Introduction
Colorectal cancer (CRC) is one of the most
aggressive cancers in the world and is associated with a high
mortality rate (1–3). Notably, an increased rate of CRC has
been reported in many countries (4–6).
Despite improvements in diagnostic and treatment techniques, the
5-year relative survival rates are still low in CRC patients
(7,8). Therefore, a better understanding of
the molecular mechanisms of CRC initiation and progression may
promote the development of new treatments for CRC patients.
Recently, several of long non-coding RNAs (lncRNAs)
have been regarded as important modulators in cancer progression
and development (9–12). It has been reported that the lncRNA
promoter of CDKN1A antisense DNA damage-activated RNA (PANDAR) is
involved in the development and progression of many cancers,
including CRC (13–15). Moreover, lncRNA PANDAR was revealed
to inhibit CRC apoptosis and induce CRC growth via the
epithelial-mesenchymal transition (EMT) pathway (16). These results demonstrated that
lncRNAs could be used as new therapeutic targets for CRC treatment
(17,18). Hence, to investigate the molecular
mechanisms that mediate cancer development, increased efforts are
required to explain the role of lncRNAs in CRC.
Tumor suppressor candidate 7 (TUSC7) is an lncRNA
that has been revealed to be downregulated in CRC tissues (19). Furthermore, TUSC7 inhibited CRC cell
proliferation by sponging miR-211-3p (19). The expression of TUSC7 in CRC cells
was revealed to reduce cell growth, whereas the low expression of
TUSC7 indicated poor prognosis of CRC patients (20). However, little is known about the
expression and the role of TUSC7 in CRC cell migration and
invasion.
To date only a few studies have analyzed the
possible role of TUSC7 in CRC (19,20).
The aim of the present study was to analyze the precise role of
TUSC7 in CRC progression.
Materials and methods
Cell culture
The HCT116, COLO205, HT29, SW480 and CaCO-2 cells
and the normal colon epithelial cell line NCM460 were seeded in
Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine
serum (FBS), 1% penicillin G-streptomycin (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in an incubator with 5%
CO2.
RNA preparation, reverse
transcription, and quantitative real-time PCR
RNA was extracted from cells by TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA was
reverse-transcribed by a reverse transcription kit (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocol. Real-time PCR expression was assessed by
SYBR® PremixEx Taq™ (Takara Biotechnology Co., Ltd.).
The PCR primers used were as follows (19): TUSC7 forward,
5′-GGAAACAGAAGGCACCTCA-3′ and reverse, 5′-TCTCAGAGGTCAAACAGGCA-3′;
GAPDH forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′. Relative quantification of TUSC7 was
performed by the 2−∆∆Cq method (21). All reactions were repeated in
triplicate.
Overexpression of TUSC7
The overexpression vector pCDNA-TUSC7 and the empty
vector pCDNA-N1 (NC) were obtained from Genomeditech Co., Ltd.
(Shanghai, China). Cells (at ~70% confluence) were transfected with
Lipofectamine™ 3000 reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocols for 24 h.
The expression of TUSC7 was determined by qRT-PCR.
Cell proliferation assay
Cell proliferation was detected by the Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) assay following the manufacturer's protocol. In brief, the
transfected cells (1×105 cells) were seeded in 96-well
plates, and then CCK-8 solution (10 µl) was added for 2 h. The
absorbance was measured by an ELISA reader (Molecular Devices
Sunnyvale, CA, USA) at 450 nm.
Cell cycle analysis
Cells (1×106) were seeded in 6-well
plates for 24 h. Then, cells were loaded with propidium iodide (PI;
100 µl) for 30 min at 4°C in the dark. The cell-cycle distribution
was determined by flow cytometry.
Invasion and migration assay
The Transwell chamber was obtained from Corning
Inc., (Corning, NY, USA) for the cell invasion and migration
assays. For the cell invasion assay, the chamber was precoated with
30 µl of Matrigel (BD Biosciences, San Jose, USA) for 1 h. Cells
(5×105 cells) were maintained in the upper chamber, and
the lower chamber contained DMEM (500 µl) supplemented with 20% FBS
for 12 h at 37°C. The invaded cells were stained with calcein-AM,
and the invaded cells were counted under a fluorescence microscope
(magnification, ×100). For the cell migration assay, cells
(5×105 cells) were seeded into the upper chamber
containing DMEM (500 ml) supplemented with 20% FBS for 12 h at
37°C. The migrated cells were stained with calcein-AM, and the
migrated cells were counted under a fluorescence microscope
(magnification, ×100).
Wound-healing assay
Cells were cultured in 6-well plates to 90–95%
confluence. Wounds were generated with a plastic scraper, then
cells were washed with phosphate-buffered saline (PBS) twice and
incubated in a FBS free medium for 24 h, images were observed by a
light microscope.
Western blot analysis
The SW480 and CaCO-2 cells were collected and lysed
in RIPA buffer (EMD Millipore, Temecula, CA, USA). The protein
concentration was determined using the BCA method (Beyotime
Institute of Biotechnology, Beijing, China). Proteins (40 µg) were
separated by using 12% SDS-polyacrylamide gel electrophoresis
(SDS-PAGE) and were then transferred to 0.22 µm nitrocellulose
membranes (EMD Millipore). The membranes were blocked with 5%
non-fat milk for 1 h at room temperature. Then, membranes were
incubated with the E-cadherin (dilution 1:1,000; cat. no. 3195;
Cell Signaling Technology Inc., Danvers, MA, USA), vimentin
(dilution 1:1,000; cat. no. 5741; Proteintech Group, Inc., Chicago,
IL, USA) overnight at 4°C. Membranes were then incubated with a
horseradish peroxidase-linked secondary antibody (mouse anti-rabbit
IgG-FITC; dilution 1:1,000; cat. no. sc-2359; Santa Cruz
Biotechnology, Dallas, TX, USA) for 1 h at room temperature. The
blots were visualized by using enhanced chemiluminescence ECL
(Pierce Biotechnology; Thermo Fisher Scientific, Inc.).
