Introduction
Globally, breast cancer remains the leading cause of
cancer-related deaths in females. An estimated 1.7 million new
cases were diagnosed and 521,900 breast cancer patients succumbed
to this disease in 2012 (1).
Similarly, in China, a diagnosis of breast cancer between the ages
of 30 and 59 years is more common than for any other type of cancer
except that of the thyroid (2).
Breast cancer is classified into five intrinsic subtypes through
detection of progesterone receptor (PR), estrogen receptor-alpha
(ERα) and human epidermal growth factor receptor 2 (HER2), namely
luminal A-like (ERα+ and/or PR+ and
HER2−), luminal B-like (ERα+ and/or
PR+ and HER2+), HER2 overexpression
(ERα−, PR− and HER2+) and
triple-negative breast cancer (TNBC) (HER2−,
ERα− and PR−) and normal-like tumors
(3). Luminal A-like tumors have
higher expression of ER-related genes and lower expression of
proliferative genes than luminal B-like cancers (4). Growing evidence indicates that TNBC is
a highly aggressive tumor with limited treatment strategies and has
poorer survival outcomes compared with other subtypes of breast
cancer. Although TNBC patients do benefit from chemotherapy, more
effective and less toxic treatments are required in order to reduce
the risk of disease progression and improve the prognosis.
Therefore, further studies are urgently needed to screen molecular
biomarkers to determine the therapeutic efficacy of treatments, and
improve the performance of diagnosis and prognosis of TNBC.
MicroRNAs (miRNAs) are small 20–22 nucleotide
non-coding RNAs that are known to regulate the expression of genes
participating in the control of cell proliferation, apoptosis,
development and stress response by binding to the 3′ or 5′
untranslated region of target transcripts (5). Furthermore, miRNAs are also involved
in tumorigenesis by acting as either tumor suppressors or
oncogenes. This suggests that miRNAs can potentially be biomarkers
for cancer diagnosis and prognosis. It has been reported that four
miRNAs, namely hsa-miR-125b, hsa-miR-16, hsa-miR-155 and
hsa-miR-374a are significantly associated with overall survival of
TNBC patients, of which three are correlated with better prognosis
and one with worse prognosis (6).
In addition, a 4-miRNA signature defined by the expression levels
of miR-155, miR-493, miR-30e and miR-27a has both diagnostic and
prognostic value for predicting outcomes of TNBC patients most
commonly treated with chemotherapy (7). Furthermore, another 4-miRNA signature
(miR-18b, miR-103, miR-107 and miR-652) may accurately predict
tumor relapse and overall survival of TNBC patients (8). Although several miRNA signatures have
been identified which could be used for TNBC diagnosis and survival
prediction, the novel prognosis strategy used by miRNA signatures
has not been applied in clinical studies. Hence, it is useful to
screen new miRNA biomarkers for TNBC overall survival.
In the present study, in order to identify
diagnostic and prognostic miRNAs in TNBC patients, we analyzed
large scale clinical data and miRNA sequencing data from The Cancer
Genome Atlas (TCGA) datasets. After identification of
differentially expressed miRNAs in TNBC, miRNAs with diagnostic and
prognostic value were identified and a 4-miRNA signature was
determined and then used to predict overall survival.
Materials and methods
Retrieval of breast cancer clinical
and miRNA expression data
A total of 1,098 anonymized patients were identified
in the TCGA database as having breast cancer. The clinical data
were retrieved from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/version 10.0,
release time: December 21, 2017; Species: human) on January 22,
2018. Of these 1,098 patients, 155 were diagnosed as having TNBC
based on their ER, PR and Her-2 status and defined using
immunohistochemistry (IHC). Three patients who were diagnosed as
TNBC did not have miRNA expression data recorded and thus were
excluded. The other 943 patients were diagnosed as
non-triple-negative breast cancer (nTNBC) subtypes (positive
expression of ER, PR or Her-2). Finally, a total of 152 TNBC and
943 nTNBC patients were included in the present study. The detailed
clinic data is presented in Table
I. The follow-up time was different for every patient and the
longest time was 3,472 days.
 | Table I.Clinical features of all 152 TNBC
patients included in the present study. |
Table I.
Clinical features of all 152 TNBC
patients included in the present study.
| Features | N (%) |
|---|
| Age (years) |
|
<60 | 100 (65.79) |
|
≥60 | 52 (34.21) |
| Sex |
|
Female | 152 (100) |
|
Male | 0 (0) |
| Vital status |
|
Alive | 134 (88.16) |
|
Dead | 18 (11.84) |
| Pathological
stage |
| I | 29 (19.08) |
| II | 97 (63.82) |
| II | 24 (15.79) |
| IV | 2 (0.01) |
| Tumor size |
| T1 | 41 (26.97) |
| T2 | 92 (60.53) |
| T3 | 15 (9.87) |
| T4 | 4 (2.63) |
| Lymph node |
| N0 | 102 (67.10) |
| NX | 50 (32.90) |
| Metastasis
status |
| M0 | 129 (84.87) |
| MX | 23 (15.13) |
In addition, 1,207 miRNA sequencing datasets from
frozen tumor samples by Illumina HiSeq 2000 platform (Illumina,
Inc., San Diego, CA, USA) were also downloaded from the TCGA data
portal. Of these, 1,103 were tumor samples and 104 were associated
with adjacent normal tissue. A total of 200 had unknown ER, PR or
Her-2 status and were thus excluded. In total, the miRNA sequencing
data from 152 TNBC and 751 nTNBC samples were used for dysregulated
miRNA exploration.
Identification of dysregulated miRNAs
from TNBC patients
To discover differentially expressed miRNAs from
TNBC patients, the edgeR software package of the Bioconductor
project (9) using the R programming
environment with default parameter settings was utilized. miRNAs
from TNBC samples were considered dysregulated in comparison with
adjacent normal samples and nTNBC tissue. Differences were assessed
with the Mann-Whitney U test and were significant if |log FC (fold
change)|>1 and P<0.05. Furthermore, gplots (version 3.0.1;
http://cran.r-project.org/src/contrib/Archive/gplots/)
and pheatmap (version 1.0.8; http://CRAN.R-project.org/package=pheatmap) packages
were used to study the expression levels and distribution of miRNAs
expressed differentially between the TNBC and the normal samples as
well as the nTNBC tissue with default parameter settings.
Identification of miRNAs with
diagnostic value
To identify differentially dysregulated miRNAs that
had capacity for diagnosing TNBC, receiver operating characteristic
(ROC) curves were plotted to compute the sensitivity and
specificity of each miRNA associated with TNBC diagnosis using the
pROC software package (version 1.10.0; http://cran.r-project.org/web/packages/pROC/) with
default parameter settings. Sensitivity was defined as the percent
of tumor cases with a diagnostic test exceeding a criterion and
specificity as the percent of non-tumor cases less than or equal to
that criterion with a diagnostic test. Dysregulated miRNAs with an
area under curve (AUC) >0.8 were selected and miRNAs common to
two comparison groups were considered as having diagnostic
value.
Survival analysis
The entire set including 104 normal and 152 TNBC
samples were randomly divided into a training set (51 normal and 77
TNBC samples) and a validation set (53 normal and 75 TNBC samples).
To evaluate the relationship between the miRNA expression levels
and the overall survival in tumor patients, univariate Cox
regression analysis was conducted with the aim of identifying
potential miRNAs related to TNBC prognosis. miRNAs that were
clearly associated with patient survival were included when
P<0.05 and then subjected to stepwise multivariate Cox
regression analysis to construct a TNBC prognostic signature based
on the following formula:
Riskscore=exp1xβ1+exp2xβ2+…+expnxβn
where n was the number of the prognostic miRNA, β
was the regression coefficient and exp was the expression level of
that miRNA. After risk score acquisition of each patient, patients
in the training set were divided into the high-risk and the
low-risk groups using the median score as the central cut-off
point. In addition, univariate and multivariate Cox proportional
hazards regression analyses were performed to examine the
relationship between the risk score of TNBC patients and other
clinical features including age (<60 or ≥60), pathologic stage
(I–II or III–IV), stage T, stage N and stage M. ROC curves were
plotted and AUC values were calculated. Patient survival was
evaluated using the Kaplan-Meier method and log-rank tests using R
package ‘survival’ (version: 2.42–3; http://CRAN.R-project.org/package=survival) with
default parameter settings. A 95% confidence interval (CI) and
hazard ratio (HR) were calculated to evaluate the prognostic
variables related to TNBC survival.
