Introduction
Cancer has contributed to the rising rate in
disease-associated mortality in the last few decades. The demand
for early-stage diagnosis, prognosis and therapeutic targets is
increasing. The serum deprivation response (SDPR) gene has
been characterized to serve a critical role in breast cancer as a
tumor suppressor (1,2), and recently showing a characteristic
gene signature in specific types of cancer, including oral cancer,
thyroid cancer and liposarcoma (3–5).
SDPR localizes to chr2q 32–33, also known as
caveolae associated protein 2, and has been known for its role in
caveolae formation (6–8). Its encoding protein SDPR, which is
overexpressed in serum starved cells, was firstly identified as a
substrate for protein kinase C (PKC) phosphorylation, an
interaction that targets PKC in caveolae formation (9). Caveolae are plasma membrane microdomains
involved in multiple biological processes, including lipid
metabolism, endocytosis, cellular signal transduction, cell
proliferation and migration (7,10).
Previous studies have gradually implicated the
differential expression and tumor suppressor function of
SDPR in cancer progression and metastasis; reduced
SDPR expression has been observed in breast (1,2,11–13),
kidney (11,14) and prostate (11,15)
cancer. In breast cancer, SDPR was identified as a tumor suppressor
(1,2).
SDPR serves an anti-metastatic function by promoting apoptosis
(1), and depletion of SDPR induced
epithelial-mesenchymal transition (EMT) through transforming growth
factor-β (TGF-β) signaling activation (2). The reduction of SDPR expression
has been reported to be associated with significantly reduced
survival in patients with breast cancer, who underwent therapy
(1).
In addition to breast cancer, it has been suggested
that SDPR may serve as a tumor suppressor gene, with a
broader clinical relevance, in other types of cancer. In oral
cancer, it was identified that SDPR-negative patients had
high tumor progression (5), whereas
in sarcoma (SARC), a lower SDPR expression was observed in
more aggressive or dedifferentiated tumor forms (4). In addition, it was suggested that the
differently expressed SDPR gene can be used as a possible
diagnostic marker to discriminate malignant tumors from benign
formations, not only in serum from patients with kidney tumors
(14), but also in follicular thyroid
carcinomas (3). Nevertheless, gene
expression alterations of SDPR in various types of cancer
and their relevance to clinical outcomes remain unclear.
In the present study, mRNA levels of SDPR
were compared in various unique tumor tissue datasets compared with
normal tissue datasets, indicating that SDPR was
downregulated in various types of cancer. In addition,
downregulated SDPR was found to be significantly associated
with the survival of the patients, not only in previously reported
breast cancer cases, but also in brain, lung and soft tissue
tumors. However, SDPR failed to emerge as a frequent target
for gene mutational inactivation in a previous next-generation
sequencing study (16). As
SDPR has been reported to be hypermethylated and silenced in
breast cancer cell lines, it was suggested that it is likely to be
epigenetically inactivated in cancer (1). In order to examine the mechanism
underlying SDPR downregulation in cancer, the present study
analyzed and found SDPR gene methylation alteration between
cancer and normal tissues. Furthermore, the present study
investigated the methylation sites relevant to the survival of
patients with lung adenocarcinoma (LUAD) and SARC. In addition, the
potential transcription factor binding sites of the SDPR
promoter were analyzed. The potential transcription factor GATA
binding protein 2 (GATA2) was identified from the analysis of genes
that are co-expressed with SDPR. The results of the present
study provide additional insight in understanding the underlying
molecular mechanisms of SDPR in cancer, in addition to
revealing a novel approach in the clinical prognostic evaluation
and treatment of LUAD and SARC.
Materials and methods
Gene expression analysis using
oncomine platform
The profile of SDPR gene expression level in
various types of cancer was identified in Oncomine™ Platform and
the detailed datasets are available online (https://www.oncomine.org/) (17). By comparing with normal tissues, the
mRNA expression-fold of SDPR in cancer tissues was obtained
using the parameters of P-value <10−4 or 0.01,
fold-change >2, and gene ranking in the top 10%. To adjust the
false discovery rate, the P-values were corrected by using the
Benjamini-Hochberg procedure (B-H method) (18) in R, version 3.5.0 (https://www.r-project.org/).
