
applied to the log2 transformed FPKM dataset. In the bulk-cell
quantification step, Cufflinks 2.2.1 was used to generate FPKM data
with default parameter settings (22).
Cancer stemness analysis
Cancer stemness can be measured according to the
expression profiles of stem cell genes, which is a property shared
by embryonic and adult stem cells (23). The 35 known stem cell markers were
identified according to previous publications, which are listed in
Table SI. To quantify the
heterogeneity of cancer stemness, a cancer stemness index (SI) was
defined by averaging the expression values of these 35 known stem
cell markers. Heatmaps for EAC and ESCC were generated and 274
single cancer cells were clustered using the SI level.
Database for Annotation, Visualization
and Integrated Discovery (DAVID) analysis
DAVID (http://david.abcc.ncifcrf.gov) functional annotation
bioinformatics microarray analysis was used to identify
significantly enriched Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) terms among the given list of genes that
were identified to be differentially expressed in response to
curcumin. Statistically overrepresented GO and KEGG categories with
a P-value of ≤0.05 were considered significant.
Gene expression in online
database
Gene expression and survival analysis in various
cancer tissues was detected using cBioPortal (http://www.cbioportal.org) and Gene Expression
Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) according to the protocol
(http://gepia.cancer-pku.cn/example.html).
Immunohistochemical (IHC)
staining
Human tissues were provided by the Department of
Pathology, Hangzhou Cancer Hospital (Hangzhou, China) under a
protocol approved by the Institutional Review Board of the Cancer
Research Institute, Hangzhou Cancer Hospital. The tissues were
collected within 1 h of tumor resection and fixed in formalin for
24 h at room temperature. Dehydration and embedding in paraffin was
performed following routine methods. Paraffin sections were cut at
5-µm thickness, then deparaffinized and rehydrated. Once endogenous
peroxidase activity was blocked with 3% H2O2
for 10 min at room temperature, the sections were treated with
citrate buffer (pH=6.0) in a microwave oven for 10 min for antigen
retrieval. Following a pre-incubation with blocking buffer (Perkin
Elmer, Waltham, MA, USA) for 10 min, the sections were incubated
with primary antibodies in a humidified chamber for 1 h at room
temperature. The primary antibody against poly [ADP-ribose]
polymerase (PARP)4 (1:20; cat. no. ab24110; Abcam, Cambridge, MA,
USA) was diluted with blocking buffer. Once the sections were
washed with phosphate buffered saline twice for 5 min, the
antigenic binding sites were visualized using the GTVision II
Detection System (Gene Tech Co., Ltd., Shanghai, China) according
to the manufacturer's instructions. Microscope images were assessed
using Image-Pro plus 6.0 software (Media Cybernetics, Inc.,
Rockville, MD, USA), and the mean integrated optical density (IOD)
of each image was collected. Mean IOD was referred to the average
level of positive tissues.
Statistical analysis
Statistical comparisons were made with one-way
analysis of variance with Bonferroni's multiple comparison post hoc
tests. Data were collected into a spreadsheet program and then
imported into statistical software packages (GraphPad Prism version
7.1 for Windows; GraphPad Software, Inc., La Jolla, CA, USA).
Survival curves were plotted using the Kaplan-Meier method. Curves
were analyzed using the log-rank method and hazard ratios were
reported where applicable. P<0.05 was considered to indicate a
statistically significant difference.
Results
Intratumoral cancer stemness of EAC
and ESCC at single-cell level
Previously, we described the heterogeneity of cancer
tissues in 2 patients with EAC and 3 with ESCC through separating
and characterizing the expression profiles of different cell types
via single-cell transcriptome sequencing (SRP119465) (24). All the single cells were picked
arbitrarily. In the present study, it was investigated whether the
characterized single cancer cells of EAC and ESCC (150 EAC and 124
ESCC single cells) contain more malignant cancer cells representing
cancer stem cells. Thus, all the single cancer cells were evaluated
in terms of their expression of the cancer stemness-associated
signaling pathways. A small proportion of cancer cells aberrantly
overexpressing cancer stemness genes were identified. To quantify
the heterogeneity of stemness, cancer stemness analysis was used,
which defines a stemness index (SI) by averaging the expression
values of 35 known stem cell markers according to previous
publications (Table SI) (18). Overall, the distribution of SI
values was continuous in EAC and ESCC (Fig. 1). As the cancer tissues consist of
single cancer cells, the distribution of SI values may reflect
their potential resistance to cancer therapy. Compared with patient
EA02, the single cancer cells from patient EA01 demonstrated
significantly higher SI values (P<0.01), which was consistent
with the poor prognosis outcome of patient EA01 following clinical
radiotherapy (data not shown). By contrast, the single cells of
these 3 ESCC patients demonstrated uniform distribution with regard
to the distribution of cancer stemness, and in bulk level, they
exhibited moderate levels of cancer stemness.
Cell cycle signaling is associated
with high cancer stemness of EAC
Due to the deep sequencing data of single-cell
transcriptome (5–10 M reads per single cell), the detailed features
of single cancer cells with high SI values were investigated. The
SIhigh single cancer cells (top 5) were compared with
SIlow single cancer cells (top 5) in each patient with
EAC (EA01 and EA02), and the respective differentially expressed
genes (DEGs) were identified. Venny analyses (http://bioinfogp.cnb.csic.es/tools/venny/) of these
DEGs in EA01 and EA02 indicated that 131 genes were upregulated,
among which 16 genes were significantly upregulated (P<0.01;
Fig. 2A), including the previously
reported cancer stemness-associated genes aldehyde dehydrogenase 1
(ALDH1A1) (25–29), aurora kinase A (AURKA)
(30–33), Replication Timing Regulatory Factor
1 (RIF1) (34–36), stathmin 1 (STMN1) (37,38)
and Tripartite Motif Containing 59 (TRIM59) (39,40)
(Table SII). Following this, KEGG
and GO analyses was applied to these 131 genes, which indicated
that genes associated with the ‘cell cycle’ and ‘metabolic
pathways’ were significantly enriched (P=0.0025 and P=0.039,
respectively; Fig. 2B). Taken
together, unlike previous bulk measurements (41), the methods used in the present study
allowed the identification of significant genes and signaling
pathways that were associated with the cancer stemness of EAC at
the single-cell level.