Target prediction of potential miRNA
signatures and functional annotation
To gain more insight into the role of prognostic
miRNA signatures in TNBC, miRNA-target prediction programs were
used to predict the target genes of the miRNAs, including miRDB
(http://www.miRdb.org/index.html),
miRTarBase (http://miRtarbase.mbc.nctu.edu.tw/php/index.php) and
TargetScan (http://www.targetscan.org/vert_71/). Genes that were
commonly identified by the three tools were considered as target
mRNAs of a prognostic miRNA signature. To reveal the potential
roles of the target genes, clusterProfileR package (version 3.6) in
the R environment (10) with
default parameter settings was employed to perform Gene Ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses. P<0.05 was set as the cut-off.
Results
Identification of differentially
expressed miRNAs in TNBC and nTNBC patients
To identify dysregulated miRNAs in TNBC patients in
comparison with adjacent normal tissue and miRNAs in TNBC compared
with nTNBC samples, the edgeR package was utilized, with 216
upregulated and 73 downregulated miRNAs based on the cut-off of
P<0.05 and |log FC|>1 in TNBC compared with adjacent normal
samples. When compared with nTNBC samples, there were 58
upregulated and 38 downregulated miRNAs in tumor samples from TNBC
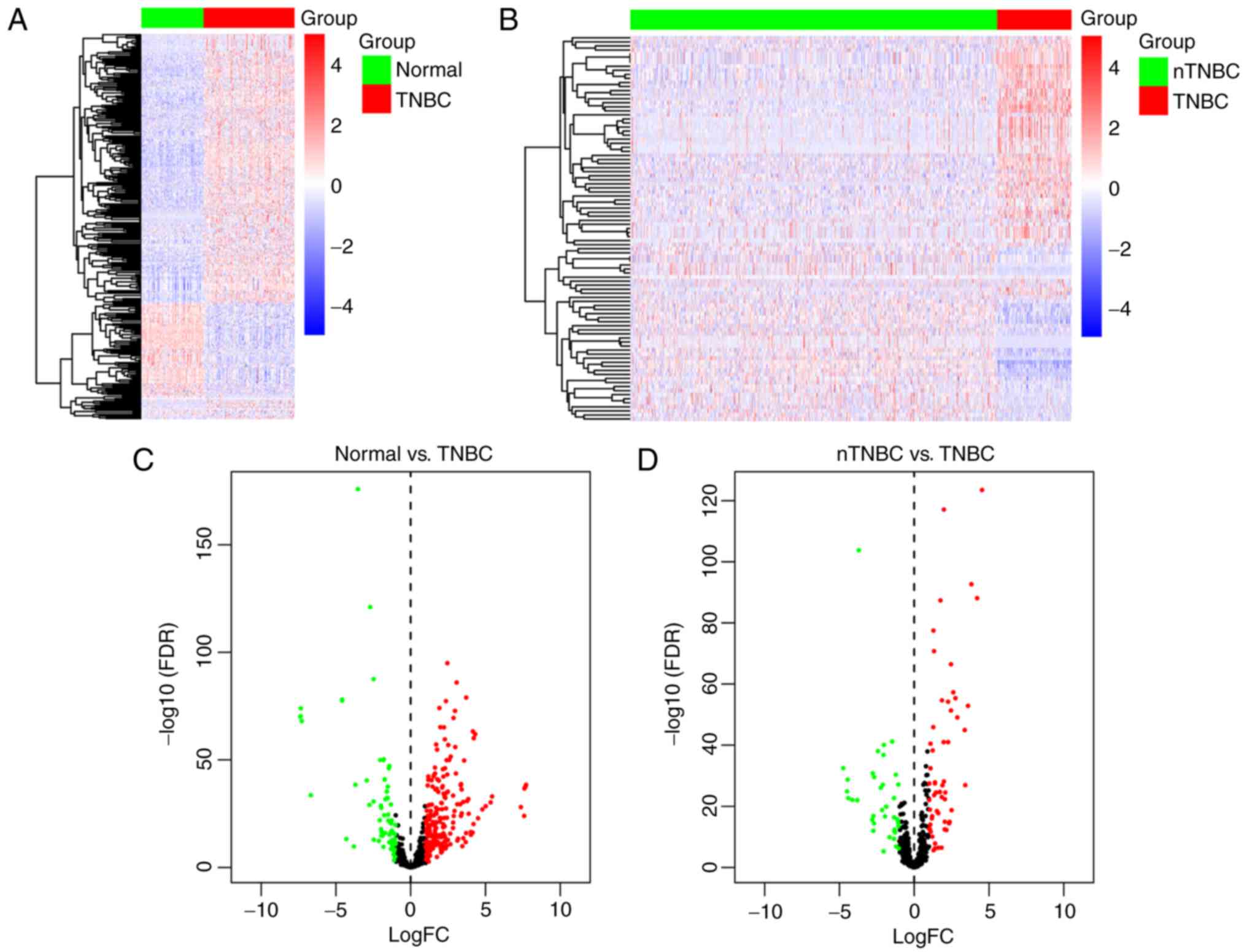
patients. Hierarchical clustering of differentially expressed
miRNAs presented clear separation in the expression profiles of
TNBC compared with normal samples (Fig.
1A) and nTNBC patients (Fig.
1B). A volcano plot was created to indicate the dysregulated
expressed miRNAs (Fig. 1C and
D).
Diagnostic value of dysregulated
miRNAs
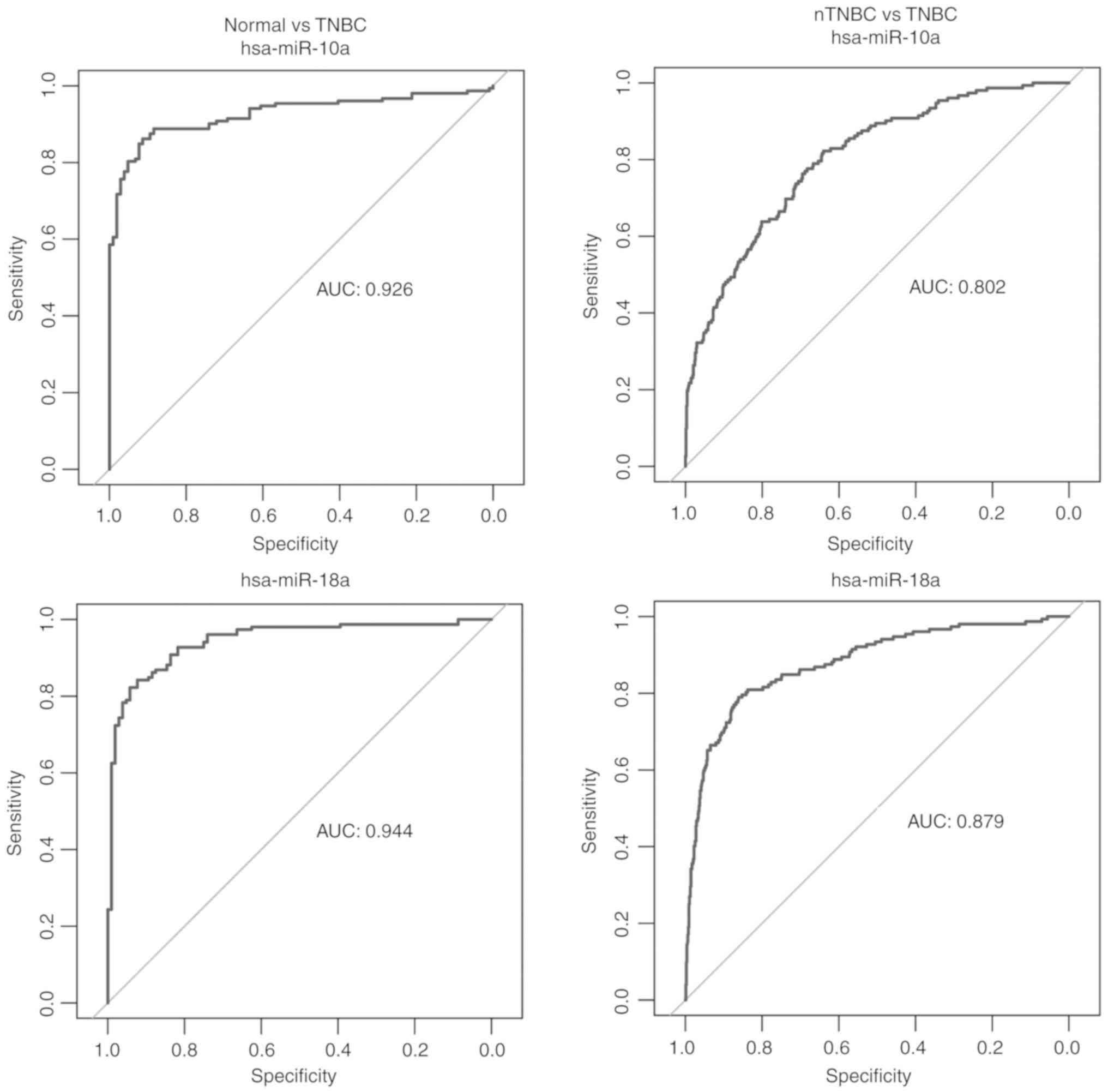
ROC analyses were performed to evaluate the possible
diagnostic capacity of each dysregulated miRNA. Differentially
expressed miRNAs with an AUC >0.8 were selected as miRNAs likely
to be useful as biomarkers in the diagnosis of TNBC. We obtained 27
miRNAs from the comparison between the TNBC and the adjacent normal
tissue and 6 miRNAs from the comparison between the TNBC and the
nTNBC samples with an AUC >0.8. There were 4 common miRNAs
between two comparisons, namely hsa-miR-10a, hsa-miR-18a,
hsa-miR-135b and hsa-miR-577 (Fig.
