Introduction
Glioblastoma multiforme (GBM) is the most malignant
primary human brain tumor. Tumors are characterized by a high
proliferation rate and chemoresistance (1).
Although the combination of radiotherapy and
chemotherapy has progressed after surgical resection, the 5-year
survival rate of WHO grade IV glioblastoma is still lower than 5%
(2,3).
Although significant progress has been made in understanding the
molecular status of this type of tumor, there is still a need for
new and effective therapeutic approaches (4–6).
Epithelial membrane protein 1 (EMP1) is a member of
the EMP family that has been implicated as a cell junction protein
on the plasma membrane (7). However,
little is known about its specific functions and mechanisms.
Recently, several members of the EMP family have been indicated to
participate in cancer progression. EMP1 has been revealed to be a
novel poor prognostic factor in pediatric leukemia and gastric
carcinoma by regulating cell proliferation, migration and invasion,
and was demonstrated to be involved in gefitinib resistance in lung
cancer (8,9). EMP1 was also identified as a novel
MYC-interacting gene with cancer-related functions in GBM (10). However, the biological role of EMP1 in
GBM remains unclear.
To the best of our knowledge, we are the first to
determine the important role of EMP1 in GBM. In the present study,
it was revealed that knockdown of EMP1 inhibited GBM cell
proliferation, migration and invasion. In addition, it was
determined that EMP1 is an independent predictor of poor prognosis
in GBM patients. To sum up, our data indicated that EMP1 is a
potential therapeutic target for the treatment of GBM.
Materials and methods
Ethical statement
All animal procedures were approved by the
Institutional Animal Care and Use Committee (IACUC) of Shandong
University (Jinan, China).
Database searches
The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/), Rembrandt (http://www.betastasis.com/glioma/rembrandt/), and the
Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) were mined for relevant
molecular data.
Cell lines and cultures
Human glioblastoma cell lines U251 (cat. no.
TCHu58), A172 (cat. no. TCHu171) and human glioblastoma of unknown
origin cell line U87MG (cat. no. TCHu138, authentication was
performed using STR Multi-Amplification Kit in Guangzhou Cellcook
Biotech Co., Ltd.) were purchased from the Chinese Academy of
Sciences Cell Bank (Shanghai, China). Human fibroblast glioblastoma
cell line T98, primary human GBM biopsy xenograft propagated tumor
cells P3 and normal human astrocytes were kindly provided by
Professor Rolf Bjerkvig, University of Bergen (Bergen, Norway).
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
cat. no. SH30022.01B; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (cat. no. 10082147; Hyclone; GE
Healthcare Life Sciences) in 5% CO2 in a humidified
incubator at 37°C.
Immunohistochemical staining
Samples were fixed in 4% formalin at 20°C for 24 h,
paraffin-embedded and sectioned (4 µm). After dewaxing and
rehydration, sections were incubated with 0.01 M citrate buffer at
95°C for 20 min for antigen retrieval. Endogenous peroxidase
activity and non-specific antigen were blocked with 3% hydrogen
peroxide and 10% normal goat serum (both from ZSGB-Bio; OriGene
Technologies, Inc.), respectively, followed by primary antibody
(EMP1; 1:100; cat. no. 63735; LifeSpan BioSciences) overnight at
4°C. Sections were washed with phosphate-buffered saline (PBS),
treated with goat anti-rabbit secondary antibody (cat. no. 1:200;
cat. no. PV-9000; ZSGB-BIO), visualized with 3,3′-diaminobenzidine
(DAB; both from ZSGB-Bio; OriGene Technologies, Inc.), and
hematoxylin (Beyotime Institute of Biotechnology). Normal mouse
serum served as a negative control.
shRNA transfections
Short hairpin (sh)-EMP1 (#1: GTT TGT TAG CAC CAT TGC
CAA TGT TTC AAG AGA ACA TTG GCA ATG GTG CTA ACA AAT TTT TT; #2: GGT
CTT TGG AAA AAC TGT ACC AAT TCA AGA GAT TGG TAC AGT TTT TCC AAA GAC
CTT TTT T; #3: GCC AGT GAA GAT G CCC TCA AGA CAT TCA AGA GAT GTC
TTG AGG GCA TCT TCA CTG GTT TTT T), were conjugated in the
lentiviral vector of pLKO.1 with a puromycin resistant region
(GenePharma). U87MG and P3 GBM cells were plated and infected with
lentiviruses expressing sh-EMP1 for 24 h, followed by puromycin
selection (2 mg/ml; Sigma-Aldrich; Merck KGaA). Western blotting
was performed to verify knockdown efficiency and cells were
allocated for different assays.
Cell viability assays
Cell viability was assessed using Cell Counting
Kit-8 (CCK-8; cat. no. CK04-500; Dojindo Molecular Technologies,
Inc.). Cells (1.0×104 cells/well) were seeded into
96-well plates and incubated at 37°C overnight. After the desired
treatment, the cells were incubated with 10 µl of CKCK-8 in 100 µl
of serum-free DMEM for a further 4 h at 37°C. The absorbance at 450
nm was measured using a microplate reader (Bio-Rad Laboratories,
Inc.).
Western blot analysis
Cell lysates (20 µg protein) were subjected to
western blot analysis, according to previously described protocols
(11). Membranes were incubated with
the following antibodies from Cell Signaling Technology: AKT
(dilutions 1:1,000, cat. no. 9272), p-Akt (Ser473, dilution
1:1,000; cat. no. 4060), mTOR (dilution 1:1,000; cat. no. 2972),
p-mTOR (Ser2448, dilution 1:1,000; cat. no. 2974), GAPDH (dilution
1:1,000; cat. no. 5174). Additional antibody was EMP1 (dilution
1:1,000; cat. no. 63735; LifeSpan BioSciences).