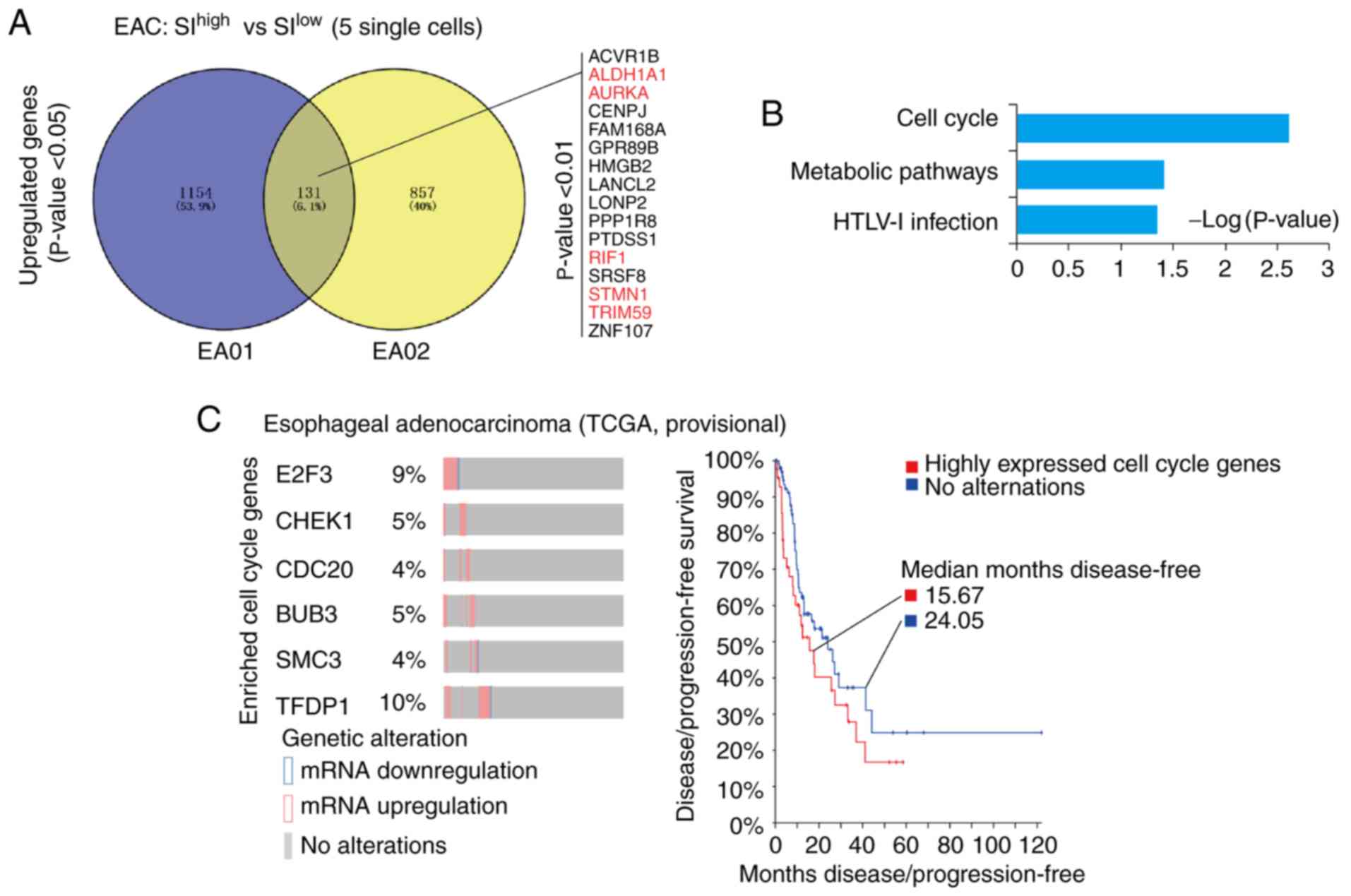 | Figure 2.Upregulated genes in the cell cycle
are responsible for the high cancer stemness of EAC. (A) Venny
analysis of upregulated genes in the cancer stemness of EA01 and
EA02. A total of 131 genes were upregulated in the high-stemness
single cancer cells of EA01 and EA02. Among these, 16 genes were
identified with P<0.01. Previously reported stemness-associated
genes were marked in red; references were listed in Table SI. (B) Database for Annotation,
Visualization and Integrated Discovery-Kyoto Encyclopedia of Genes
and Genomes analysis of upregulated genes in EAC. The signaling
pathway of cell cycle and metabolic pathways were significantly
enriched in the cancer stemness of EAC (P<0.05). (C) Dynamic
expression of EAC cancer stemness-associated cell cycle genes in
TCGA data as assessed with cBioPortal. E2F3, CHEK1, CDC20, BUB3,
SMC3 and TFDP1 (EAC cancer stemness-associated cell
cycle genes) were significantly upregulated in some patients with
EAC (percentage is given), which correlated with poor prognosis in
patients (median disease-free: 15.67 vs. 24.05 months). EAC,
esophageal adenocarcinoma; SI, stemness index; TCGA, The Cancer
Genome Atlas; E2F3, E2F transcription factor 3; CHEK1, checkpoint
kinase 1; CDC20, cell division cycle 20; SMC3, structural
maintenance of chromosomes protein 3; TRDP1, transcription factor
Dp-1; HTLV-1, human T-lymphotropic virus type 1; ACVR1B, activin A
receptor type 1B; ALDH1A1, aldehyde dehydrogenase 1 family member
A1; AURKA, aurora kinase A; RIF1, replication timing regulatory
factor 1; STMN1, stathmin 1; TRIM59, tripartite motif containing
59; CENPJ, centromere protein J; FAM168A, family with sequence
similarity 168 member A; GPR89B, G protein-coupled receptor 89B;
HMGB2, high mobility group box 2; LANCL2, lanthionine synthetase
C-like 2; LONP2, lon peptidase 2, peroxisomal; PPP1R8, protein
phosphatase 1 regulatory subunit 8; PTDSS1, phosphatidylserine
synthase 1; SRSF8, serine and arginine rich splicing factor 8;
ZNF107, zinc finger protein 107. |
Cancer stem cells are believed to constitute a
principal cellular source for tumor progression and therapeutic
drug resistance (42). Therefore,
the small proportion of ‘stem cell-like’ carcinoma cells likely
represents the true cancer stem cells of esophageal cancer. In
cBioPortal of TCGA-EAC (http://www.cbioportal.org), it was indicated that the
cell cycle-associated genes identified in the present single-cell
data were significantly upregulated in some patients with EAC
(TCGA, EAC provisional). In particular, 6 genes [E2F Transcription
Factor 3 (E2F3), checkpoint kinase 1 (CHEK1), Cell
Division Cycle 20 (CDC20), BUB3, Structural