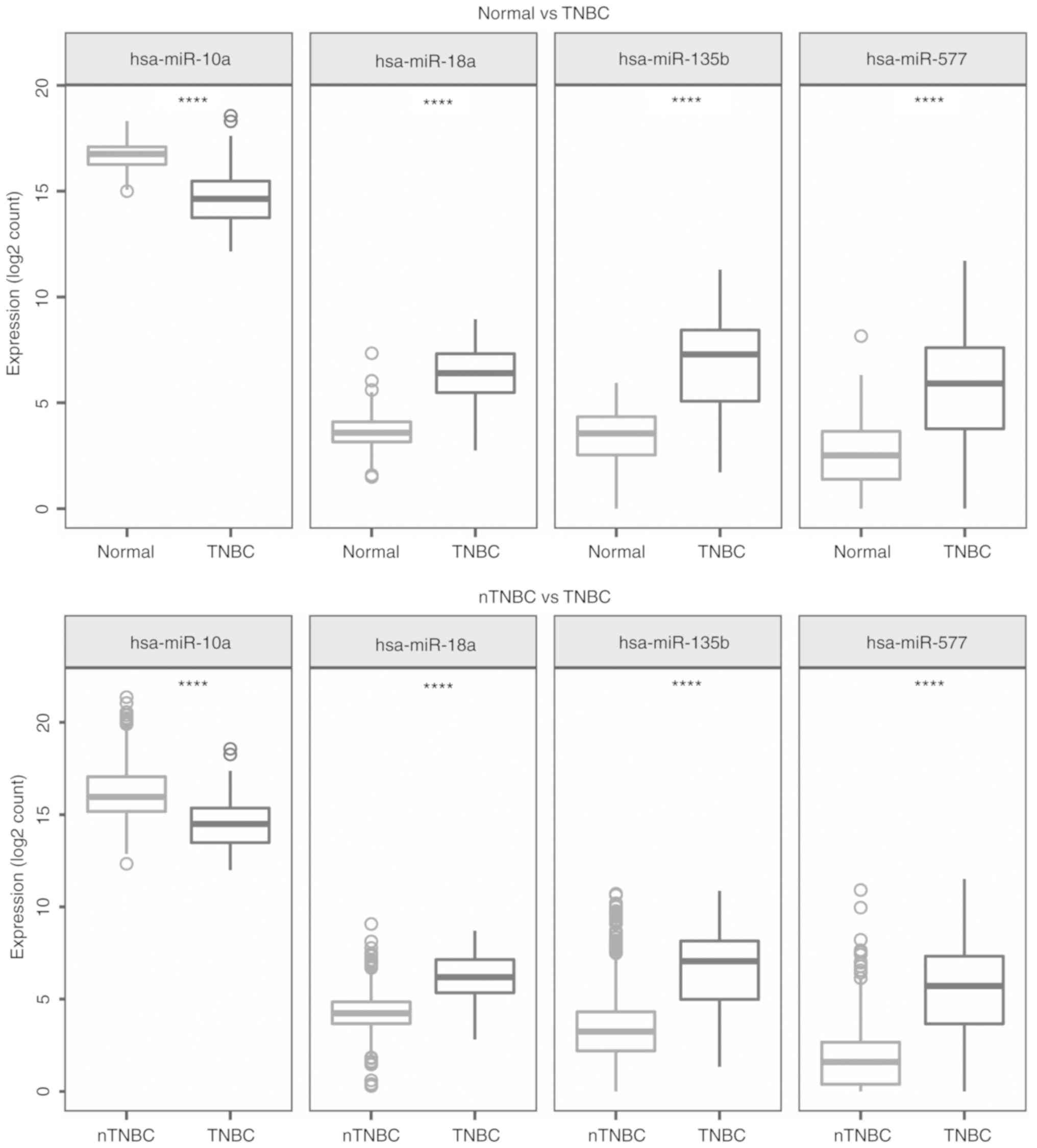
2). The expression levels of these 4 miRNAs in TNBC, adjacent
normal samples and nTNBC are presented in Fig. 3. Compared with the adjacent normal
breast tissues and nTNBC, the expression of hsa-miR-10a was lower
in the TNBC tumors however the expression of the other three miRNAs
was higher (|log2FC>1|, P<0.001). The results
indicated that these miRNAs were specifically upregulated and
downregulated in TNBC and could be used as diagnostic biomarkers
for the diagnosis of TNBC.