Cell migration and invasion
assays
The wound healing assay was used to assess cell
migration. U87MG and P3 GBM cells were seeded into 6-well flat
bottom plates and incubated overnight at 37°C. The cell-free space
was created by scraping with a 200-µl pipette tip. Wound closure
areas were monitored at different time-points under a microscope
and quantified using ImageJ software (National Institutes of
Health). Cell invasion assays were performed in uncoated and
Matrigel-coated Transwell chambers (8-µm pore size inserts;
Corning, Inc.). Cells (2×104) in medium containing 1%
FBS (200 µl) were seeded in the top chamber. The lower chamber was
filled with medium containing 30% FBS (600 µl). The cells that
invaded the lower surface were fixed with 4% paraformaldehyde
(Solarbio; Beijing, China) at 20°C for 15 min, stained with 0.1%
crystal violet (Solarbio) at 20°C for 15 min and counted under a
bright field microscope. Images were captured from 5 random fields
in each well and the number of cells was determined using Kodak MI
SE 5.0 software (Carestream Health). Each experiment was repeated
three times.
Intracranial xenograft model
Athymic mice (male; 4 weeks old; 20–30 g) were
provided by Shanghai SLAC Laboratory Animal Co., Ltd. The mice were
anesthetized with 5% chloral hydrate and secured on a stereotactic
frame. A longitudinal incision was made in the scalp and a 1-mm
diameter hole was drilled 2.5 mm lateral to the bregma.
Luciferase-stable P3 GBM cells (2×105) in 20 µl of
serum-free DMEM were implanted 2.5 mm into the right striatum using
a Hamilton syringe. Mice were monitored by bioluminescence imaging
every week. Briefly, mice were injected with 100 mg of luciferin
(Caliper; PerkinElmer, Inc.) while anesthetized with 3% isoflurane,
followed by a cooled charge-coupled device camera (IVIS-200;
Xenogen; Alameda, CA, USA). Bioluminescence values of tumors were
quantitated using the Living Image 2.5 software package (Xenogen
Corp.). Mice were sacrificed by CO2 inhalation and
dislocation of neck after 30 days or when they developed
neurological symptoms such as rotational behavior, reduced activity
or displayed grooming and dome head. The brains were extracted,
perfused with 4% paraformaldehyde in PBS and coronally sectioned
for immunohistochemistry assays.
Statistical analysis
Three independent experiments were performed, and
results were expressed as the mean ± the standard deviation (SD).
Data were compared using paired Student t-tests or one-way ANOVA
followed by Bonferroni tests in GraphPad Prism 6 software (GraphPad
Software, Inc.). P-values determined from different comparisons
<0.05 were considered statistically significant and are
indicated as follows: *P<0.05; **P<0.01; ***P<0.001.
Results
EMP1 expression is positively
correlated with tumor grade
To begin, to determine whether EMP1 was
differentially expressed between glioblastoma and normal tissues,
microarray data from patient samples were extensively examined in
the Oncomine database. A meta-analysis of five independent
glioblastoma data sets including 829 human glioblastoma samples and
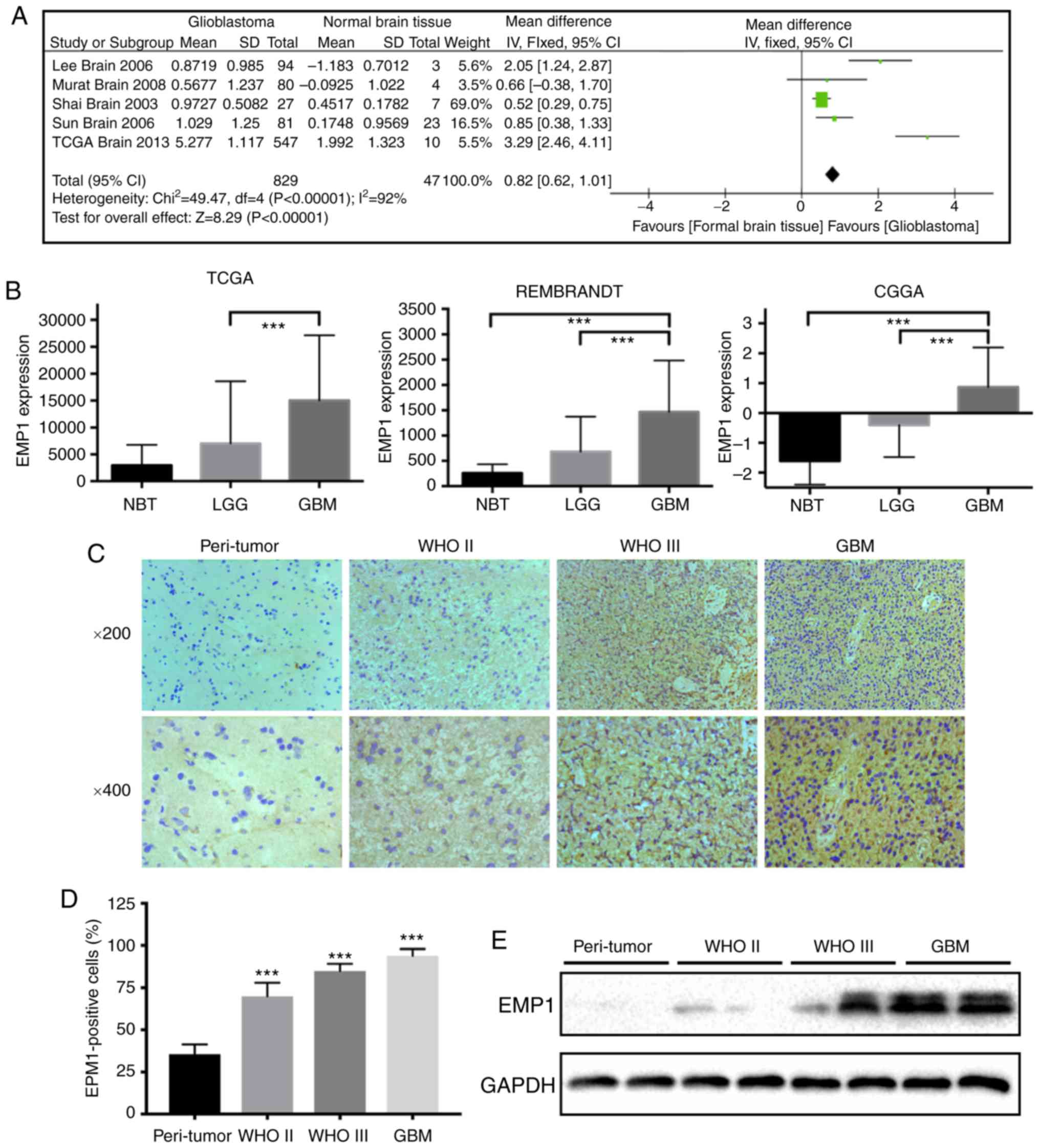
47 normal brain tissues revealed that EMP1 was significantly and
consistently present in glioblastoma in all data sets (Fig. 1A). To further verify the level of EMP1
in normal brain tissues and different grade glioma tissues, the
publicly available databases, The Cancer Genome Atlas (TCGA),
Rembrandt, and the Chinese Glioma Genome Atlas (CGGA) were
systematically retrieved and a relatively higher mRNA level of EMP1
was revealed in high grade gliomas compared to low grade gliomas
(P<0.01; Fig. 1B).