Maintenance of Chromosomes protein 3 (SMC3) and
Transcription Factor Dp-1 (TFDP1)] were upregulated in 4–10%
of patients with EAC, as indicated in Fig. 2C. Notably, the patients with EAC who
expressed a high level of these cell cycle-associated genes
exhibited poorer prognosis with regard to disease/progression-free
survival compared with patients without altered expression of these
genes (median months disease-free: 15.67 vs. 24.06; Fig. 2C).
DNA replication and DNA damage
repair-associated genes are enriched in high-cancer stemness cells
in ESCC
Cancer stemness analysis was performed using SI
values on single cancer cells in the single-cell data of ESCC. Due
to the few single cancer cells in patient ESC01, this patient was
excluded in further analysis. The DEGs of SIhigh single
cancer cells (top 5) and SIlow single cancer cells (top
5) in patient ESC02 and ESC03 were identified. Venny analyses of
these DEGs indicated that 338 genes were upregulated, among which
35 genes were significantly upregulated (P<0.01; Fig. 3A), including previously reported
cancer stemness-associated genes cell division cycle-associated
protein 7 (CDCA7) (43),
Kinesin Family Member 11 (KIF11) (44–46),
Wnt ligand secretion mediator (WLS) (47,48)
and zinc finger MYM-type containing 2 (ZMYM2) (49,50)
(Table SIII). These 338 genes were
further analyzed with KEGG and GO analyses, which indicated that
they were significantly enriched in ‘DNA replication’
(P=1.25×10−04), ‘Fanconi anemia pathway’ (P=0.006), ‘RNA
transport’ (P=0.0097) and ‘nucleotide excision repair’ (P=0.027;
Fig. 3B).
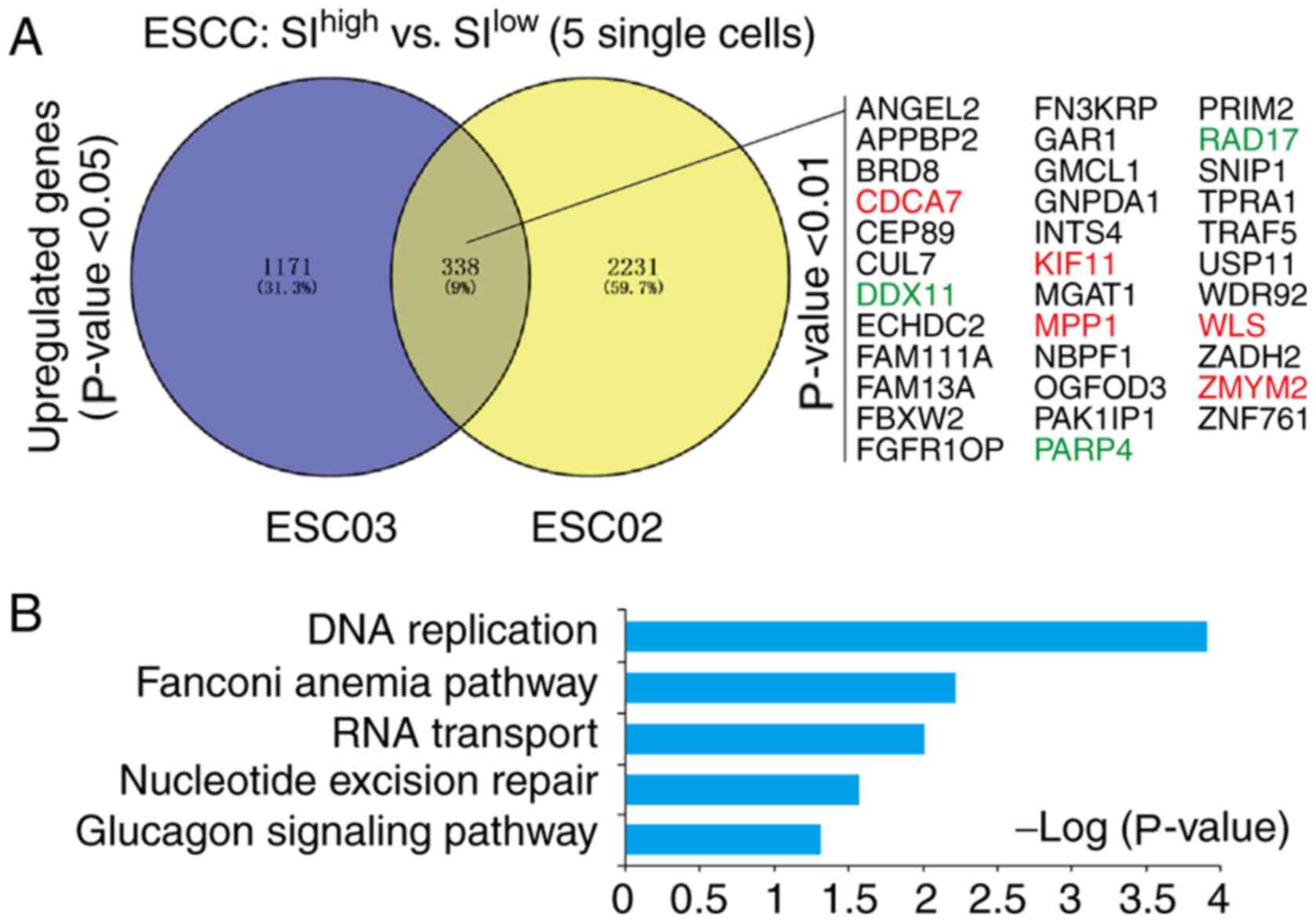 | Figure 3.Signature genes and signaling
pathways responsible for the high cancer stemness of ESCC. (A)
Venny analysis of upregulated genes in the cancer stemness of ESC02
and ESC03. A total of 338 upregulated genes were identified. Among
these, a total of 35 significantly upregulated genes were listed
(P<0.01). Previously reported stemness-associated genes were
marked in red; the references were given in Table SII. (B) Database for Annotation,
Visualization and Integrated Discovery-Kyoto Encyclopedia of Genes
and Genomes analysis of upregulated genes in ESCC. The signaling
pathways of DNA replication, Fanconi anemia pathway, RNA transport
and nucleotide excision repair were significantly enriched in the
cancer stemness of ESCC (P<0.05). ESCC, esophageal squamous cell
carcinoma; SI, stemness index; ANGEL2, angel homolog 2; APPBP2,
amyloid β precursor protein binding protein 2; BRD8, bromodomain
containing 8; CDCA7, cell division cycle associated 7; CEP89,
centrosomal protein 89; CUL7, cullin 7; DDX11, DEAD/H-Box Helicase