Identification of miRNAs associated
with TNBC prognosis
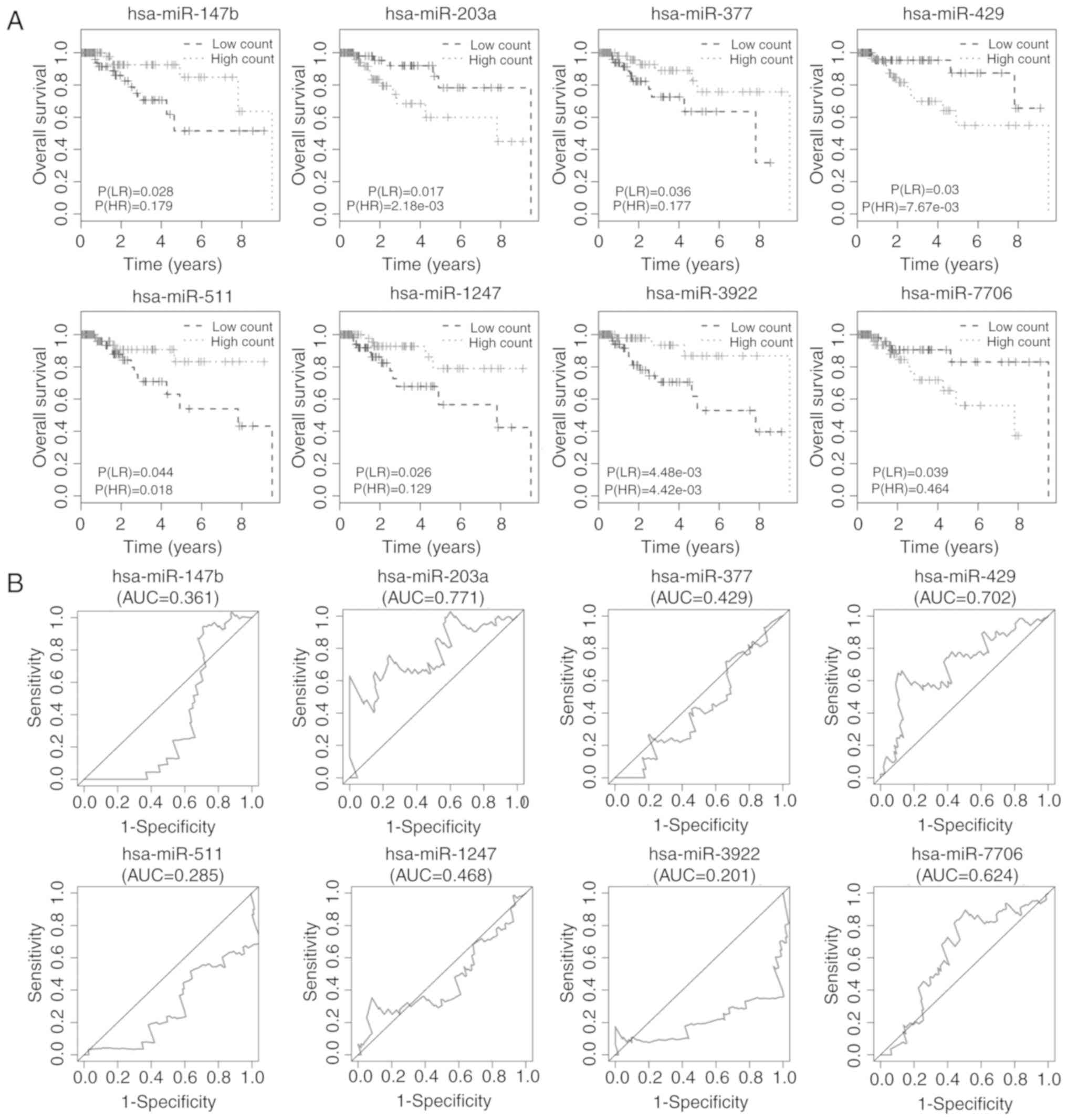
Kaplan-Meier plots and log-rank tests were used to
identify miRNAs related to overall survival of TNBC patients with a
cut-off log-rank (LR) P<0.05. A total of 8 miRNAs were
associated with overall survival, namely: hsa-miR-147b,
hsa-miR-203a, hsa-miR-377, hsa-miR-429, hsa-miR-511, hsa-miR-1247,
hsa-miR-3922 and hsa-miR-7706 (Fig.
4A). ROC analysis was then performed to assess the prognostic
capacity of the miRNAs to predict survival. The AUCs of the 8
miRNAs are displayed in Fig. 4B,
and indicated that 2 miRNAs, hsa-miR-203a and hsa-miR-429, may have
prognostic value. The AUCs for the 2 miRNAs predicting 5-year
survival were 0.771 and 0.702, respectively.
The predictive capacity of a 4-miRNA
signature in TNBC
A univariate Cox's proportional hazards regression
model was fitted to the entire set and 5 miRNAs (hsa-miR-148b,
hsa-miR-203a, hsa-miR-203b, hsa-miR-3922 and hsa-miR-429) were
identified as being associated with overall survival with P (HR)
<0.01. A group of miRNAs that could be defined as a signature,
namely hsa-miR-148b, hsa-miR-203a, hsa-miR-203b and hsa-miR-3922
was identified after stepwise multivariate Cox's regression model
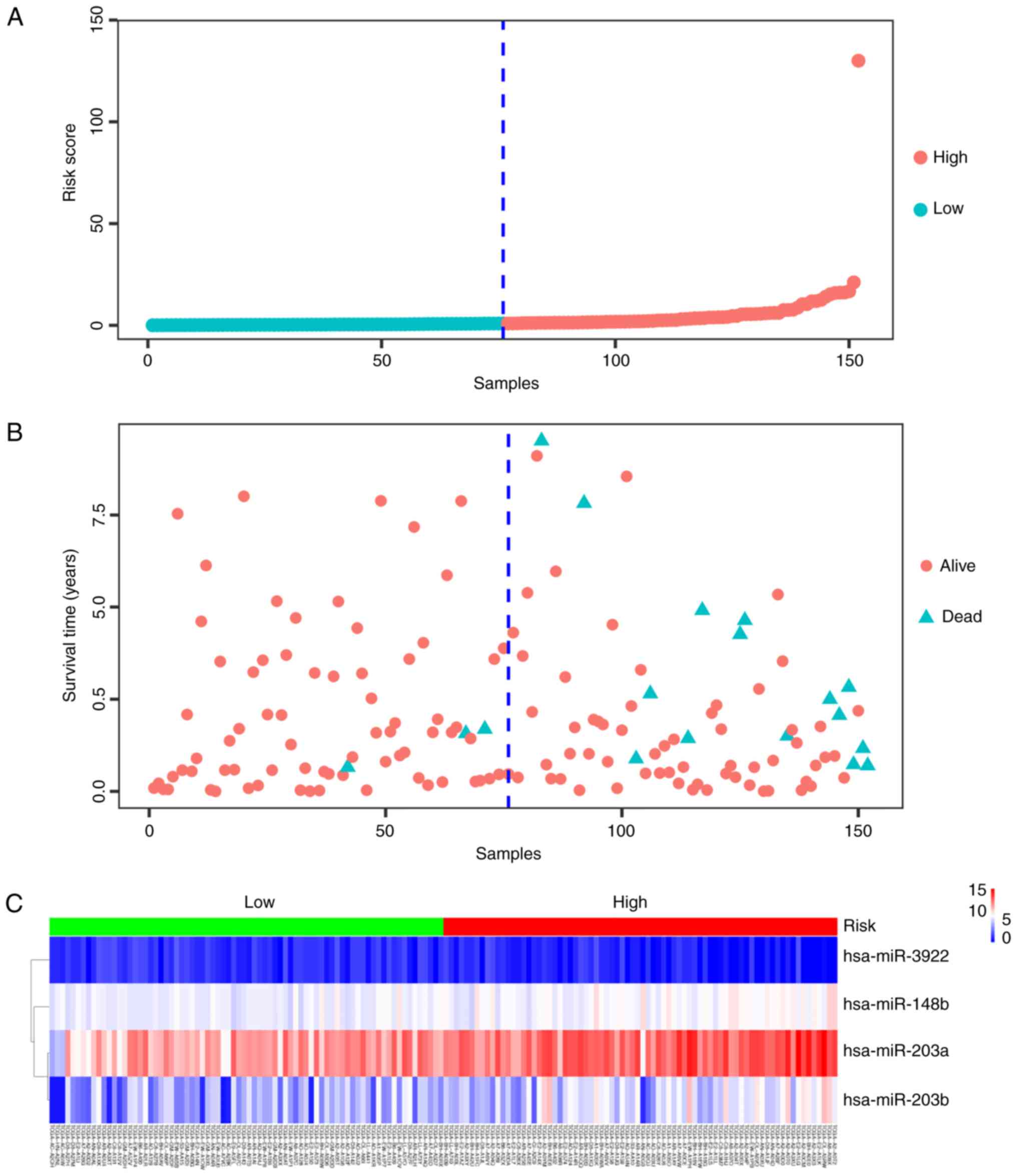
analysis by calculating the prognostic risk score. TNBC patients
were divided into a low- or a high-risk group depending on their
score relative to that of the median risk score (Fig. 5A). The mortality rate of the
high-risk group was 19.74% and that of the low-risk group was
3.95%, the difference being significant (P<0.05; Fig. 5B). The heatmap presented in Fig. 5C demonstrated that the 4-miRNA
signature was expressed differently in the low- and the high-risk
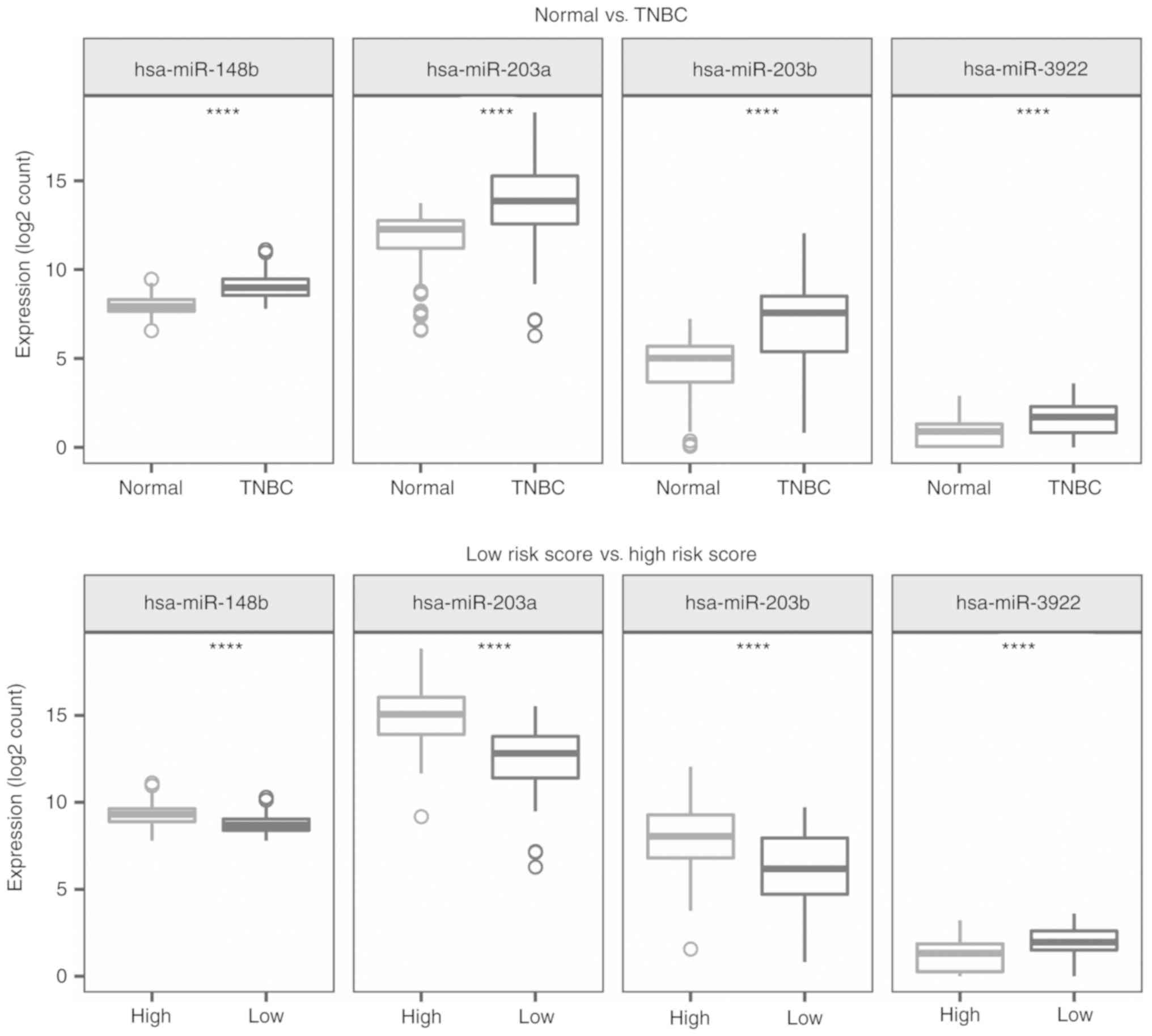