Statistical analysis
All data were obtained from three independent
experiments. Data were analyzed using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). The statistical significance of the differences
between groups was determined using t-tests. Data are expressed as
the means ± standard deviations (SD). Differences with P-values
<0.05 were considered to indicate a statistically significant
difference.
Results
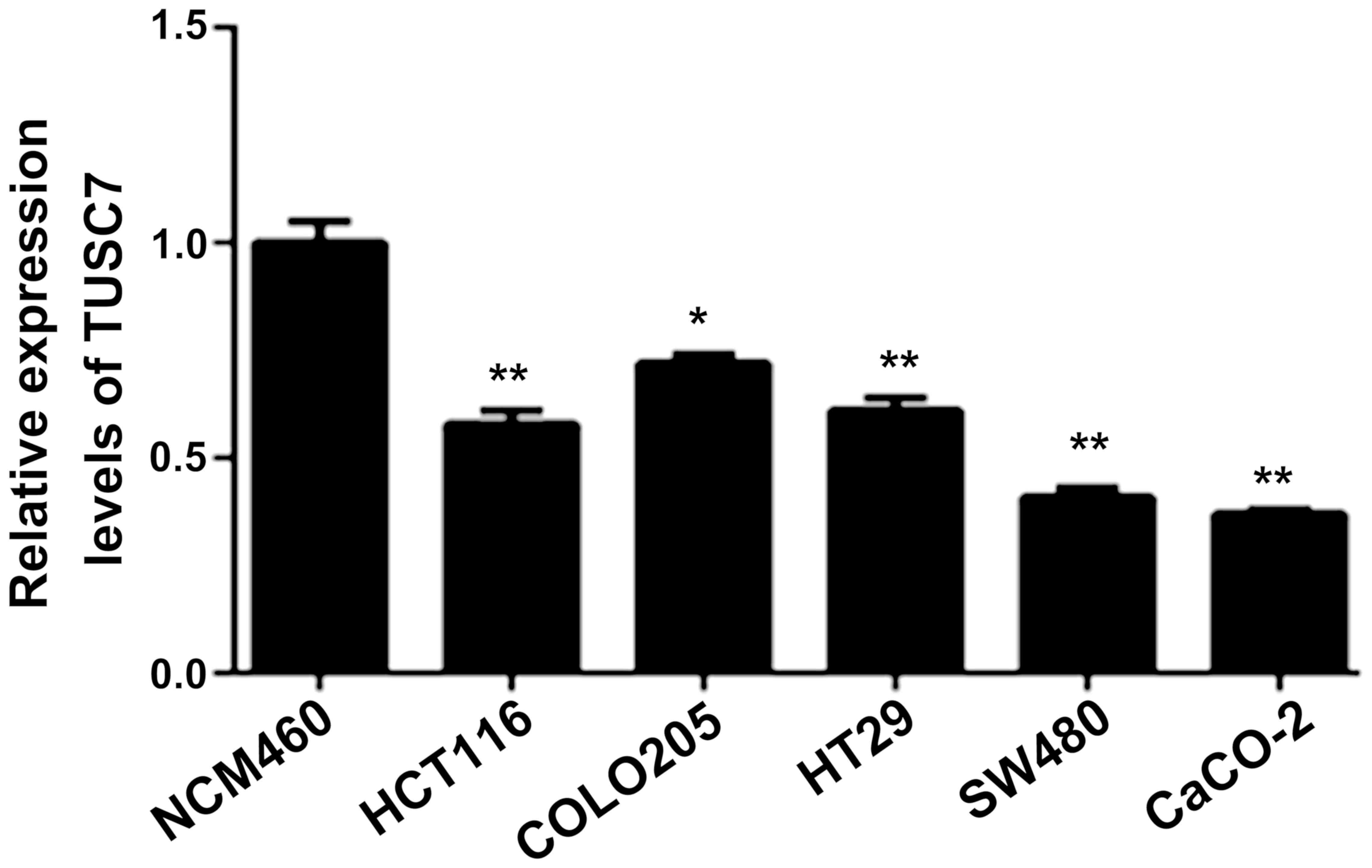
The expression of TUSC7 decreases in
CRC cells
The expression of TUSC7 in HCT116, COLO205, HT29,
SW480 and CaCo-2 cells was lower than that in the NCM460 cells
(P<0.05; Fig. 1), which
indicated that TUSC7 plays a key role in CRC progression. Since
TUSC7 expression in SW480 and CaCO-2 cells was lower than the other
cell lines, SW480 and CaCO-2 cells were selected in the subsequent
experiments.