11; ECHDC2, enoyl-CoA hydratase domain containing 2; FAM111A,
family with sequence similarity 111 member A; FAM13A, family with
sequence similarity 13 member A; FBXW2, F-Box and WD repeat domain
containing 2; FGFR10P, FGFR1 oncogene partner; FN3KRP, fructosamine
3 kinase related protein; GAR1, GAR1 ribonucleoprotein; GMCL1, germ
cell-less, spermatogenesis associated 1; GNPDA1,
glucosamine-6-phosphate deaminase 1; INTS4, integrator complex
subunit 4; KIF11, kinesin family member 11; MGAt1, mannosyl
(alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase;
MPP1, membrane palmitoylated protein 1; NBPF1, NBPF member 1;
OGFOD3, 2-oxoglutarate and iron dependent oxygenase domain
containing 3; PAK1IP1, p21-activated protein kinase-interacting
protein 1; PARP4, poly(ADP-ribose) polymerase family member 4;
PRIM2, DNA primase subunit 2; RAD17, RAD17 checkpoint clamp loader
component; SNIP1, Smad nuclear interacting protein 1; TPRA1,
transmembrane protein adipocyte associated 1; TRAF5, TNF receptor
associated factor 5; USP11, ubiquitin specific peptidase 11; WDR92,
WD repeat domain 92; WLS, Wntless Wnt ligand secretion mediator;
ZADH2, zinc binding alcohol dehydrogenase domain containing 2;
ZMYM2, zinc finger MYM-type containing 2; ZNF761, zinc finger
protein 761. |
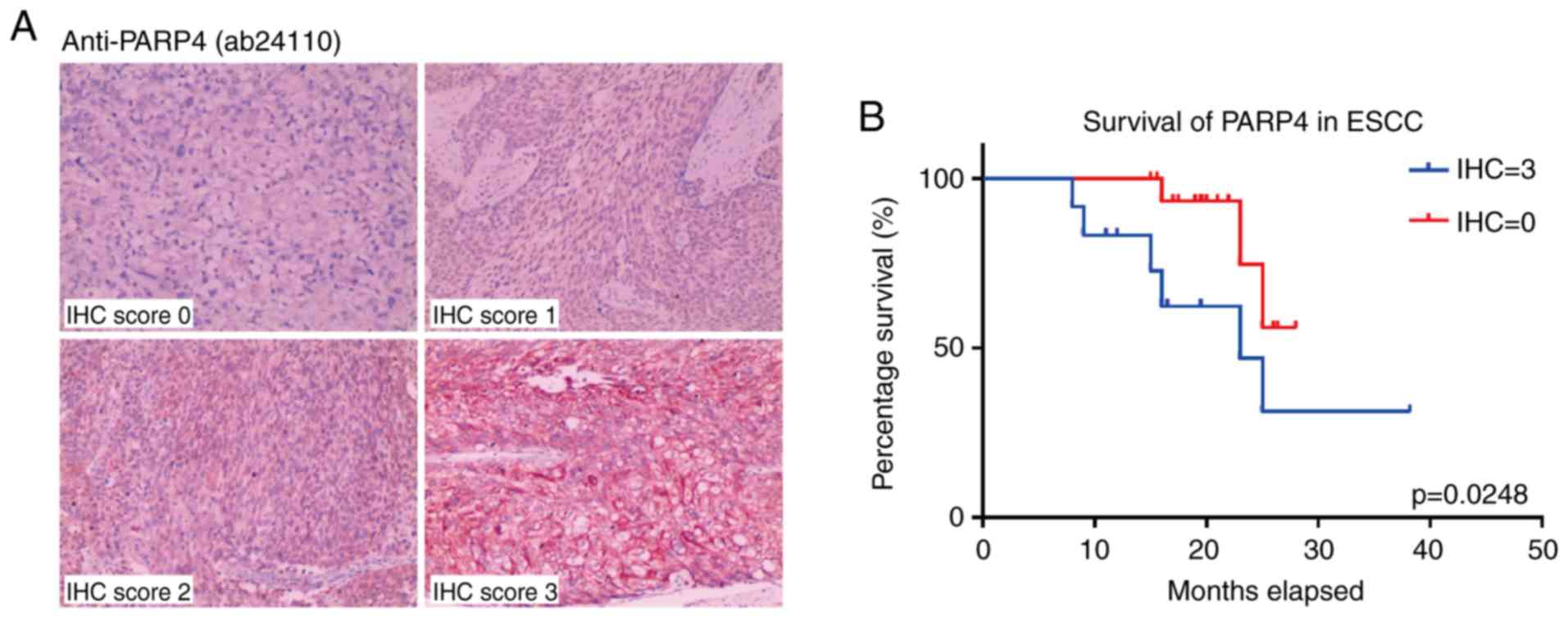
Identification of PARP4 as a potential
marker of high cancer stemness for ESCC
Among the cancer stemness features of ESCC, it was
indicated that several DNA damage repair-associated genes
(DDX11, RAD17 and PARP4) were significantly
upregulated within the more stringently upregulated genes (35
genes) (Fig. 3A; Fig. S1A). Furthermore, in the GEPIA
database (http://gepia.cancer-pku.cn), it was
identified that PARP4 was upregulated in various human
cancers (Fig. S1B), and highly
expressed PARP4 was correlated with poor prognosis in
cervical squamous cell carcinoma and lung squamous cell carcinoma,
but not EAC (Fig. S2).
In order to validate the correlation between the
expression level of PARP4 and the prognosis of these patients with
ESCC, an ESCC cohort with 121 patients was assessed in the present
study (Table SIV).
Immunohistochemical scoring for the cancer tissues from these
patients indicated that high PARP4 expression was associated with
poorer survival (Fig. 4A),
particularly in patients with higher PARP4 scores (12 patients, IHC
score=3; P=0.0248; Fig. 4B).
Notably, the majority of patients with ESCC exhibited moderate
expression of PARP4 (55 patients, IHC score=1; 36 patients, IHC
score=2; Fig. S3A), and their
overall survival was longer than that of patients with high PARP4
(IHC score=3) but shorter than that of patients with low PARP4 (IHC