groups. The expression pattern of the 4-miRNA signature in TNBC and
normal samples, and the low- and the high-risk groups are presented
in Fig. 6. These 4 miRNAs were
significantly upregulated in TNBC patients compared with the normal
controls (P<0.001). Similarly, three miRNAs, hsa-miR-148b,
hsa-miR-203a and hsa-miR-203b, were expressed at a higher level in
the high-risk patients than in samples from the low-risk group
(P<0.001). However, the expression of hsa-miR-3922 showed the
opposite expression pattern (P<0.001).
The 4-miRNA signature is an
independent prognostic factor associated with overall survival
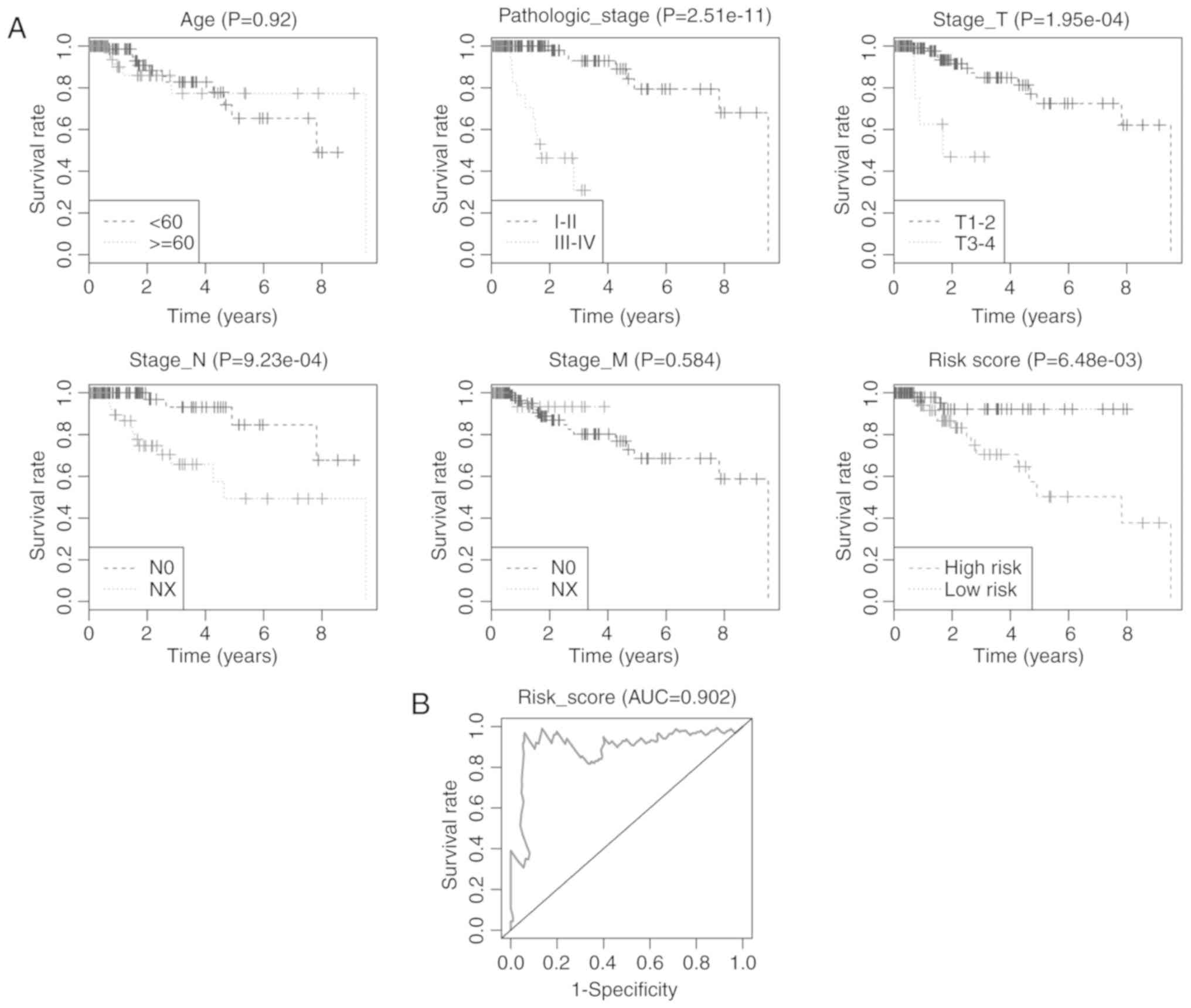
Finally, both univariate and multivariate Cox's
proportional hazards regression model analyses were performed to
evaluate the prognostic power of the 4-miRNA signature. The
univariate Cox's regression model demonstrated that pathological
stage, N stage, T stage and Risk score based on the 4-miRNA
signature were significantly correlated with overall survival of
TNBC patients (P<0.05), while the multivariate Cox's regression
model revealed that only the pathological stage and Risk score were
independent prognostic factors associated with overall survival
(P=0.002 and 0.021, respectively; Table II). Kaplan-Meier curves of the
clinical characteristics and risk score are displayed in Fig. 7A. The highest survival rate was
found within the low-risk group in comparison with the high-risk
group (P=0.0065). The AUC for the 4-miRNA signature predicting
5-year survival of TNBC patients was 0.902 (Fig. 7B). Our results suggest that the
4-miRNA signature may have prognostic value for predicting the
overall survival of TNBC patients.
 | Table II.The predictive values of clinical
features and risk score. |
Table II.