Immunohistochemistry results revealed that increased EMP1 levels
were associated with increased glioma grade. EMP1 was highly
expressed in grade II/III astrocytoma and glioblastoma (P<0.01),
whereas staining was weak or absent in peri-tumor tissues (Fig. 1C and D). Western blotting results
corroborated these results. EMP1 protein levels were increased in
glioma cases relative to peri-tumor tissues (Fig. 1E). Collectively, these results
demonstrated that EMP1 levels were increased in glioma compared to
normal brain tissues.
EMP1 expression is inversely
associated with glioblastoma patient prognosis
The differences in expression levels of EMP1 in
glioma and normal brain tissues prompted us to further investigate
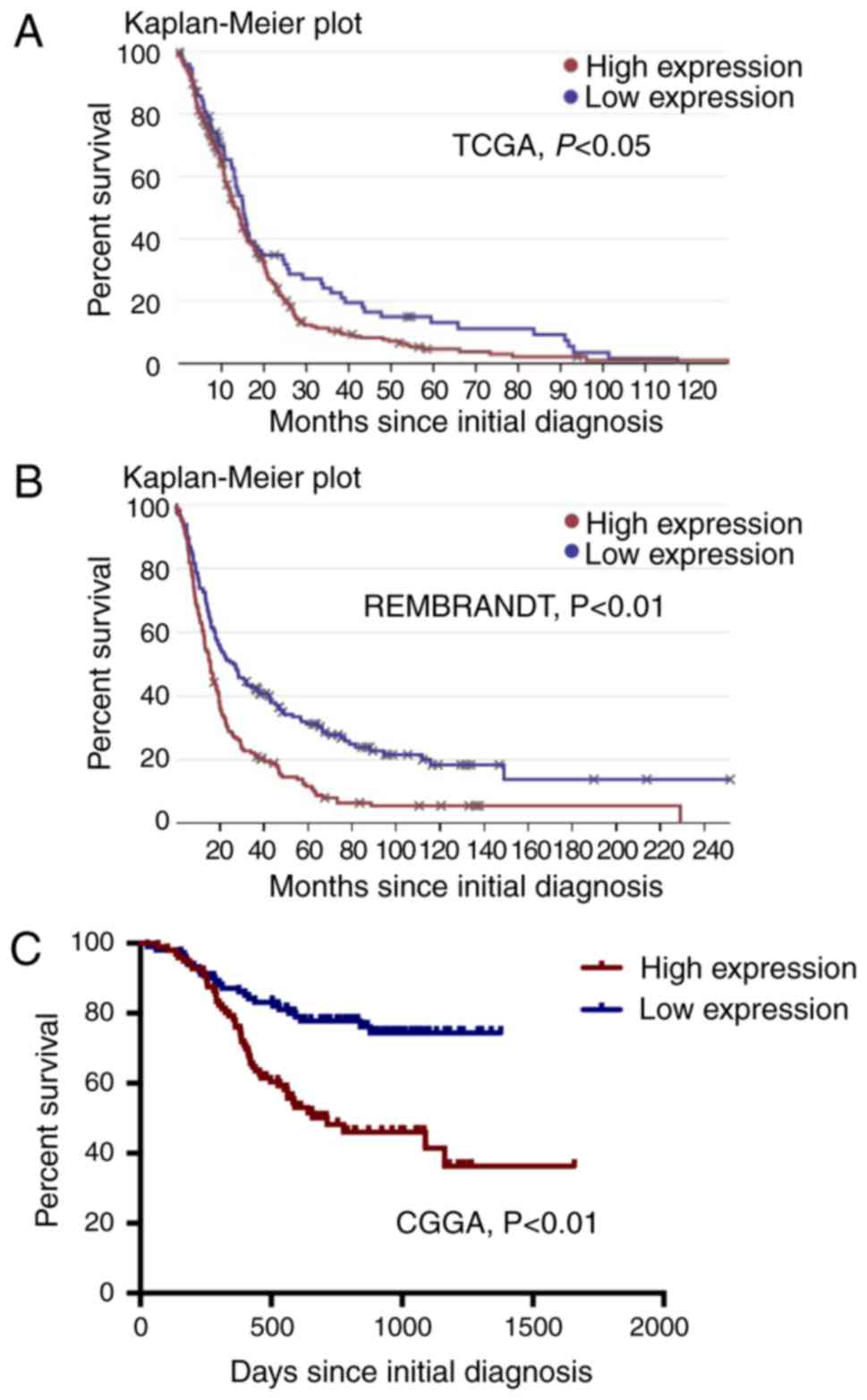
whether EMP1 can be used as a prognostic marker in glioma patients.
Data from the TCGA, Rembrandt, and CGGA databases was used to
determine the relationship between EMP1 levels and overall survival
(OS) in glioma patients. Each sample was classified as EMP1-high
expression if the signal was above the median expression for the
population. These data revealed significant differences in OS and
progression-free survival (PFS) between glioma patients with low
EMP1 expression and those with high expression (Fig. 2).