score=0; Fig. S3B).
Discussion
Current cancer treatments ultimately fail owing to
metastasis and relapse, which is primarily due to the heterogeneity
of cancer tissues (51). Novel
methodologies to evaluate and design strategies to reduce the
therapeutic resistance in cancer tissues are urgently required for
advancing cancer treatment in the clinic (3). Although our increased understanding of
the genomic, transcriptomic and proteomic complexity of tumor
heterogeneity further highlights the extreme heterogeneity of
cancer cells, it has been indicated that the heterogeneity of
intratumoral cancer cells was considerably more complicated than
originally conjectured. Previous results have indicated that
targeting cancer stemness can be used as a strategy to develop the
next generation of cancer therapeutics to suppress cancer relapse
and metastasis (8). To this end, in
the present study, single-cell transcriptome sequencing was
utilized to describe the detailed expression profiles of
intratumoral cancer stemness in EAC and ESCC at single-cell
level.
With the combined expression of potential cancer
stemness genes, the SI was generated for each of the single cells,
and the heterogeneity of cancer cells was characterized to separate
the potential cancer relapse-associated cancer stem cells (18). With the deep sequencing of
single-cell transcriptome, detailed features of these potential
cancer stem cells could be investigated. This approach can identify
a large number of potential cancer stem cell-associated genes and
signaling pathways, which may be useful therapeutic targets for
future clinical trials.
In EAC, a number of upregulated genes in
high-stemness cancer cells were indicated, including previously
reported cancer stemness-associated genes ALDH1A1 (25–29),
AURKA (30–33), RIF1 (34–36),
STMN1 (37,38) and TRIM59 (39,40).
These genes were significantly enriched in the ‘cell cycle
signaling pathway’. In particular, AURKA and CDC20
genes were significantly enriched in the ‘cell cycle signaling
pathway’. This finding holds considerable promise, as in the clinic
numerous compounds are already utilized that can target components
of cell cycle signaling (52),
including cyclin-dependent kinase (CDK)4/CDK6 (palbociclib,
ribociclib and abemaciclib), aurora kinases (AT9283 and MLN8237),
Wee1 kinase (MK-1775), KSP (ispinesib) and tubulin (taxanes and
vinca alkaloids). Based on the characterization of intratumoral
cancer stemness in EAC provided in the present study, it was
considered that combining such cell cycle inhibitors with current
standard treatments might be worthwhile to reduce the potential
cancer stemness-associated therapeutic resistance in EAC.