The predictive values of clinical
features and risk score.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | Patients (N) | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) |
|
<60/≥60 | 100/52 | 0.95
(2.7–0.33) | 0.920 | 0.5
(0.15–1.67) | 0.259 |
| Pathological
stage |
|
I–II/III–IV | 126/26 | 22.21
(81.6–6.05) | 0.000 | 16.56
(2.79–98.38) | 0.002 |
| Stage T |
|
T1-T2/T3-T4 | 133/19 | 6.99
(22.99–2.13) | 0.001 | 2.28
(0.58–8.98) | 0.239 |
| Stage N |
|
N0/NX | 102/50 | 5.43
(16.67–1.77) | 0.003 | 2.51
(0.51–12.46) | 0.259 |
| Stage M |
|
M0/MX | 129/23 | 0.57
(4.39–0.07) | 0.589 | 0.12
(0.01–1.27) | 0.079 |
| Risk score |
|
Low/high | 76/76 | 9.32
(41.07–2.11) | 0.003 | 6.75
(1.33–34.21) | 0.021 |
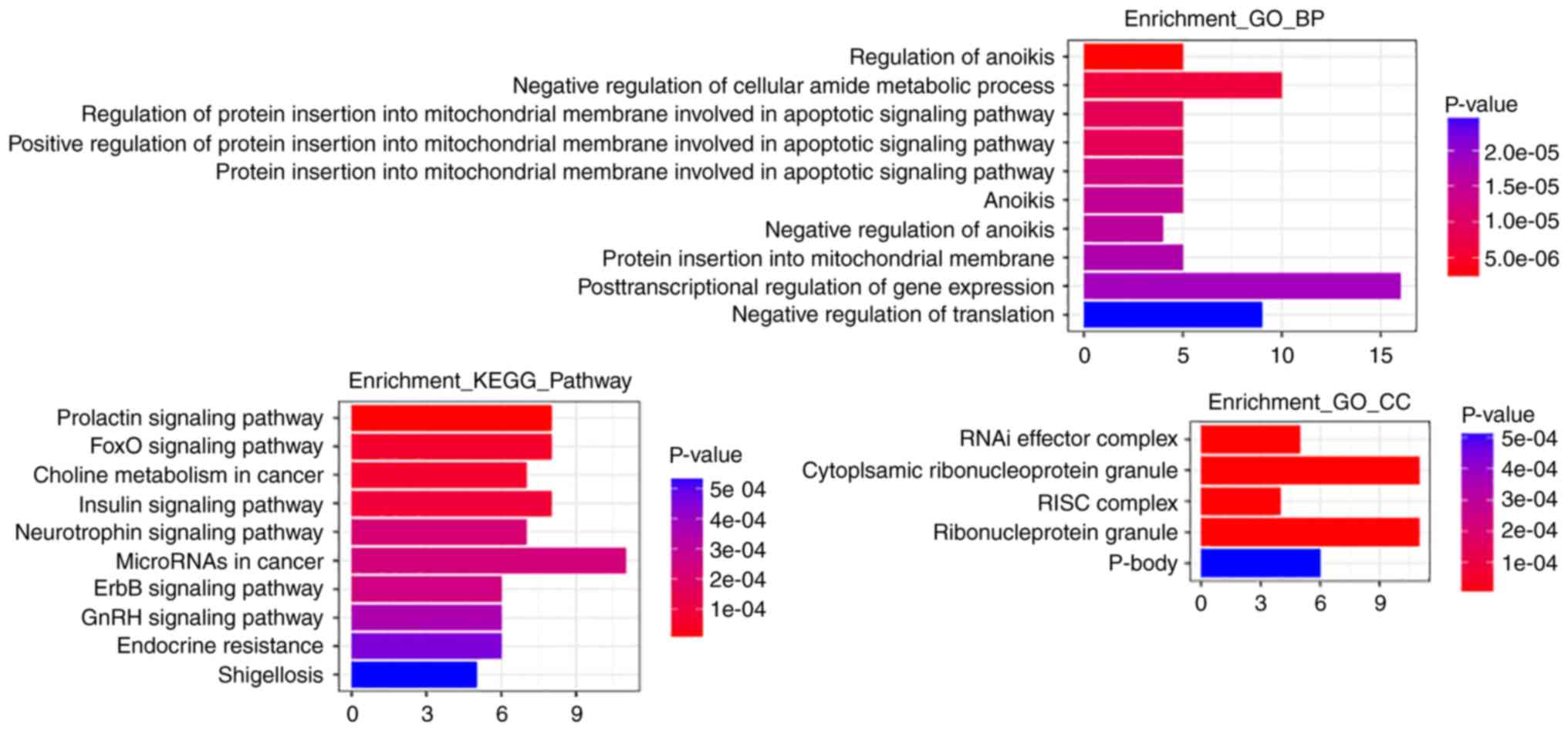
Functional annotation of the target
genes of the miRNA signature
After target prediction of the 4 miRNAs
(hsa-miR-148b, hsa-miR-203a, hsa-miR-203b and hsa-miR-3922) using
miRDB, miRTarBase and TargetScan, GO function and KEGG pathway
enrichment analysis of the target genes was performed using the R
clusterProfiler software package. Details of target genes of the 4
miRNAs are presented in Table
III. According to the results of the GO functional enrichment
analysis, regulation of anoikis, negative regulation of cellular
amide metabolic process and regulation of protein insertion into
mitochondrial membranes involved in apoptotic signaling were the
most significantly enriched biological processes. The most clearly
enriched cellular components were RNAi effector complex,
ribonucleoprotein granule, RNA-induced silencing complex (RISC) and
ribonucleoprotein granule. The most significantly enriched
signaling pathways determined through KEGG analysis were the
prolactin signaling pathway, the FOXO signaling pathway, and the
ErbB and Insulin signaling pathways, and miRNAs and choline
metabolism in cancer. The results of enrichment analysis are
displayed in Fig. 8.
 | Table III.miRNAs targeting mRNAs of TNBC. |
Table III.
miRNAs targeting mRNAs of TNBC.
| miRNA | mRNA |
|---|
| hsa-mir-148b | SYNCRIP, TNRC6A,
WASL, MLEC, BTBD3, YWHAB, PPP6R1, USP33, NPTX1, CUL5, C1GALT1,
AGO1, SECISBP2L, MTMR10, ATP6AP2, ZCCHC2, DNMT1, PRKAA1, DICER1,
RTN4, CCT6A, ZFYVE26, ARL8B, DLG2, ATP7A, SESTD1, ACVR1, ALCAM,
OTUD4, FBXO28, ITSN2, KLF6, CDK19, NPEPL1, CLCN3, MAP3K9, CYB5R4,
ZDHHC17, EOGT, SIK1, RAB14, PAPD4, TBL1XR1, RAB34, GLRX5, CEP55,
NRAS, NCKIPSD, FAM104A, SOS2, C3orf58, PRNP, DCP2, STARD13,
OSBPL11, DDX6, FLOT2, ABCB7, BMP3, MARCH2, RAB12, JARID2, USP48,
AP4E1, ITGA5, TXNIP, NSD1 |
| hsa-mir-203a | TFAM, SHOC2, ERI2,
SOCS5, GAN, CACNB2, BTG2, TSC22D2, YWHAE |
| hsa-mir-203b | SPTY2D1, VEZF1,
EFHD2, TIPARP |
Discussion
Growing evidence indicates that triple-negative
breast cancer (TNBC) is a heterogeneous disease comprising several
distinct disorders with clearly different clinical behavior and
molecular characteristics (11,12).