Pathway analysis of EMP1 and
co-regulated genes
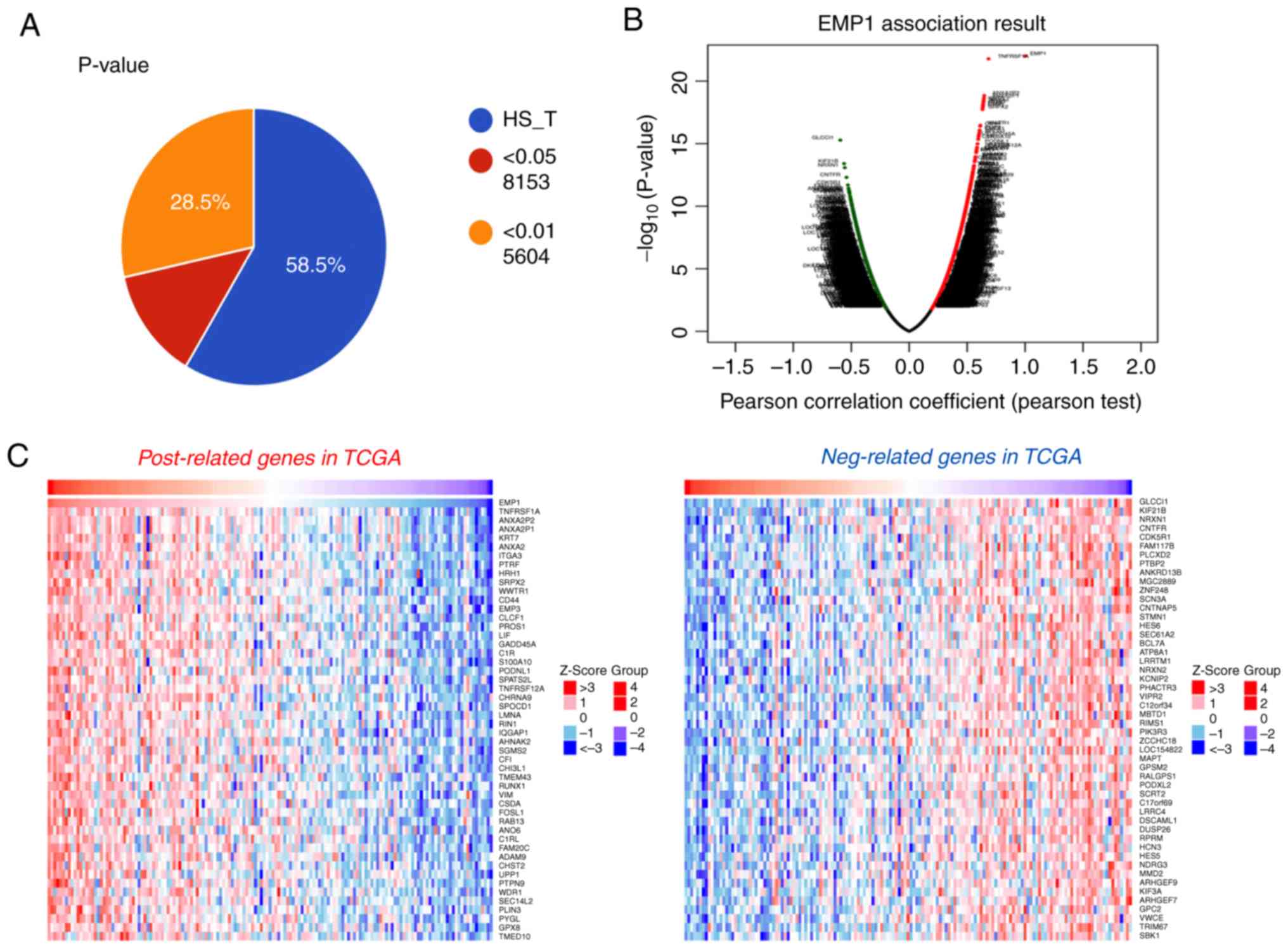
Next, to further understand the biological
implications of EMP1 in gliomas, correlation analysis of EMP1
expression in whole genome gene profiling was performed in TCGA.
The results revealed that 5,604 genes were correlated with EMP1
expression in TCGA database (P<0.01; Fig. 3A). As illustrated in the volcano plot
and heatmaps, these significant correlated genes were separated
into positively correlated and negatively correlated genes
(Fig. 3B and C). Subsequently, GO
analysis revealed that EMP1 positively-correlated genes were
strongly associated with biological processes including positive
regulation of cell proliferation, negative regulation of apoptotic
process, cell adhesion and extracellular matrix organization
(Fig. 3D). In KEGG analysis,
EMP1-correlated genes were enriched in pathways in cancer,
especially in the PI3K/AKT signaling pathway (Fig. 3D).