In ESCC, with the exception of the previously
reported cancer stemness-associated genes CDCA7 (43), KIF11 (44–46),
WLS (47,48) and ZMYM2 (49,50),
the present study identified the cancer stemness-associated
signaling pathways of DNA replication and DNA damage repair;
therefore, these pathways may represent the features of cancer
stemness in single ESCC cancer cells. Furthermore, the DNA damage
repair-associated factor PARP4 was identified as a novel potential
cancer stemness marker in ESCC, the expression of which correlated
with patient prognosis. PARP4 may thus serve as a novel therapeutic
target for the inhibition of cancer stemness in ESCC. Notably,
several compounds are also currently utilized in the clinic that
can target PARP-1 (PARP-1 inhibitors) (53). Considering that these have been used
to treat various types of cancer (54–56),
with excellent outcomes reported in non-small cell lung cancer
(57), such PARP-1 inhibitors may
serve as effective supplementary therapy to conventional anticancer
agents for ESCC (58,59).
In conclusion, through transcriptomic analyses of
single cancer cells, the present research demonstrated the features
of intratumoral cancer stemness in EAC and ESCC. The diversity of
cancer stemness within and between different types of cancer
implied that the treatment of EAC and ESCC should be approached
differently in future clinical therapy. Furthermore, the detailed
profiles of intratumoral cancer stemness in EAC and ESCC provided
several indications for applying treatment strategies against
specific cancer stemness features in future cancer therapy clinical
trials.
Supplementary Material
Supporting Data
Acknowledgements
The authors thank the Novogene Company for providing
the sequencing support. The sequenced data were analyzed by Xingbao
Health Co. Ltd.
Funding
The present work was supported by grants from the
Hangzhou Health Science and Technology Project (grant no. 2017A28),
Zhejiang Health Science and Technology Project (grant no.
20180533B64), National Natural Science Foundation of China (grant
nos. 81672994 and 31860306), Zhejiang Provincial Foundation for
Natural Sciences (grant nos. LY14H160005 and LZ15H220001) and the
Zhejiang Provincial Medical Scientific Research Foundation of China
(grant nos. 2015KYB325 and 2015PYA009).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The sequencing RAW data
analyzed during the current study are available in the NCBI SRA
database (https://www.ncbi.nlm.nih.gov/sra), with accession no.
SRP119465.
Authors' contributions
HW conceived the study. YL, HW and QH performed the
experiments. RZ and ZL performed the IHC score analysis. JY and HW
analyzed the data. HW, JY and ZL prepared the manuscript. MJ and SW
reviewed the results, participated in the discussion of the
manuscript and were also involved in the conception of the study.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Human tissues were provided by the Department of
Pathology, Hangzhou Cancer Hospital following approval by the
Institutional Review Board of the Cancer Research Institute,
Hangzhou Cancer Hospital. Written, informed consent was obtained
from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DEG
|
differentially expressed gene
|
|
EAC
|
esophageal adenocarcinoma
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
FPKM
|
fragments per kilobase of transcript
per million mapped reads
|
|
GO
|
Gene Ontology
|
|
IHC
|
immunohistochemistry
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ScRNA-Seq
|
single-cell RNA sequencing
|
|
SI
|
stemness index
|
|
SRA
|
Sequence Read Archive
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Wong DJ, Segal E and Chang HY: Stemness,
cancer and cancer stem cells. Cell Cycle. 7:3622–3624. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monteiro J and Fodde R: Cancer stemness
and metastasis: Therapeutic consequences and perspectives. Eur J
Cancer. 46:1198–1203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khosrotehrani K and Roy E: Tell me about
your stemness. I'll give your cancer risk! Cell Death Differ.
24:6–7. 2017. View Article : Google Scholar
|
|
4
|
Ghisolfi L, Keates AC, Hu X, Lee DK and Li
CJ: Ionizing radiation induces stemness in cancer cells. PLoS One.
7:e436282012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu X, Ghisolfi L, Keates AC, Zhang J,
Xiang S and Lee DK: Induction of cancer cell stemness by
chemotherapy. Cell Cycle. 11:2691–2698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Z, Duan Q, Zhao H, Liu T, Wu H, Shen
Q, Wang C and Yin T: Gemcitabine treatment promotes pancreatic
cancer stemness through the Nox/ROS/NF-kappaB/STAT3 signaling
cascade. Cancer Lett. 382:53–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu L, Yang L, Yan W, Zhai J, Pizzo DP,
Chu P, Chin AR, Shen M, Dong C, Ruan X, et al: Chemotherapy induces
breast cancer stemness in association with dysregulated
monocytosis. Clin Cancer Res. 24:2370–2382. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Rogoff HA, Keates S, Gao Y,
Murikipudi S, Mikule K, Leggett D, Li W, Pardee AB and Li CJ:
Suppression of cancer relapse and metastasis by inhibiting cancer
stemness. Proc Natl Acad Sci USA. 112:1839–1844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang ZM, Chen Y and Luo ML: Targeting
stemness: Implications for precision medicine in breast cancer. Adv
Exp Med Biol. 1026:147–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fodde R and Brabletz T: Wnt/beta-catenin
signaling in cancer stemness and malignant behavior. Curr Opin Cell
Biol. 19:150–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang X, Lou Y, Wang H, Zheng X, Dong Q,
Sun J and Han B: Wnt signaling regulates the stemness of lung
cancer stem cells and its inhibitors exert anticancer effect on
lung cancer SPC-A1 cells. Med Oncol. 32:952015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin L, Vu T, Yuan G and Datta PK: STRAP
promotes stemness of human colorectal cancer via epigenetic
regulation of the NOTCH pathway. Cancer Res. 77:5464–5478. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu T, Li Z, Yang Y, Ji W, Yu Y, Niu X,
Zeng Q, Xia W and Lu S: The Hippo/YAP1 pathway interacts with FGFR1
signaling to maintain stemness in lung cancer. Cancer Lett.