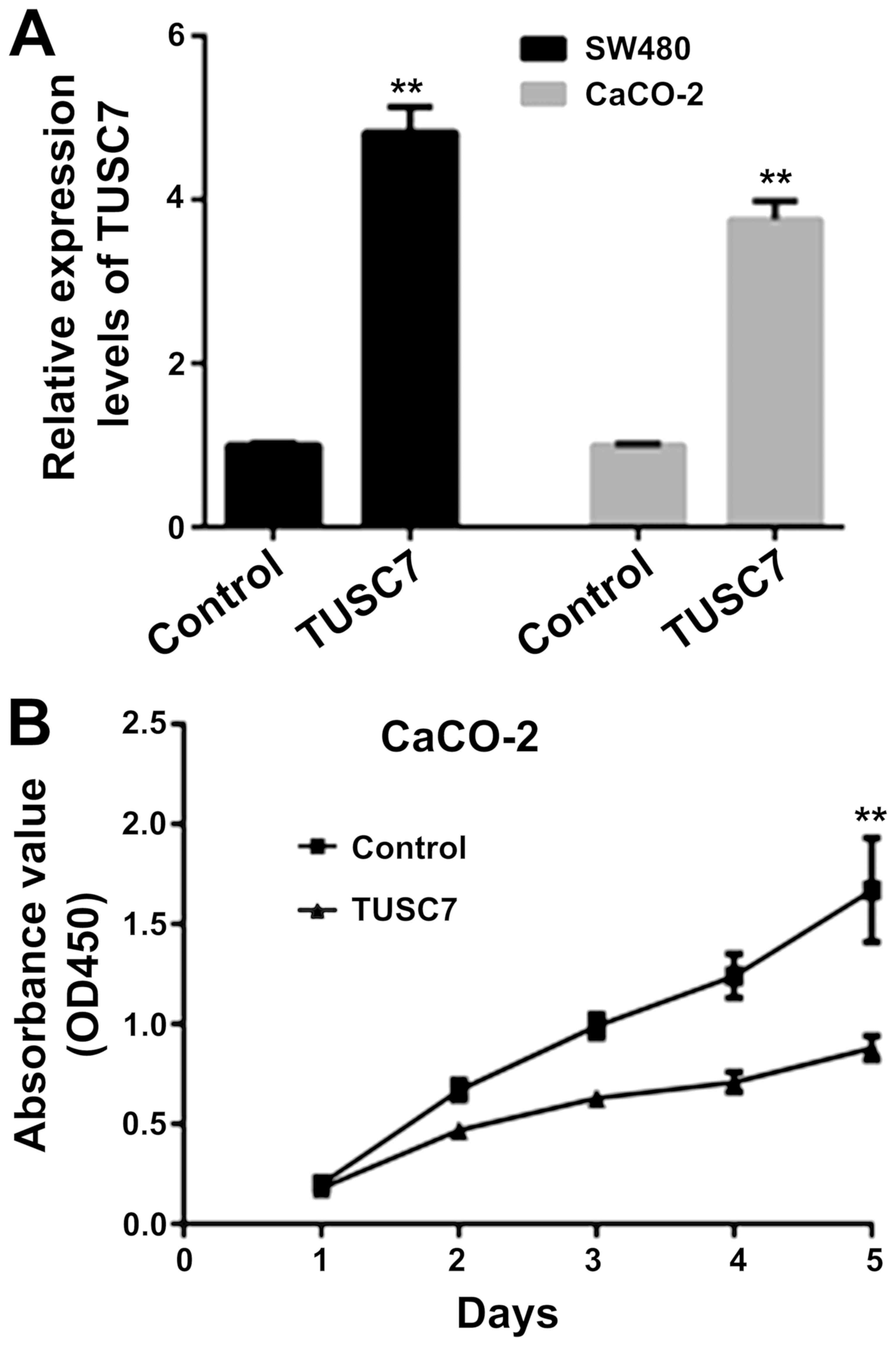
TUSC7 inhibits SW480 and CaCO-2 cell
proliferation
To overexpress TUSC7, a lentiviral vector was used
in SW480 and CaCO-2 cells. As revealed in Fig. 2A, TUSC7 expression was increased in
SW480 and CaCO-2 cells. The CCK-8 assay revealed that
overexpression of TUSC7 decreased the proliferation rate of both
SW480 (data not shown) and CaCO-2 cells (Fig. 2B).
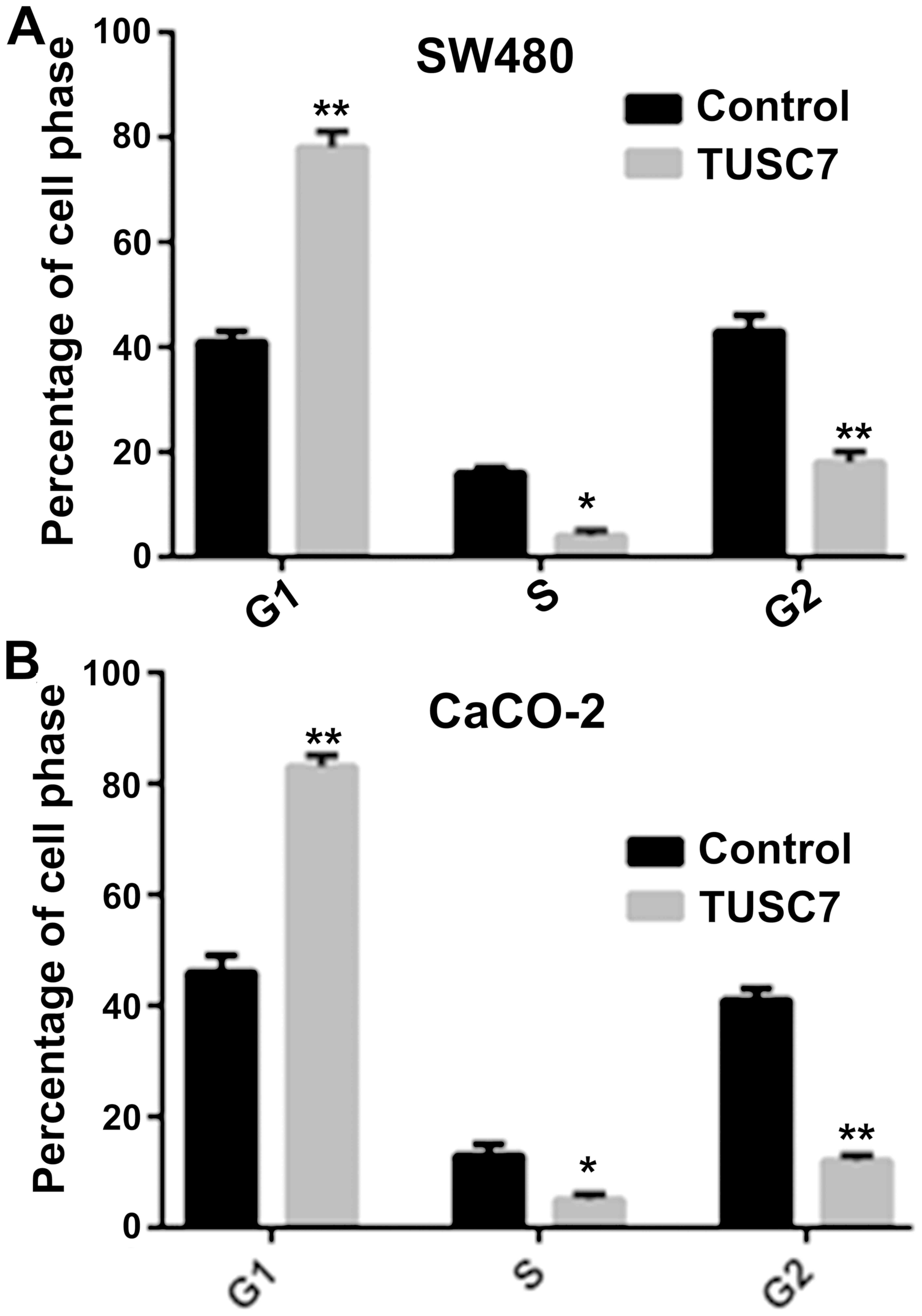
TUSC7 induces SW480 and CaCO-2 cell
cycle arrest
To explore whether TUSC7 regulates the cell cycle of