Knockdown of EMP1 inhibits
proliferation, migration and invasion in glioma cells in vitro
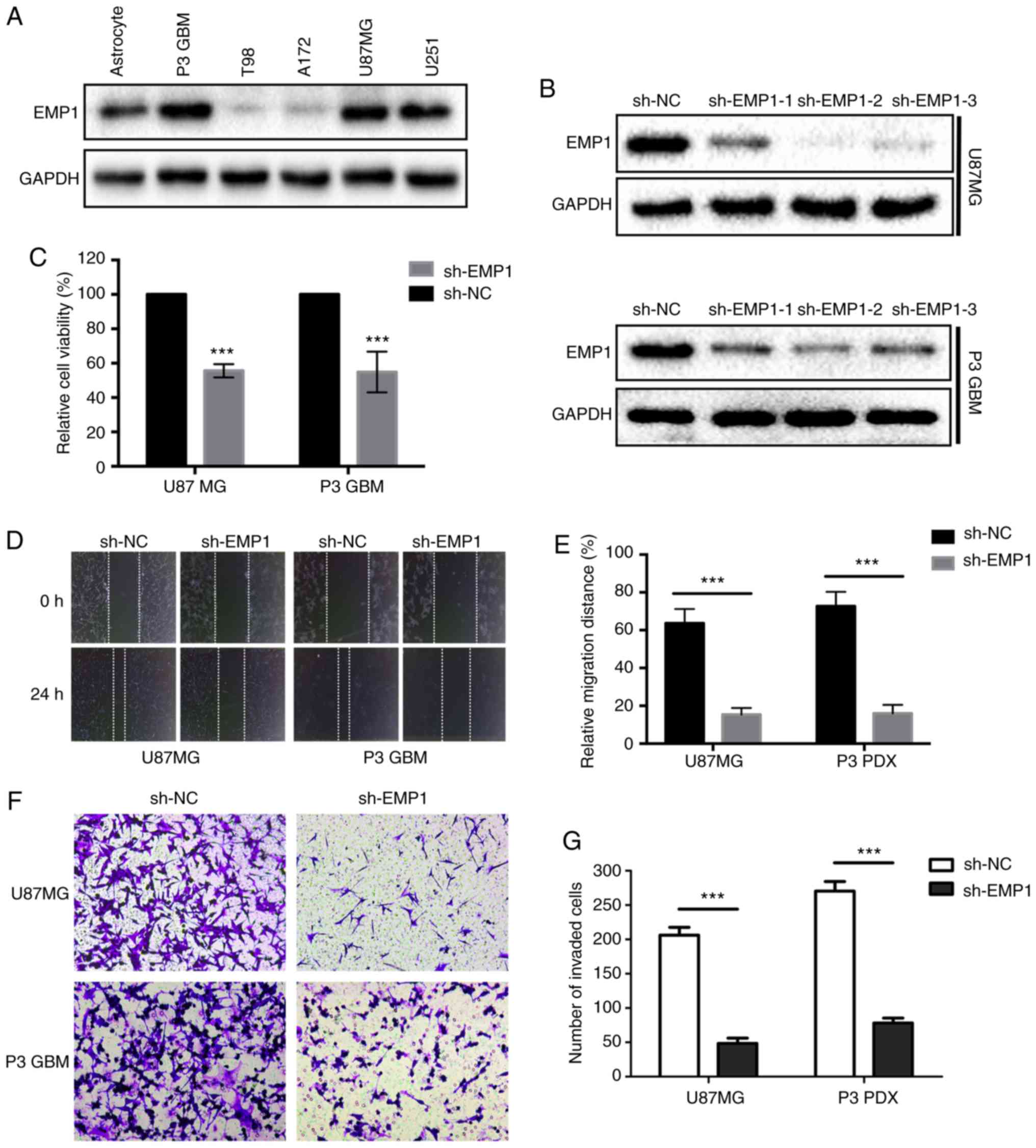
To determine the biological roles in glioma, the
expression level of EMP1 was first verified in several glioma cell
lines. Western blots results confirmed that the expression levels
of EMP1 protein were increased in several glioma cell lines,
especially in P3 GBM, which was derived from a primary GBM through
orthotopic passage in mice (12–14), and
U87MG cells (Fig. 4A). shRNA
targeting EMP1 lentiviral constructs were designed for stable
knockdown of expression. EMP1 protein levels in U87MG and P3 GBM
cells were significantly downregulated after infection with three
different shRNAs against EMP1 compared to NC constructs, especially
sh-EMP1-2 (Fig. 4B). Therefore, this
shRNA was selected for the subsequent functional assays.
It was then determined whether EMP1 knockdown may be
effective against GBM, using the cell viability assay, CCK-8.
Knockdown of EMP1 led to significant decreases in cell viability in
both U87MG and P3 GBM cells (P<0.05; Fig. 4C). Having observed a marked change in
cell morphology and retraction of pseudopodia after knockdown of
EMP1, a wound healing assay was used to examine whether knockdown
of EMP1 affected migration in GBM cells. Knockdown of EMP1 led to a
significant lower migratory rate both in U87MG and P3 GBM cells
(P<0.01; Fig. 4D and E). Transwell
analysis was further applied to assess the inhibitory effect of
EMP1 knockdown on cell invasion. In order to mimic the
extracellular matrix around glioma, U87GBM and P3 GBM cells were
plated in the upper chambers of a Transwell system coated with
Matrigel. The results revealed that the invasive ability of GBM
cell was significantly decreased after knockdown of EMP1 (Fig. 4F and G).
EMP1 promotes human glioma progression
through activation of the PI3K/AKT/mTOR signaling pathway