423:36–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yun Z and Lin Q: Hypoxia and regulation of
cancer cell stemness. Adv Exp Med Biol. 772:41–53. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Li F, Huang Q, Zhang Z, Zhou L,
Deng Y, Zhou M, Fleenor DE, Wang H, Kastan MB and Li CY:
Self-inflicted DNA double-strand breaks sustain tumorigenicity and
stemness of cancer cells. Cell Res. 27:764–783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carné Trécesson S, Souazé F, Basseville A,
Bernard AC, Pécot J, Lopez J, Bessou M, Sarosiek KA, Letai A,
Barillé-Nion S, et al: BCL-XL directly modulates RAS
signalling to favour cancer cell stemness. Nat Commun. 8:11232017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sottoriva A, Spiteri I, Piccirillo SG,
Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C and Tavaré
S: Intratumor heterogeneity in human glioblastoma reflects cancer
evolutionary dynamics. Proc Natl Acad Sci USA. 110:4009–4014. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Courtois ET, Sengupta D, Tan Y, Chen
KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al: Reference
component analysis of single-cell transcriptomes elucidates
cellular heterogeneity in human colorectal tumors. Nat Genet.
49:708–718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morgan M, Anders S, Lawrence M, Aboyoun P,
Pages H and Gentleman R: ShortRead: A bioconductor package for
input, quality assessment and exploration of high-throughput
sequence data. Bioinformatics. 25:2607–2608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trapnell C, Cacchiarelli D, Grimsby J,
Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS and
Rinn JL: The dynamics and regulators of cell fate decisions are
revealed by pseudotemporal ordering of single cells. Nat
Biotechnol. 32:381–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with tophat and cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dalerba P, Cho RW and Clarke MF: Cancer
stem cells: Models and concepts. Annu Rev Med. 58:267–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu H, Yu J, Li Y, Hou Q, Zhou R, Zhang N,
Jing Z, Jiang M, Li Z, Hua Y, et al: Single-cell RNA sequencing
reveals diverse intratumoral heterogeneities and gene signatures of
two types of esophageal cancers. Cancer Lett. 438:133–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kulsum S, Sudheendra HV, Pandian R,
Ravindra DR, Siddappa G, R N, Chevour P, Ramachandran B, Sagar M,
Jayaprakash A, et al: Cancer stem cell mediated acquired
chemoresistance in head and neck cancer can be abrogated by
aldehyde dehydrogenase 1 A1 inhibition. Mol Carcinog. 56:694–711.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu X, Wang L, Cui W, Yuan X, Lin L, Cao
Q, Wang N, Li Y, Guo W, Zhang X, et al: Targeting ALDH1A1 by
disulfiram/copper complex inhibits non-small cell lung cancer
recurrence driven by ALDH-positive cancer stem cells. Oncotarget.
7:58516–58530. 2016.PubMed/NCBI
|
|
27
|
Meng E, Mitra A, Tripathi K, Finan MA,
Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA,
Palle K, et al: ALDH1A1 maintains ovarian cancer stem cell-like
properties by altered regulation of cell cycle checkpoint and DNA
repair network signaling. PLoS One. 9:e1071422014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thomas ML, de Antueno R, Coyle KM, Sultan
M, Cruickshank BM, Giacomantonio MA, Giacomantonio CA, Duncan R and
Marcato P: Citral reduces breast tumor growth by inhibiting the
cancer stem cell marker ALDH1A3. Mol Oncol. 10:1485–1496. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang L, Ren Y, Yu X, Qian F, Bian BS, Xiao
HL, Wang WG, Xu SL, Yang J, Cui W, et al: ALDH1A1 defines invasive
cancer stem-like cells and predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Mod Pathol. 27:775–783. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen C, Song G, Xiang J, Zhang H, Zhao S
and Zhan Y: AURKA promotes cancer metastasis by regulating
epithelial-mesenchymal transition and cancer stem cell properties
in hepatocellular carcinoma. Biochem Biophys Res Commun.
486:514–520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eterno V, Zambelli A, Villani L, Tuscano
A, Manera S, Spitaleri A, Pavesi L and Amato A: AurkA controls
self-renewal of breast cancer-initiating cells promoting wnt3a
stabilization through suppression of miR-128. Sci Rep. 6:284362016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang N, Wang C, Wang Z, Zona S, Lin SX,
Wang X, Yan M, Zheng FM, Li SS, Xu B, et al: FOXM1 recruits nuclear
aurora kinase A to participate in a positive feedback loop
essential for the self-renewal of breast cancer stem cells.
Oncogene. 36:3428–3440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng F, Yue C, Li G, He B, Cheng W, Wang
X, Yan M, Long Z, Qiu W, Yuan Z, et al: Nuclear AURKA acquires
kinase-independent transactivating function to enhance breast
cancer stem cell phenotype. Nat Commun. 7:101802016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
GursesCila HE, Acar M, Barut FB, Gunduz M,
Grenman R and Gunduz E: Investigation of the expression of RIF1
gene on head and neck, pancreatic and brain cancer and cancer stem
cells. Clin Invest Med. 39:275002016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li P, Ma X, Adams IR and Yuan P: A tight
control of Rif1 by Oct4 and Smad3 is critical for mouse
embryonic stem cell stability. Cell Death Dis. 6:e15882015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mei Y, Peng C, Liu YB, Wang J and Zhou HH:
Silencing RIF1 decreases cell growth, migration and increases
cisplatin sensitivity of human cervical cancer cells. Oncotarget.