CRC cells, PI staining and flow cytometric analysis were used. As
indicated in Fig. 3, overexpression
of TUSC7 notably increased G1 phase cell cycle arrest, whereas the
S/G2 phase was reduced in both SW480 and CaCO-2 cells.
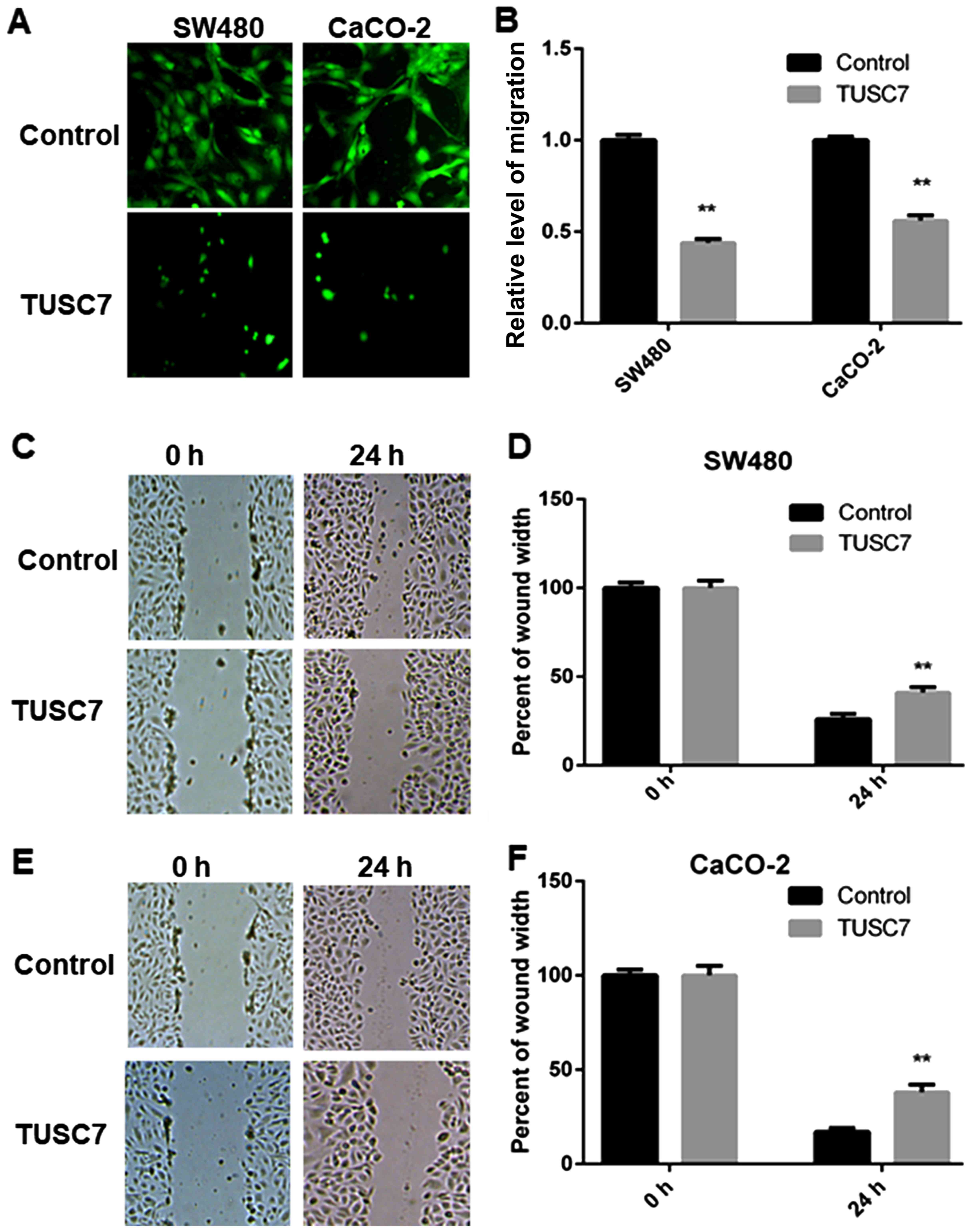
TUSC7 inhibits SW480 and CaCO-2 cell
migration
To explain the role of TUSC7 in CRC cell metastasis,
Transwell and wound-healing assays were employed. Compared with the
control group, overexpression of TUSC7 significantly reduced the
migration ability of both SW480 and CaCO-2 cells (Fig. 4A and B).