However, no specific and well-defined molecular targets have thus
far been defined in TNBCs, and therefore few therapeutic strategies
can be utilized as treatments, or are on the development horizon.
In the present study, we found 4 miRNAs (hsa-miR-10a, hsa-miR-18a,
hsa-miR-135b and hsa-miR-577) with significant value in TNBC
diagnosis. We conducted a detailed analysis of a 4-miRNA signature
which was comprised of hsa-miR-148b, hsa-miR-203a, hsa-miR-203b and
hsa-miR-3922 and exhibited capacity for predicting TNBC overall
survival.
In previous studies, hsa-miR-10a belonging to the
miR-10 family has been revealed to be dysregulated in several types
of cancers (13), such as breast
(14), glioblastoma (15), lung cancer (16) and chronic lymphocytic leukemia
(17). Another member of the miR-10
family, miR-10b, has been demonstrated to functionally contribute
to tumor invasion and metastasis in breast cancer (18). hsa-miR-10a and miR-10b deviate only
one nucleotide located at the center of their sequence and the
primary hsa-miR-10b transcript may be equivalent to a
promoter-associated RNA that could be targeted by miR-10a,
suggesting the important role of miR-10a in breast cancer
progression (19). Liu et al
demonstrated that miR-18a prevented ER-α expression blocking the
protective effects of estrogen and promoting the development of
hepatocellular carcinoma (20). A
previous study revealed that the expression of miR-18a-5p was
enhanced in TNBC compared with luminal A (21). Aberrant upregulation of miR-18a
could enhance autophagy in TNBC cells via inhibition of the mTOR
signaling pathway (22) and
decrease Dicer expression as well as increase paclitaxel resistance
(23). Moreover, hsa-miR-135b was
revealed to be upregulated in TNBC tissue which targeted estrogen
receptor 1 (ESR1)-related proteins (24). Notably, miR-135b has been proposed
as an oncogene involved in the pathogenesis of TNBC and the
differential expression of miR-135b in blood could predict overall
survival in follow-up of basal-like TNBC patients (25,26).
High expression of its family member, hsa-miR-135a, has been
demonstrated to be associated with good prognosis in ER-positive
tumors (27). Furthermore,
hsa-miR-577 has been revealed to be dysregulated in several cancer
types including gastric (28),
bladder (29) and breast cancer
(30). In addition, miR-577 has
been demonstrated to suppress epithelial-mesenchymal transition and
metastasis by inhibiting Rab25 expression in breast cancer
(31). Our results demonstrated
that hsa-miR-10a, hsa-miR-18a, hsa-miR-135b and hsa-miR-577 were
significantly differentially expressed in the TNBC group and are
potential candidate diagnostic markers of TNBC.
A 4-miRNA signature was identified after univariate
and multivariate Cox's proportional hazards regression model
analysis that was significantly correlated with the overall
survival of TNBC patients. miR-148b, a tumor suppressor, has been
reported to be dysregulated in pancreatic (32), non-small cell lung cancer (33) and hepatocellular carcinoma (34) through suppression of cell
proliferation and invasion by targeting the AMPKα1 and
WNT1/β-catenin pathways. A previous study indicated that
downregulation of miR-148b may be a molecular biomarker for the
early detection of hepatocellular carcinoma and a prognostic marker
(35). Increasing evidence
indicates that miR-203 is involved in several cancers, including
hepatocellular carcinoma (36),
prostate (37), breast (38), gastric and colorectal cancers
(39) through control of tumor cell
proliferation, migration and invasive potential by interaction with
target genes. Notably, dysregulated expression of miR-203 has been
revealed to be associated with poorer survival of pancreatic tumors
(40) and adenocarcinoma (41). As members of the miR-203 family,
hsa-miR-203a and hsa-miR-203b may exert important roles in TNBC.
Our results found 4 miRNAs (hsa-miR-148b, hsa-miR-203a,
hsa-miR-203b and hsa-miR-3922) which may play a role in TNBC
prognosis.
To further explore the molecular mechanisms of the
miRNA signature in TNBC, target genes of the miRNAs were predicted
and functional annotation of targets was performed. The results of
functional annotation of target genes revealed that regulation of
anoikis, negative regulation of cellular amide metabolic process
and protein insertion into mitochondrial membrane involved in the
apoptotic signaling pathway were significant enriched GO terms.
Anoikis is defined as apoptosis that is induced by inadequate or
inappropriate cellular interaction with the extracellular matrix
(42). Recently, more studies have
confirmed that the breakdown of anoikis leads to the malignancy of
mammary and colon cancers (43).
Meanwhile, anoikis-resistance is a hallmark of metastasis (44). Therefore, these miRNAs may be
involved in TNBC metastasis. According to pathway enrichment
analysis of the miRNA signature targets, miRNAs in cancer, and the
ErbB and prolactin signaling pathways were clearly enriched terms
that encompassed most genes. Increasing evidence suggests that
miRNAs participate in almost all aspects of cancer, including
proliferation, apoptosis, angiogenesis and invasion/metastasis
(45). Therefore, identification of
clear diagnostic and prognostic miRNA biomarkers can contribute to
cancer evaluation and treatment. Additionally, dimerization of ErbB
receptors leads to induction of kinase activity that activates
downstream MAPK and PI3K/AKT pathways which have significant
involvement in tumor cell proliferation and survival (46). Notably, ErbB-2 overexpression has
adverse prognostic value in breast cancer and thus, ErbB-directed
strategies have been developed and used as treatments (47). The peptide hormone prolactin,
synthesized by human breast cancer cells in culture, has been found
to stimulate cell proliferation in an autocrine manner (48). Given the ability of prolactin to
stimulate the proliferation of human breast cancer cells and the
aberrant expression of its active receptors in breast carcinomas,
it is fully consistent that prolactin plays a key role in breast
cancer. Consequently, miRNAs in cancer, and the ErbB and prolactin
signaling pathways may be significantly implicated in TNBC and
inhibition of these pathways may be potential therapeutic
strategies for TNBC patients.
However, there is also one limitation in the present
study. The percentage of TNBC patients in breast cancer is so small
that it is difficult to collect enough TNBC samples with follow-up
information in a short time to verify the function of the
identified miRNAs. Information of TNBC patients is now collected,
however, just a few cases were obtained. The biological roles of
these miRNAs in TNBC are still not clear and will be investigated
in further experimental studies when enough TNBC samples are
obtained.
In conclusion, the present study identified 4
aberrantly expressed miRNAs including hsa-miR-10a, hsa-miR-18a,
hsa-miR-135b and hsa-miR-577 with diagnostic value for early
diagnosis of TNBC patients, and subsequently, a 4-miRNA signature
composed of hsa-miR-148b, hsa-miR-203a, hsa-miR-203b and
hsa-miR-3922 was identified that may be a prognostic biomarker for
predicting the overall survival of TNBC patients. However, further
studies are required to validate these findings and the underlying
molecular mechanisms of these miRNAs also require exploitation in
combating TNBC in future.
Acknowledgements
We wish to express our warm thanks to Dr Lei Ma
(Department of Breast Surgery, China-Japan Union Hospital of Jilin
University, Changchun, China) who provided valuable guidance to our
research.
Funding
No funding was received.
Availability of data and materials
All raw miRNA RNA-seq and clinical data of TNBC can
be downloaded from TCGA data portal.