It is well known that abnormal activation of the
PI3K/AKT/mTOR signaling pathway promotes tumorigenesis, and in KEGG
analysis, EMP1-correlated genes were enriched in the PI3K/AKT
signaling pathway (Fig. 3D).
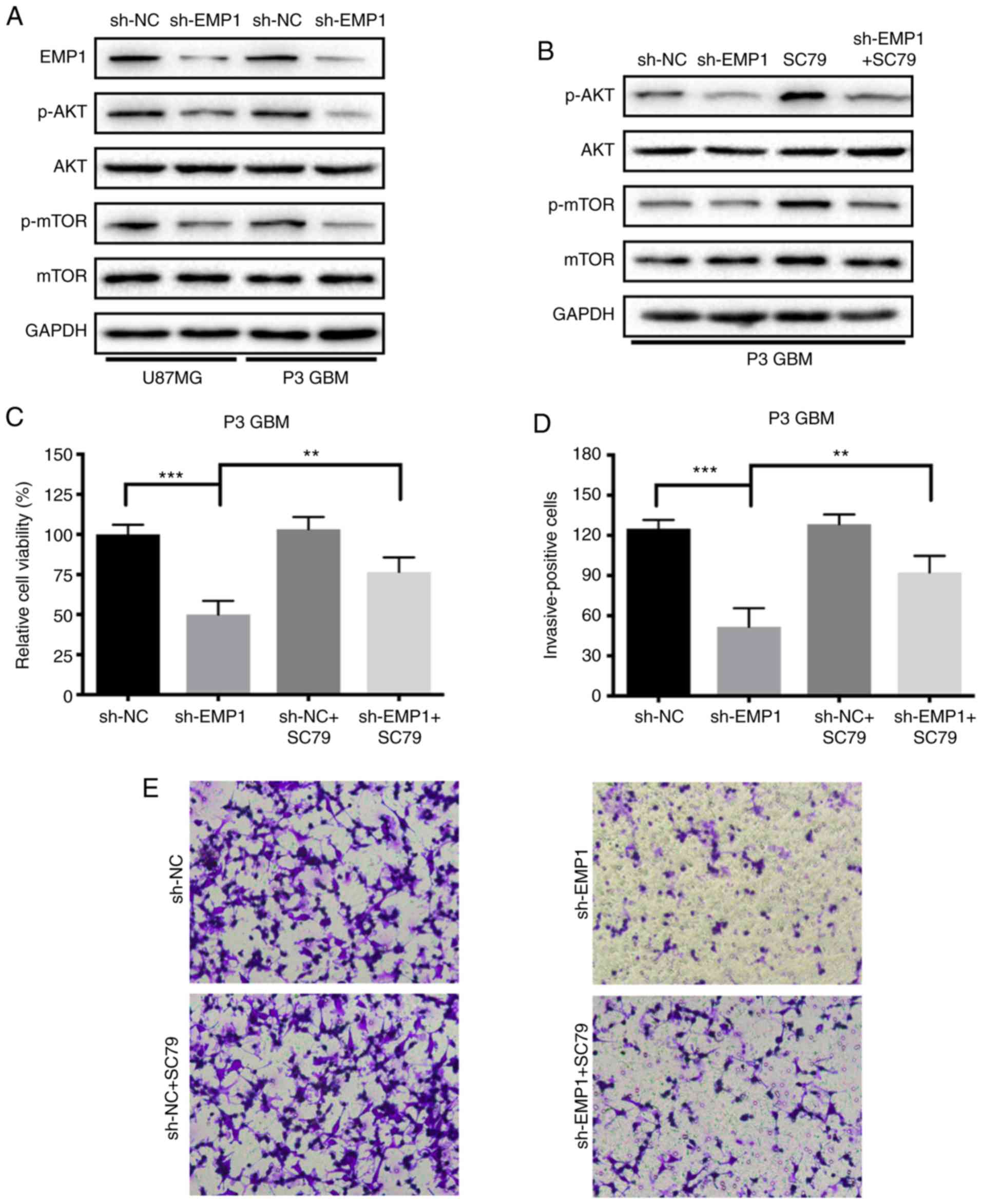
Therefore, phosphorylation status of AKT and mTOR proteins after
knockdown of EMP1 was examined by western blotting to determine
whether the observed responses could be due to a decrease in
PI3K/AKT/mTOR signaling. Phosphorylated AKT and mTOR decreased
compared to sh-NC following knockdown of EMP1 in modified cells,
demonstrating that the PI3K/AKT/mTOR signaling pathway was
inhibited after EMP1 knockdown (Fig.
5A). After being treated simultaneously with the novel AKT
activator SC79, the activation of the PI3K/AKT/mTOR signaling
pathway in GBM cells was partially restored. SC79 increased
phosphorylation of AKT and mTOR in EMP1-knockdown GBM cells, and
therefore further confirmed PI3K/AKT/mTOR signaling as a molecular
target for EMP1 knockdown of GBM growth (Fig. 5B). CCK-8 and Transwell invasion assays
were repeated to assess whether SC79 could restore proliferation
and invasion in EMP1-knockdown GBM cells. The results demonstrated
that SC79 increased proliferation (~50 vs. 70%; sh-EMP1 vs.
sh-EMP1+SC79; P<0.05) and invasion (~50 vs. 90%; sh-EMP1 vs.
sh-EMP1+SC79; P<0.05) in EMP1-knockdown GBM cells (Fig. 5C-E).
EMP1 enhances growth of GBM cells in
vivo
Considering the heterogeneity of GBM, P3 GBM, which
is an in vivo-propagated primary GBM tumor cell line, was
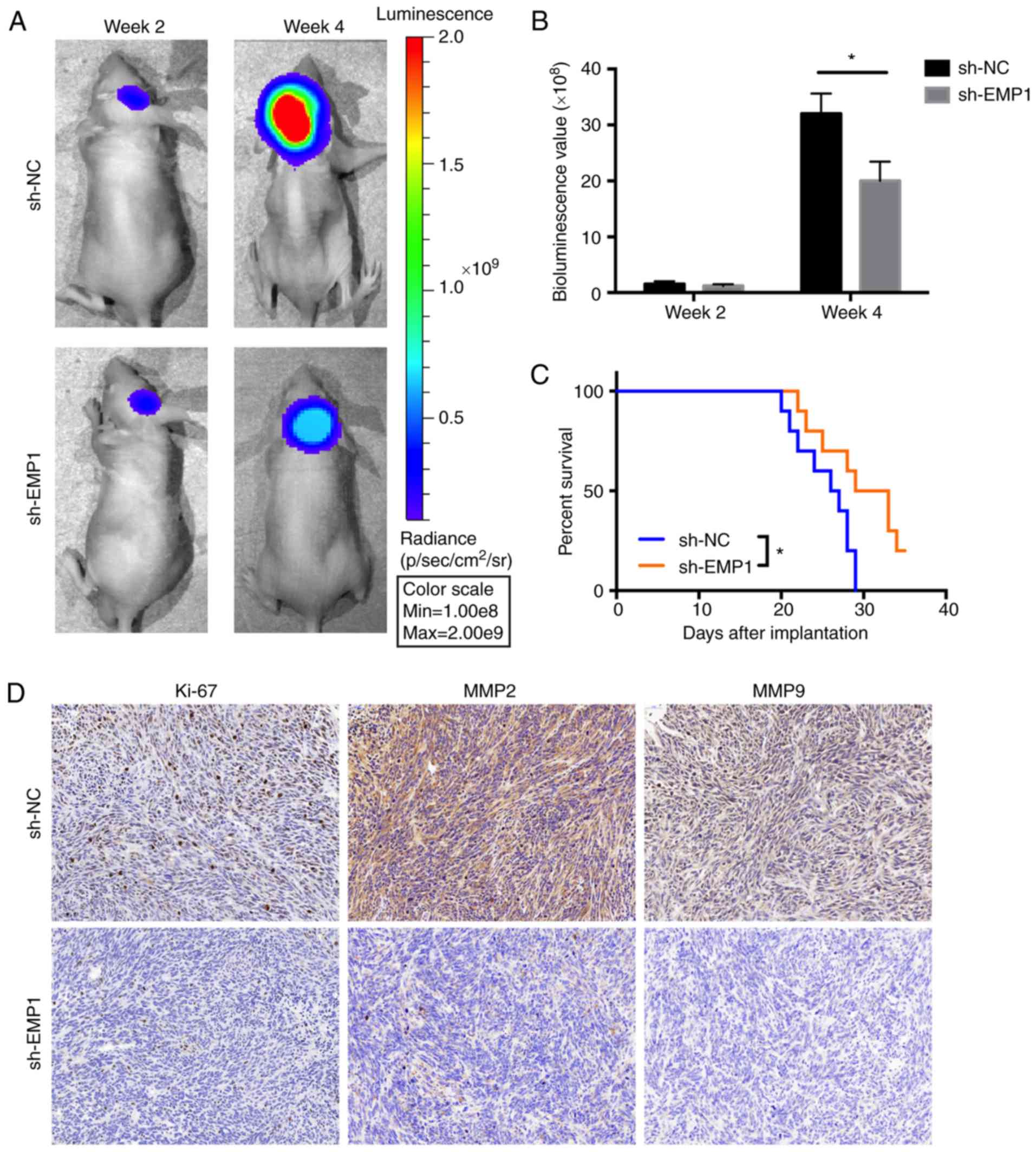
applied to investigate EMP1 function. Athymic nude mice
(n=10) were implanted with luciferase-stable P3 cells in an
intracranial tumor model and tumor growth was monitored over time
using bioluminescence values. The results demonstrated that
knockdown of EMP1 significantly reduced tumor growth
(~35×108 vs. ~20×108 photons/sec, sh-NC vs.