The migration abilities of CRC cells were also
assessed by a wound-healing assay. The wound-healing assay
exhibited similar results. Compared with the control group,
overexpression of TUSC7 resulted in significantly decreased
migration abilities of both SW480 (Fig.
4C and D) and CaCO-2 cells (Fig. 4E
and F), suggesting that overexpression of TUSC7 inhibited the
migration of CRC cells.
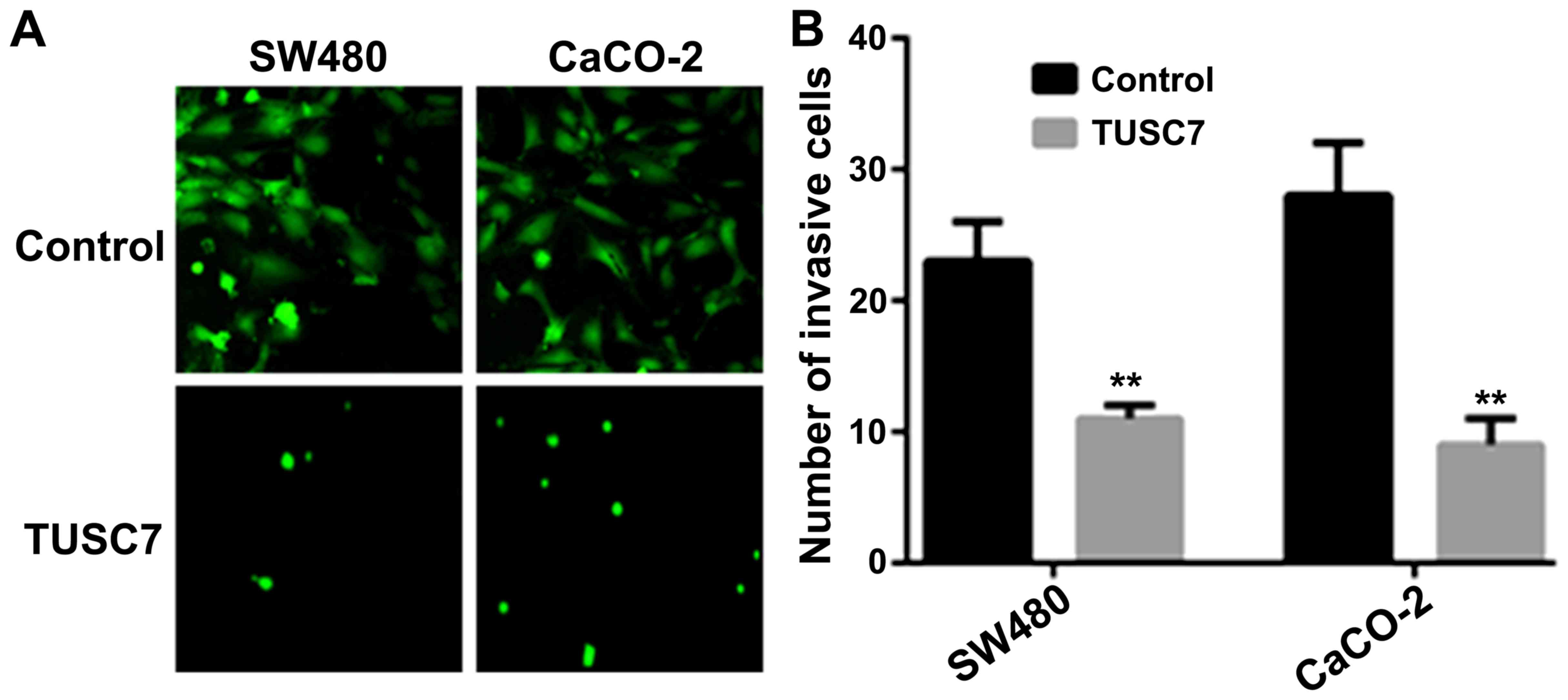
TUSC7 inhibits SW480 and CaCO-2 cell
invasion
The invasion ability of CRC cells was evaluated by a
Transwell invasion assay. Compared with the control group,
overexpression of TUSC7 markedly reduced the invasion ability of
CRC cells (Fig. 5). These results
suggested that TUSC7 plays a key role in CRC metastasis.