Authors' contributions
NL conceived and designed the study. CF analyzed the
data and wrote the manuscript. Both authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
ROC
|
receiver operating characteristic
|
|
TCGA
|
The Cancer Genome Atlas
|
|
AUC
|
area under curve
|
|
miRNA
|
microRNA
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the Carolina
Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cascione L, Gasparini P, Lovat F, Carasi
S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et
al: Integrated microRNA and mRNA signatures associated with
survival in triple negative breast cancer. PLoS One. 8:e559102013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gasparini P, Cascione L, Fassan M, Lovat
F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, et
al: microRNA expression profiling identifies a four microRNA
signature as a novel diagnostic and prognostic biomarker in triple
negative breast cancers. Oncotarget. 5:1174–1184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kleivi Sahlberg K, Bottai G, Naume B,
Burwinkel B, Calin GA, Børresen-Dale AL and Santarpia L: A serum
microRNA signature predicts tumor relapse and survival in
triple-negative breast cancer patients. Clin Cancer Res.
21:1207–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A BioconductoR package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dawson SJ, Provenzano E and Caldas C:
Triple negative breast cancers: Clinical and prognostic
implications. Eur J Cancer. 45 (Suppl 1):S27–S40. 2009. View Article : Google Scholar
|
|
12
|
Santarpia L, Qi Y, Stemke-Hale K, Wang B,
Young EJ, Booser DJ, Holmes FA, O'Shaughnessy J, Hellerstedt B,
Pippen J, et al: Mutation profiling identifies numerous rare drug
targets and distinct mutation patterns in different clinical
subtypes of breast cancers. Breast Cancer Res Treat. 134:333–343.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X,
Guo X, He S and Chen R: Transcriptional inhibiton of Hoxd4
expression by miRNA-10a in human breast cancer cells. BMC Mol Biol.
10:122009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan Y, Wang Q, Yan XL, Zhang Y, Li W, Tang
F, Li X and Yang P: miR-10a controls glioma migration and invasion
through regulating epithelial-mesenchymal transition via
EphA8. FEBS Lett. 589:756–765. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gaur A, Jewell DA, Liang Y, Ridzon D,
Moore JH, Chen C, Ambros VR and Israel MA: Characterization of
microRNA expression levels and their biological correlates in human
cancer cell lines. Cancer Res. 67:2456–2468. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma L: Role of miR-10b in breast cancer
metastasis. Breast Cancer Res. 12:2102010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han J, Kim D and Morris KV:
Promoter-associated RNA is required for RNA-directed
transcriptional gene silencing in human cells. Proc Natl Acad Sci
USA. 104:12422–12427. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin
CY, Chen DS and Chen PJ: MicroRNA-18a prevents estrogen
receptor-alpha expression, promoting proliferation of
hepatocellular carcinoma cells. Gastroenterology. 136:683–693.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR and Baracat
EC: Triple-negative and luminal A breast tumors: Differential
expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol.
35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan YX, Dai YZ, Wang XL, Ren YQ, Han JJ
and Zhang H: MiR-18a upregulation enhances autophagy in triple
negative cancer cells via inhibiting mTOR signaling pathway. Eur
Rev Med Pharmacol Sci. 20:2194–2200. 2016.PubMed/NCBI
|
|
23
|
Sha LY, Zhang Y, Wang W, Sui X, Liu SK,
Wang T and Zhang H: MiR-18a upregulation decreases dicer expression
and confers paclitaxel resistance in triple negative breast cancer.
Eur Rev Med Pharmacol Sci. 20:2201–2208. 2016.PubMed/NCBI
|
|
24
|
Dai X, Chen A and Bai Z: Integrative
investigation on breast cancer in ER, PR and HER2-defined subgroups
using mRNA and miRNA expression profiling. Sci Rep. 4:65662014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uva P, Cossu-Rocca P, Loi F, Pira G,
Murgia L, Orrù S, Floris M, Muroni MR, Sanges F, Carru C, et al:
miRNA-135b contributes to triple negative breast cancer molecular
heterogeneity: Different expression profile in basal-like versus
non-basal-like phenotypes. Int J Med Sci. 15:536–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paszek S, Gablo N, Barnas E, Szybka M,
Morawiec J, Kołacińska A and Zawlik I: Dysregulation of microRNAs
in triple-negative breast cancer. Ginekol Pol. 88:530–536. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan HW, Li SC and Tsai KW: MicroRNA
dysregulation in gastric cancer. Curr Pharm Des. 19:1273–1284.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tatarano S, Chiyomaru T, Kawakami K,
Enokida H, Yoshino H, Hidaka H, Yamasaki T, Kawahara K, Nishiyama
K, Seki N, et al: miR-218 on the genomic loss region of chromosome
4p15.31 functions as a tumor suppressor in bladder cancer. Int J
Oncol. 39:13–21. 2011.PubMed/NCBI
|
|
30
|
Kolacinska A, Morawiec J, Fendler W,
Malachowska B, Morawiec Z, Szemraj J, Pawlowska Z, Chowdhury D,
Choi YE, Kubiak R, et al: Association of microRNAs and pathologic
response to preoperative chemotherapy in triple negative breast
cancer: Preliminary report. Mol Biol Rep. 41:2851–2857. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin C, Mou Q, Pan X, Zhang G, Li H and Sun
Y: MiR-577 suppresses epithelial-mesenchymal transition and
metastasis of breast cancer by targeting Rab25. Thorac Cancer.
9:472–479. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao G, Zhang JG, Liu Y, Qin Q, Wang B,
Tian K, Liu L, Li X, Niu Y, Deng SC, et al: miR-148b functions as a
tumor suppressor in pancreatic cancer by targeting AMPKalpha1. Mol
Cancer Ther. 12:83–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu
RX, Liu SH, Yi QT, Li J and Song CH: Serum miR-152, miR-148a,
miR-148b, and miR-21 as novel biomarkers in non-small cell lung
cancer screening. Tumour Biol. 36:3035–3042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang JG, Shi Y, Hong DF, Song M, Huang D,
Wang CY and Zhao G: MiR-148b suppresses cell proliferation and
invasion in hepatocellular carcinoma by targeting WNT1/β-catenin
pathway. Sci Rep. 5:80872015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ziari K, Zarea M, Gity M, Fayyaz AF,
Yahaghi E, Darian EK and Hashemian AM: Downregulation of miR-148b
as biomarker for early detection of hepatocellular carcinoma and
may serve as a prognostic marker. Tumour Biol. 37:5765–5768. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saini S, Majid S, Yamamura S, Tabatabai L,
Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y and Dahiya R:
Regulatory role of mir-203 in prostate cancer progression and
metastasis. Clin Cancer Res. 17:5287–5298. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z
and Dong JT: Epigenetic silencing of miR-203 upregulates
SNAI2 and contributes to the invasiveness of malignant
breast cancer cells. Genes Cancer. 2:782–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chiang Y, Song Y, Wang Z, Chen Y, Yue Z,
Xu H, Xing C and Liu Z: Aberrant expression of miR-203 and its
clinical significance in gastric and colorectal cancers. J
Gastrointest Surg. 15:63–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Greither T, Grochola LF, Udelnow A,
Lautenschlager C, Wurl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hezova R, Kovarikova A, Srovnal J,
Zemanova M, Harustiak T, Ehrmann J, Hajduch M, Svoboda M, Sachlova
M and Slaby O: Diagnostic and prognostic potential of miR-21,
miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell
carcinoma of esophagus. Diagn Pathol. 10:422015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Streuli CH and Gilmore AP:
Adhesion-mediated signaling in the regulation of mammary epithelial
cell survival. J Mammary Gland Biol Neoplasia. 4:183–191. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Park SH, Riley P IV and Frisch SM:
Regulation of anoikis by deleted in breast cancer-1 (DBC1) through
NF-κB. Apoptosis. 18:949–962. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garofalo M, Leva GD and Croce CM:
MicroRNAs as anti-cancer therapy. Curr Pharm Des. 20:5328–5335.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
DiGiovanna MP, Stern DF, Edgerton SM,
Whalen SG, Moore D II and Thor AD: Relationship of epidermal growth
factor receptor expression to ErbB-2 signaling activity and
prognosis in breast cancer patients. J Clin Oncol. 23:1152–1160.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vonderhaar BK: Prolactin involvement in
breast cancer. Endocr Relat Cancer. 6:389–404. 1999. View Article : Google Scholar : PubMed/NCBI
|