sh-EMP1; Fig. 6A and B). Kaplan-Meier
analysis of the survival data demonstrated a statistically
significant difference between sh-NC and sh-EMP1 mice (P<0.05,
~29 vs. >32 days, sh-NC vs. sh-EMP1; Fig. 6C). Immunohistochemistry (IHC) was
performed on tissue sections from animals to examine proliferation
and invasion. Ki-67, a marker for proliferation, MMP2 and MMP9,
markers for invasion were decreased after EMP1 knockdown (Fig. 6D).
Discussion
Molecular-targeting therapy has become a promising
therapeutic strategy for extending the survival time of cancer
patients. Therefore, identifying novel therapeutic targets is
critical for the design of more effective tumor specific strategies
(15). Recently, several members of
the EMP family have been indicated to participate in cancer
progression (16–19). For instance, it has been reported that
EMP1 is an oncogene of resistance to gefitinib in lung cancer, and
it contributes to prednisolone resistance in patients with acute
lymphoblastic leukemia. EMP2 was reported to be a biomarker in
endometrial and ovarian cancer patients. Overexpression of EMP3 in
breast cancer is significantly associated with high expression
level of HER-2 which is one of the most important biomarkers of
progression and metastasis-free survival for urothelial carcinoma
of the upper urinary tract patients (19). Ramnarain et al revealed that
the expression of EGFRvIII resulted in upregulation of a small
group of genes including EMP1 in glioma cell lines (20). Bredel et al revealed that EMP1
is one of the novel MYC-responsive genes in gliomas (10). These studies indicated that EMP1 may
be an oncogene in GBM, however, the biological role and underlying
mechanism of EMP1 in GBM remains unclear.
In the present study, EMP1 was identified as a
potential target of GBM molecular-targeting therapy. According to
the mRNA microarray of TCGA, Rembrandt and CGGA, it was revealed
that the mRNA level of EMP1 was increased in glioma compared to
normal brain tissues. The IHC and western blot results of GBM or
normal brain tissues further verified this view. Moreover, glioma
patients with low EMP1 expression level had improved overall
survival. Collectively, these data indicated that EMP1 could be
associated with the malignancy of GBM and may serve as a novel
prognostic indicator in clinical practice.
Abnormal cell proliferation, migration and invasion
are hallmark characteristics of human gliomas. Many genetic changes
lead to uncontrolled growth through dysregulation of proteins
directly involved in cell cycle progression and cell invasion
(21). GO analyses revealed that
EMP1-assosiated genes exhibited significant enrichment mainly in
processes related to cell proliferation, adhesion and extracellular
matrix organization. The in vitro and in vivo data
supported this analysis. EMP1 knockdown decreased the
proliferation, migration and invasion in glioma cells and reduced
tumor growth in orthotopic xenografts. Inhibition of cell
proliferation can be the cause of senescence, apoptosis and
autophagic cell death. Recent studies revealed that EMP1 also
participated in nasopharyngeal and gastric cancer cell apoptosis
(9,22). Therefore, the cross-talk between EMP1
and senescence, apoptosis and autophagic cell death requires
further exploration.
Glioma progression is a dynamic process in which the
PI3K/AKT signaling pathway is a key event driving abnormal
proliferation, differentiation and invasion of tumor cells
(23). Mutations in the PTEN
tumor-suppressor gene, a key regulator of the PI3K/AKT signaling
pathway, lead to misaligned pathways in tumor cells. Results of
KEGG analysis revealed that EMP1-associated upregulated genes were
mainly enriched in the PI3K-AKT pathway. In the present in
vitro study, the expression levels of p-AKT and p-mTOR were
both decreased in EMP1-knockdown GBM cells. However, in the
presence of an AKT activator, SC79, p-AKT and p-mTOR levels as well
as functional activities including proliferation, migration, and
invasion were partially restored. In conclusion, the PI3K/AKT/mTOR
signaling pathway provides a molecular basis for the inhibition of
tumor growth through EMP1 knockdown. However, the precise molecular
mechanisms of the cross-talk between EMP1 and PI3K/AKT/mTOR
signaling in gliomas require further investigation.
In summary, EMP1 facilitates proliferation,
migration and invasion of GBM cells both in vitro and in
vivo potentially by activation of the PI3K/AKT/mTOR signaling
pathway. These results raise the possibility that targeting EMP1
may represent a promising strategy for the treatment of GBM.
Acknowledgements
The authors thank Professor Rolf Bjerkvig for
providing human fibroblast glioblastoma cell line T98, primary
human GBM biopsy xenograft propagated tumor cells P3 and normal
human astrocytes, and Dr Janice Nigro for critical comments on the
manuscript.