8:107044–107051. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li M, Yang J, Zhou W, Ren Y, Wang X, Chen
H, Zhang J, Chen J, Sun Y, Cui L, et al: Activation of an
AKT/FOXM1/STMN1 pathway drives resistance to tyrosine kinase
inhibitors in lung cancer. Br J Cancer. 117:974–983. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Obayashi S, Horiguchi J, Higuchi T,
Katayama A, Handa T, Altan B, Bai T, Bao P, Bao H, Yokobori T, et
al: Stathmin1 expression is associated with aggressive phenotypes
and cancer stem cell marker expression in breast cancer patients.
Int J Oncol. 51:781–790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sang Y, Li Y, Song L, Alvarez AA, Zhang W,
Lv D, Tang J, Liu F, Chang Z, Hatakeyama S, et al: TRIM59 promotes
gliomagenesis by inhibiting TC45 dephosphorylation of STAT3. Cancer
Res. 78:1792–1804. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Malta TM, Sokolov A, Gentles AJ,
Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J,
Omberg L, Gevaert O, et al: Machine learning identifies stemness
features associated with oncogenic dedifferentiation. Cell.
173:338–354.e15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guiu J, Bergen DJ, De Pater E, Islam AB,
Ayllon V, Gama-Norton L, Ruiz-Herguido C, González J, López-Bigas
N, Menendez P, et al: Identification of Cdca7 as a novel Notch
transcriptional target involved in hematopoietic stem cell
emergence. J Exp Med. 211:2411–2423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Imai T, Oue N, Sentani K, Sakamoto N,
Uraoka N, Egi H, Hinoi T, Ohdan H, Yoshida K, Yasui W, et al: KIF11
is required for spheroid formation by oesophageal and colorectal
cancer cells. Anticancer Res. 37:47–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang M, Zhuang H, Xia R, Gan L, Wu Y, Ma
J, Sun Y and Zhuang Z: KIF11 is required for proliferation and
self-renewal of docetaxel resistant triple negative breast cancer
cells. Oncotarget. 8:92106–92118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Venere M, Horbinski C, Crish JF, Jin X,
Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al: The
mitotic kinesin KIF11 is a driver of invasion, proliferation, and
self- renewal in glioblastoma. Sci Transl Med. 7:304ra1432015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Augustin I, Dewi DL, Hundshammer J,
Erdmann G, Kerr G and Boutros M: Autocrine wnt regulates the
survival and genomic stability of embryonic stem cells. Sci Signal.
10:eaah68292017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Augustin I, Dewi DL, Hundshammer J, Rempel
E, Brunk F and Boutros M: Immune cell recruitment in teratomas is
impaired by increased wnt secretion. Stem Cell Res. 17:607–615.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ren M and Cowell JK: Constitutive notch
pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas
associated with atypical myeloproliferative disease. Blood.
117:6837–6847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ren M, Qin H, Wu Q, Savage NM, George TI
and Cowell JK: Development of ZMYM2-FGFR1 driven AML in human CD34+
cells in immunocompromised mice. Int J Cancer. 139:836–840. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rubben A and Araujo A: Cancer
heterogeneity: Converting a limitation into a source of biologic
information. J Transl Med. 15:1902017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mills CC, Kolb EA and Sampson VB: Recent
advances of cell-cycle inhibitor therapies for pediatric cancer.
Cancer Res. 77:6489–6498. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lord CJ and Ashworth A: PARP inhibitors:
Synthetic lethality in the clinic. Science. 355:1152–1158. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Weaver AN, Cooper TS, Rodriguez M,
Trummell HQ, Bonner JA, Rosenthal EL and Yang ES: DNA double strand
break repair defect and sensitivity to poly ADP-ribose polymerase
(PARP) inhibition in human papillomavirus 16-positive head and neck
squamous cell carcinoma. Oncotarget. 6:26995–277007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nasuno T, Mimaki S, Okamoto M, Esumi H and
Tsuchihara K: Effect of a poly(ADP-ribose) polymerase-1 inhibitor
against esophageal squamous cell carcinoma cell lines. Cancer Sci.
105:202–210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wurster S, Hennes F, Parplys AC, Seelbach
JI, Mansour WY, Zielinski A, Petersen C, Clauditz TS, Münscher A,
Friedl AA and Borgmann K: PARP1 inhibition radiosensitizes HNSCC
cells deficient in homologous recombination by disabling the DNA
replication fork elongation response. Oncotarget. 7:9732–9741.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ramalingam SS, Blais N, Mazieres J, Reck
M, Jones CM, Juhasz E, Urban L, Orlov S, Barlesi F, Kio E, et al:
Randomized, placebo-controlled, phase II study of veliparib in
combination with carboplatin and paclitaxel for advanced/metastatic
non-small cell lung cancer. Clin Cancer Res. 23:1937–1944. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhan L, Qin Q, Lu J, Liu J, Zhu H, Yang X,
Zhang C, Xu L, Liu Z, Cai J, et al: Novel poly (ADP-ribose)
polymerase inhibitor, AZD2281, enhances radiosensitivity of both
normoxic and hypoxic esophageal squamous cancer cells. Dis
Esophagus. 29:215–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yasukawa M, Fujihara H, Fujimori H,
Kawaguchi K, Yamada H, Nakayama R, Yamamoto N, Kishi Y, Hamada Y
and Masutani M: Synergetic effects of PARP inhibitor AZD2281 and
cisplatin in oral squamous cell carcinoma in vitro and in vivo. Int
J Mol Sci. 17:2722016. View Article : Google Scholar : PubMed/NCBI
|