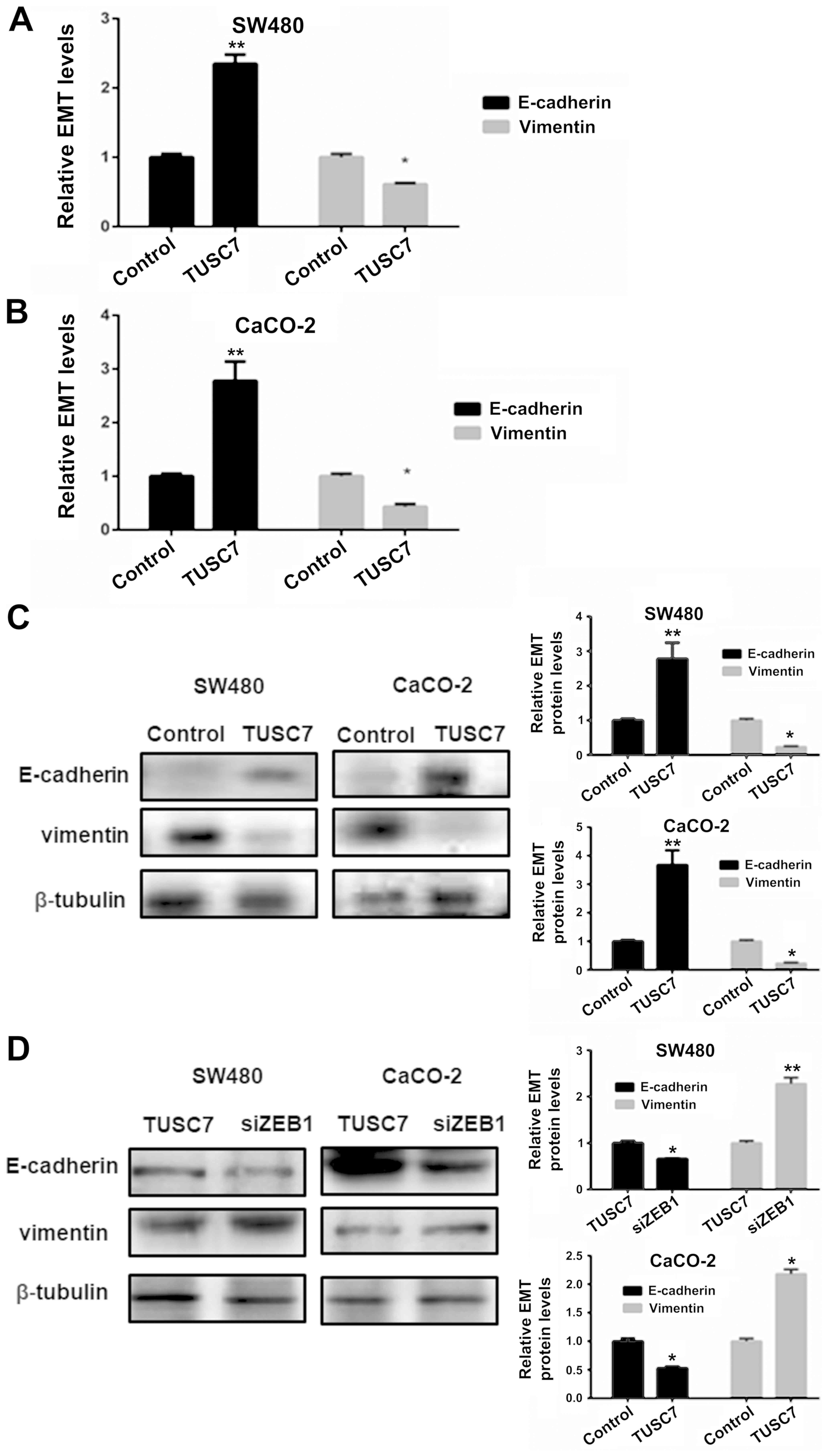
TUSC7 suppresses EMT in SW480 and
CaCO-2 cells
It is well known that EMT plays a key role in CRC
cell migration and invasion. Therefore, we investigated whether
TUSC7 affects the EMT of CRC. The results of the qRT-PCR (Fig. 6A and B) and western blot analysis
(Fig. 6C) revealed that E-cadherin
expression was increased and vimentin expression was suppressed in
the cells overexpressing TUSC7 compared to the expression levels of
the control group. Hence, this finding revealed that TUSC7
inhibited the EMT of CRC.
TUSC7 inhibits EMT in SW480 and CaCO-2
cells via activation of ZEB1
To determine whether ZEB1 has a mediatory effect on
the TUSC7-dependent suppression of EMT, we used siRNA to reduce the
expression of ZEB1. The data revealed that the expression of
E-cadherin and vimentin were restored by siZEB1, which indicated
the initiation of the EMT of CRC (Fig.
6D). These results indicated that TUSC7 inhibited the EMT of
CRC via ZEB1 expression.
Discussion
A significant association between aberrant tumor
suppressor candidate 7 (TUSC7) expression and cancer development,
including squamous cell carcinoma (22), glioma (23), gastric cancer (24) and other types of cancer (25,26)
has been demonstrated. In a previous study, TUSC7 increased
colorectal cancer (CRC) cell proliferation by sponging miR-211-3p
(19). To the best of our
knowledge, there are no previous studies that have focused on the
role of TUSC7 expression in CRC cell migration and invasion.
The expression level of TUSC7 has been associated
with cancer development and progression (19). Consistent with these previous
findings, we demonstrated that TUSC7 was downregulated in CRC cells
compared to its expression in NCM460 cells. To explore the role of
TUSC7 in CRC cells, we used an overexpression assay in both SW480
and CaCo-2 cells. As predicted, overexpression of TUSC7 suppressed
CRC cells, as revealed by the CCK-8 assay. Moreover, overexpression
of TUSC7 inhibited CRC cells to the S/G2 phase and increased the
number of CRC cells in the G1 phase. These data have also been
reported in a study by Xu et al (19). Thus, our findings indicated that
TUSC7 decreased CRC cell proliferation.
It has been revealed that the migration ability of
cancer is associated with TUSC7 expression, suggesting that TUSC7
works as a key tumor suppressor in many types of cancer (25,27,28).
In the present study, we found that overexpression of TUSC7
significantly reduced CRC cell migration. These data revealed that
TUSC7 was involved in the migration ability of CRC cells.
EMT is a key process that stimulates the mesenchymal
state and induces the migration and invasion of cancer cells
(29,30). Recent findings have revealed that
lncRNAs are involved in the EMT (31–33).
Some long non-coding RNAs (lncRNAs) promote EMT whereas some
lncRNAs suppress EMT. lncRNA CPS1-IT1 was revealed to inhibit EMT
and the migration of CRC by inactivating HIF-1α in vivo and
in vitro (34,35). lncRNA AB073614 stimulated the EMT of
CRC cells via the JAK/STAT3 signaling pathway (36). In this study, we detected EMT
biomarkers of CRC cells by qRT-PCR and western blot analysis.