Funding
The present study was supported by the Natural
Science Foundation of China Grants (81572487, 81402060 and
81472353), the Special Foundation for Taishan Scholars (ts20110814,
tshw201502056 and tsqn20161067), the Department of Science and
Technology of Shandong Province (2015ZDXX0801A01 and 2014kjhm0101),
the Shandong Province Natural Science Foundation (ZR2014HM074), the
Shandong Provincial Outstanding Medical Academic Professional
Program, the Health and Family Planning Commission of Shandong
province (2017WS673), the Fundamental Research Funds of Shandong
University (2016JC019), the University of Bergen and the K.G.
Jebsen Brain Tumor Research Centre.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XL and PZ conceived and designed the experiments;
LM, ZJ, JW, NY, QQ, WZ and ZF performed the experiments; LM and ZJ
analyzed the data; WL, QZ, BH, AC and DZ contributed to the
reagents/materials/analysis tools. All authors were involved in the
writing of the manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the
Institutional Animal Care and Use Committee (IACUC) of Shandong
University (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kalpathy-Cramer J, Gerstner ER, Emblem KE,
Andronesi OC and Rosen B: Advanced magnetic resonance imaging of
the physical processes in human glioblastoma. Cancer Res.
74:4622–4637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van den Bent M, Chinot OL and Cairncross
JG: Recent developments in the molecular characterization and
treatment of oligodendroglial tumors. Neuro Oncol. 5:128–138. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garrido W, Rocha JD, Jaramillo C,
Fernandez K, Oyarzun C, San Martin R and Quezada C: Chemoresistance
in high-grade gliomas: Relevance of adenosine signalling in
stem-like cells of glioblastoma multiforme. Curr Drug Targets.
15:931–942. 2014.PubMed/NCBI
|
|
5
|
Lee CY: Strategies of temozolomide in
future glioblastoma treatment. Onco Targets Ther. 10:265–270. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uribe D, Torres A, Rocha JD, Niechi I,
Oyarzún C, Sobrevia L, San Martín R and Quezada C: Multidrug
resistance in glioblastoma stem-like cells: Role of the hypoxic
microenvironment and adenosine signaling. Mol Aspects Med.
55:140–151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bangsow T, Baumann E, Bangsow C, Jaeger
MH, Pelzer B, Gruhn P, Wolf S, von Melchner H and Stanimirovic DB:
The epithelial membrane protein 1 is a novel tight junction protein
of the blood-brain barrier. J Cereb Blood Flow Metab. 28:1249–1260.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aries IM, Jerchel IS, van den Dungen RE,
van den Berk LC, Boer JM, Horstmann MA, Escherich G, Pieters R and
den Boer ML: EMP1, a novel poor prognostic factor in pediatric
leukemia regulates prednisolone resistance, cell proliferation,
migration and adhesion. Leukemia. 28:1828–1837. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun G, Zhao G, Lu Y, Wang Y and Yang C:
Association of EMP1 with gastric carcinoma invasion, survival and
prognosis. Int J Oncol. 45:1091–1098. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bredel M, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Qi Q, Feng Z, Zhang X, Huang B,
Chen A, Prestegarden L, Li X and Wang J: Berberine induces
autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway.
Oncotarget. 7:66944–66958. 2016.PubMed/NCBI
|
|
12
|
Fack F, Espedal H, Keunen O, Golebiewska
A, Obad N, Harter PN, Mittelbronn M, Bähr O, Weyerbrock A, Stuhr L,
et al: Bevacizumab treatment induces metabolic adaptation toward
anaerobic metabolism in glioblastomas. Acta Neuropathol.
129:115–131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fack F, Tardito S, Hochart G, Oudin A,
Zheng L, Fritah S, Golebiewska A, Nazarov PV, Bernard A, Hau AC, et
al: Altered metabolic landscape in IDH-mutant gliomas affects
phospholipid, energy, and oxidative stress pathways. EMBO Mol Med.
9:1681–1695. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tardito S, Oudin A, Ahmed SU, Fack F,
Keunen O, Zheng L, Miletic H, Sakariassen PØ, Weinstock A, Wagner
A, et al: Glutamine synthetase activity fuels nucleotide
biosynthesis and supports growth of glutamine-restricted
glioblastoma. Nat Cell Biol. 17:1556–1568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rajesh Y, Pal I, Banik P, Chakraborty S,
Borkar SA, Dey G, Mukherjee A and Mandal M: Insights into molecular
therapy of glioma: Current challenges and next generation
blueprint. Acta Pharmacol Sin. 38:591–613. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsieh YH, Hsieh SC, Lee CH, Yang SF, Cheng
CW, Tang MJ, Lin CL, Lin CL and Chou RH: Targeting EMP3 suppresses
proliferation and invasion of hepatocellular carcinoma cells
through inactivation of PI3K/Akt pathway. Oncotarget.
6:34859–34874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai S, Wang G, Cao X, Li Z, Hu J and Wang
J: EMP-1 promotes tumorigenesis of NSCLC through PI3K/AKT pathway.
J Huazhong Univ Sci Technolog Med Sci. 32:834–838. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang HT, Kong JP, Ding F, Wang XQ, Wang
MR, Liu LX, Wu M and Liu ZH: Analysis of gene expression profile
induced by EMP-1 in esophageal cancer cells using cDNA Microarray.
World J Gastroenterol. 9:392–398. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang YW, Cheng HL, Ding YR, Chou LH and
Chow NH: EMP1, EMP 2, and EMP3 as novel therapeutic targets in
human cancer. Biochim Biophys Acta. 1868:199–211. 2017.
|
|
20
|
Ramnarain DB, Park S, Lee DY, Hatanpaa KJ,
Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et
al: Differential gene expression analysis reveals generation of an
autocrine loop by a mutant epidermal growth factor receptor in
glioma cells. Cancer Res. 66:867–874. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun GG, Lu YF, Fu ZZ, Cheng YJ and Hu WN:
EMP1 inhibits nasopharyngeal cancer cell growth and metastasis
through induction apoptosis and angiogenesis. Tumour Biol.
35:3185–3193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manning BD and Toker A: AKT/PKB signaling:
Navigating the network. Cell. 169:381–405. 2017. View Article : Google Scholar : PubMed/NCBI
|