Notably, the data revealed that the expression level of TUSC7 was
positively associated with the expression of E-cadherin and
negatively associated with the expression of vimentin in CRC cells.
It was revealed that TUSC7 suppressed the EMT in CRC cells. ZEB1 is
a key regulator in the invasion and metastasis of cancer cells by
inducing the EMT of CRC. Aberrant levels of ZEB1 have been found in
different types of tumors, including hepatocellular carcinoma
(37), breast cancer (38) and CRC (39). The association of ZEB1 with CRC
development has been widely investigated, and this study suggested
that knockdown of ZEB1 suppressed the inhibition of the EMT of CRC
associated with TUSC7.
TUSC7 inhibited CRC cell proliferation, migration,
invasion and EMT, indicating that TUSC7 could be a potential tumor
suppressor in CRC. Although our results provide important insights
into these processes, the roles of TUSC7 in in vivo studies
are required to further confirm these data.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets supporting the conclusions of this
article are included within the article.
Authors' contributions
HZ, YS and CY conceived of the idea, designed the
study, performed research, analyzed and interpreted data and wrote
the manuscript. CY and XW analyzed data. CY confirmed statistical
analyses. XW revised the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Tongji Hospital Affiliated with Tongji
University.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Berian JR, Cuddy A, Francescatti AB,
O'Dwyer L, Nancy You Y, Volk RJ and Chang GJ: A systematic review
of patient perspectives on surveillance after colorectal cancer
treatment. J Cancer Surviv. 11:542–552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fuccio L, Repici A, Hassan C, Ponchon T,
Bhandari P, Jover R, Triantafyllou K, Mandolesi D, Frazzoni L,
Bellisario C, et al: Why attempt en bloc resection of
non-pedunculated colorectal adenomas? A systematic review of the
prevalence of superficial submucosal invasive cancer after
endoscopic submucosal dissection. Gut. 67:1464–1474. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gkegkes ID, Minis EE and Iavazzo C:
Dermatomyositis and colorectal cancer: A systematic review. Ir J
Med Sci. 187:615–620. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fang JY, Dong HL, Sang XJ, Xie B, Wu KS,
Du PL, Xu ZX, Jia XY and Lin K: Colorectal cancer mortality
characteristics and predictions in China, 1991–2011. Asian Pac J
Cancer Prev. 16:7991–7995. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang HY, Shi JF, Guo LW, Bai YN, Liao XZ,
Liu GX, Mao AY, Ren JS, Sun XJ, Zhu XY, et al: Expenditure and
financial burden for the diagnosis and treatment of colorectal
cancer in China: A hospital-based, multicenter, cross-sectional
survey. Chin J Cancer. 36:412017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ivanova JI, Saverno KR, Sung J, Duh MS,
Zhao C, Cai S, Vekeman F, Peevyhouse A, Dhawan R and Fuchs CS:
Real-world treatment patterns and effectiveness among patients with
metastatic colorectal cancer treated with ziv-aflibercept in
community oncology practices in the USA. Med Oncol. 34:1932017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Väyrynen JP, Tuomisto A, Väyrynen SA,
Klintrup K, Karhu T, Mäkelä J, Herzig KH, Karttunen TJ and Mäkinen
MJ: Preoperative anemia in colorectal cancer: Relationships with
tumor characteristics, systemic inflammation, and survival. Sci
Rep. 8:11262018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tabung FK, Liu L, Wang W, Fung TT, Wu K,
Smith-Warner SA, Cao Y, Hu FB, Ogino S, Fuchs CS, et al:
Association of dietary inflammatory potential with colorectal
cancer risk in men and women. JAMA Oncol. 4:366–373. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H and
Teng L: lncRNAs: emerging biomarkers in gastric cancer. Future
Oncol. 11:2427–2441. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma N, Li S, Zhang Q, Wang H, Qin H and
Wang S: Long non-coding RNA GAS5 inhibits ovarian cancer cell
proliferation via the control of microRNA-21 and SPRY2 expression.
Exp Ther Med. 16:73–82. 2018.PubMed/NCBI
|
|
11
|
Ma Z, Peng P, Zhou J, Hui B, Ji H, Wang J
and Wang K: Long non-coding RNA SH3PXD2A-AS1 promotes cell
progression partly through epigenetic silencing P57 and KLF2 in
colorectal cancer. Cell Physiol Biochem. 46:2197–2214. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeong G, Bae H, Jeong D, Ham J, Park S,
Kim HW, Kang HS and Kim SJ: A Kelch domain-containing KLHDC7B and a
long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer
cell proliferation via the interferon signaling pathway. Sci Rep.
8:129222018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou Y, Zhong Y, Wu J, Xiao H, Zhang X,
Liao X, Li J, Mao X, Liu Y and Zhang F: Long non-coding PANDAR as a
novel biomarker in human cancer: A systematic review. Cell Prolif.
51:e124222018. View Article : Google Scholar
|
|
14
|
Peng W and Fan H: Long non-coding RNA
PANDAR correlates with poor prognosis and promotes tumorigenesis in
hepatocellular carcinoma. Biomed Pharmacother. 72:113–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rivandi M, Pasdar A, Hamzezadeh L,
Tajbakhsh A, Seifi S, Moetamani-Ahmadi M, Ferns GA and Avan A: The
prognostic and therapeutic values of long noncoding RNA PANDAR in
colorectal cancer. J Cell Physiol. 234:1230–1236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu M, Liu Z, Li B, Wang G, Li D and Zhu Y:
The high expression of long non-coding RNA PANDAR indicates a poor
prognosis for colorectal cancer and promotes metastasis by EMT
pathway. J Cancer Res Clin Oncol. 143:71–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li M, Zhao LM, Li SL, Li J, Gao B, Wang
FF, Wang SP, Hu XH, Cao J and Wang GY: Differentially expressed
lncRNAs and mRNAs identified by NGS analysis in colorectal cancer
patients. Cancer Med. 7:4650–4664. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dai M, Chen X, Mo S, Li J, Huang Z, Huang
S, Xu J, He B, Zou Y, Chen J, et al: Meta-signature lncRNAs serve
as novel biomarkers for colorectal cancer: Integrated
bioinformatics analysis, experimental validation and diagnostic
evaluation. Sci Rep. 7:465722017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Zhang R and Zhao J: The Novel Long
Noncoding RNA TUSC7 inhibits proliferation by sponging miR-211 in
colorectal cancer. Cell Physiol Biochem. 41:635–644. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren W, Chen S, Liu G, Wang X, Ye H and Xi
Y: TUSC7 acts as a tumor suppressor in colorectal cancer. Am J
Transl Res. 9:4026–4035. 2017.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang ZW, Jia YX, Zhang WJ, Song LJ, Gao
M, Li MJ, Zhao RH, Li J, Zhong YL, Sun QZ, et al:
lncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal
squamous cell carcinoma by competitively regulating DESC1. J Exp
Clin Cancer Res. 37:562018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma XL, Zhu WD, Tian LX, Sun WD, Shang F,
Lin QT and Zhang HQ: Long non-coding RNA TUSC7 expression is
independently predictive of outcome in glioma. Eur Rev Med
Pharmacol Sci. 21:3605–3610. 2017.PubMed/NCBI
|
|
24
|
Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan
C, Weng WW, Sheng WQ, Zhou XY and Du X: Reciprocal repression
between TUSC7 and miR-23b in gastric cancer. Int J Cancer.
137:1269–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y,
Ding L, Jia Y, Zhang H, Li Q, et al: Long non-coding RNA TUSC7 acts
a molecular sponge for miR-10a and suppresses EMT in hepatocellular
carcinoma. Tumour Biol. 37:11429–11441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z, Jin Y, Ren H, Ma X, Wang B and
Wang Y: Downregulation of the long non-coding RNA TUSC7 promotes
NSCLC cell proliferation and correlates with poor prognosis. Am J
Transl Res. 8:680–687. 2016.PubMed/NCBI
|
|
27
|
Li N, Shi K and Li W: TUSC7: A novel tumor
suppressor long non-coding RNA in human cancers. J Cell Physiol.
233:6401–6407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shang C, Guo Y, Hong Y and Xue YX: Long
non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing
roles in human gliomas. Front Cell Neurosci. 10:2352016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li T, Huang H, Shi G, Zhao L, Li T, Zhang
Z, Liu R, Hu Y, Liu H, Yu J, et al: TGF-β1-SOX9 axis-inducible
COL10A1 promotes invasion and metastasis in gastric cancer via
epithelial-to-mesenchymal transition. Cell Death Dis. 9:8492018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song N, Zhong J, Hu Q, Gu T, Yang B, Zhang
J, Yu J, Ma X, Chen Q, Qi J, et al: FGF18 Enhances Migration and
the epithelial-mesenchymal transition in breast Cancer by
regulating Akt/GSK3β/β-catenin signaling. Cell Physiol Biochem.
49:1019–1032. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu YW, Sun M, Xia R, Zhang EB, Liu XH,
Zhang ZH, Xu TP, De W, Liu BR and Wang ZX: LincHOTAIR
epigenetically silences miR34a by binding to PRC2 to promote the
epithelial-to-mesenchymal transition in human gastric cancer. Cell
Death Dis. 6:e18022015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan C, Yao G, Liu B, Ma T, Xia Y, Wei K,
Wang J, Xu J, Chen L and Chen Y: Long noncoding RNA FAL1 promotes
cell proliferation, invasion and epithelial-mesenchymal transition
through the PTEN/AKT signaling axis in non-small cell lung cancer.
Cell Physiol Biochem. 43:339–352. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Wang YM and Song YL: Knockdown of
long noncoding RNA AB073614 inhibits glioma cell proliferation and
migration via affecting epithelial-mesenchymal transition. Eur Rev
Med Pharmacol Sci. 20:3997–4002. 2016.PubMed/NCBI
|
|
34
|
Zhang W, Yuan W, Song J, Wang S and Gu X:
lncRna CPS1-IT1 suppresses cell proliferation, invasion and
metastasis in colorectal cancer. Cell Physiol Biochem. 44:567–580.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT,
Liang KH, Shieh TM, Chen CY and Hsueh C: Long noncoding RNA
CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by
regulating HIF-1α activity and inhibiting epithelial-mesenchymal
transition. Oncotarget. 7:43588–43603. 2016.PubMed/NCBI
|
|
36
|
Xue J, Liao L, Yin F, Kuang H, Zhou X and
Wang Y: lncRNA AB073614 induces epithelial- mesenchymal transition
of colorectal cancer cells via regulating the JAK/STAT3 pathway.
Cancer Biomark. 21:849–858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao S, Zhang Y, Zheng X, Tu X, Li H, Chen
J, Zang Y and Zhang J: Loss of MicroRNA-101 promotes epithelial to
mesenchymal transition in hepatocytes. J Cell Physiol.
230:2706–2717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
de Barrios O, Győrffy B, Fernández-Aceñero
MJ, Sánchez-Tilló E, Sánchez-Moral L, Siles L, Esteve-Arenys A,
Roué G, Casal JI, Darling DS, et al: ZEB1-induced tumourigenesis
requires senescence inhibition via activation of DKK1/mutant
p53/Mdm2/CtBP and repression of macroH2A1. Gut. 66:666–682. 2017.
View Article : Google Scholar : PubMed/NCBI
|