Prognoscan database analysis
The association between SDPR expression and
survival in different types of cancer was analyzed using the
PrognoScan database (http://www.abren.net/PrognoScan/) (19), and presented as a Kaplan-Meier plot,
in which survival curves for high (red) and low (blue) expression
groups dichotomized at the optimal cut-point were plotted. The
P-values were adjusted for multiple correlation testing using the
Miller and Siegmund formula (20),
according to Prognoscan database and shown as a corrected P-value
(Pcor). The threshold was adjusted to corrected P-values
at <0.05.
MethHC database analysis
The MethHC database was used in the analysis of
SDPR DNA methylation alternation in cancer. MethHC
(http://MethHC.mbc.nctu.edu.tw/) is a
systematic database integrating DNA methylation data from The
Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/abouttcga/policies/informedconsent),
which includes >6,000 DNA methylation data generated by Illumina
HumanMethylation450K BeadChip in 18 types of cancer. The
methylation status of DNA was represented as β-values (0–1)
(21), and the average β-value of
SDPR was presented as a boxplot by comparing the transcript
expression in tumor samples and matched normal samples in all 18
types of cancer. To adjust the false discovery rate, the P-values
were corrected using the B-H method in R software.
Methsurv database analysis
Methsurv database (https://biit.cs.ut.ee/methsurv) was utilized for
survival analysis in different types of cancer based on SDPR
methylation patterns. In Methsurv, the gene methylation data was
from TCGA Genome Data Analysis Center Firehose (http://gdac.broadinstitute.org/) (22), using the HM450K array, which covers
486,428 CpGs. The methylation status of DNA was represented as
β-values (0–1) (23). The methylation
pattern was annotated by probes indicating subregions of the query
gene, according to the annotation file (Human genome build 27)
provided by Illumina [TSS to-200 nucleotides upstream of TSS
(‘TS200’); covering-200 to-1500 nucleotides upstream of TSS
(‘TSS1500’; first exon (‘1st exon’); ‘5′UTR’, ‘body’ and ‘3′UTR’].
Clustering analysis was plotted and visualized using a heatmap by
integrating Methsurv settings with ClustVis (https://biit.cs.ut.ee/clustvis/) (24). The survival analysis of each type of
cancer between the low-methylated and the high-methylated groups in
specific methylation sites was visualized using Kaplan-Meier plots.
Multivariable survival analysis was performed using a Cox
proportional-hazards model. Age and sex were used as covariates in
the multivariable prediction models. The hazard ratio (HR) with 95%
CI was derived from Cox fitting. The goodness-of-fit of the Cox
proportional hazard model was assessed using a likelihood-ratio
(LR) test, and presented as an LR P-value. The methylation status
of SDPR in different LUAD clinical stage samples was shown
as violin plots after grouping samples according to stage.
Identification of potential
transcription factor for SDPR
The transcription start site (TSS) of SDPR
was indicated by the University of California, Santa Cruz (UCSC)
Genome Browser (http://genome.ucsc.edu), and the DNA sequence from
2,000 bp nucleotides upstream and 500 bp nucleotides downstream of
the TSS was used as a potential promoter sequence for SDPR.
PROMO at the ALGGEN server was subsequently used to identify the
putative transcription factor binding sites in this promoter
sequence (25). The co-expression
profiles of the SDPR gene in LUAD were identified and
presented as the pattern of a heat map using the Oncomine database
(17), in which the node correlation
value is computed as the average of all pair-wise correlations
among genes. The node correlation value >0.5 was used to define
significant co-expressing genes. Finally, the intersection of the
above two profiles was investigated in order to identify the
potential transcription factor regulating SDPR
expression.
Results
Downregulation of SDPR in various
types of cancer
The expression of SDPR is nearly ubiquitous
in normal tissues and increased expression levels have been
reported in the heart and lungs, while lower expression levels have
been indicated in the kidney, the brain, the pancreas, skeletal
muscle and the liver (8). To explore
the gene expression alteration of SDPR in tumor tissues, the
present study compared the mRNA levels of SDPR in various
unique tumor tissue datasets compared with normal tissue datasets
from the Oncomine™ Platform (17).
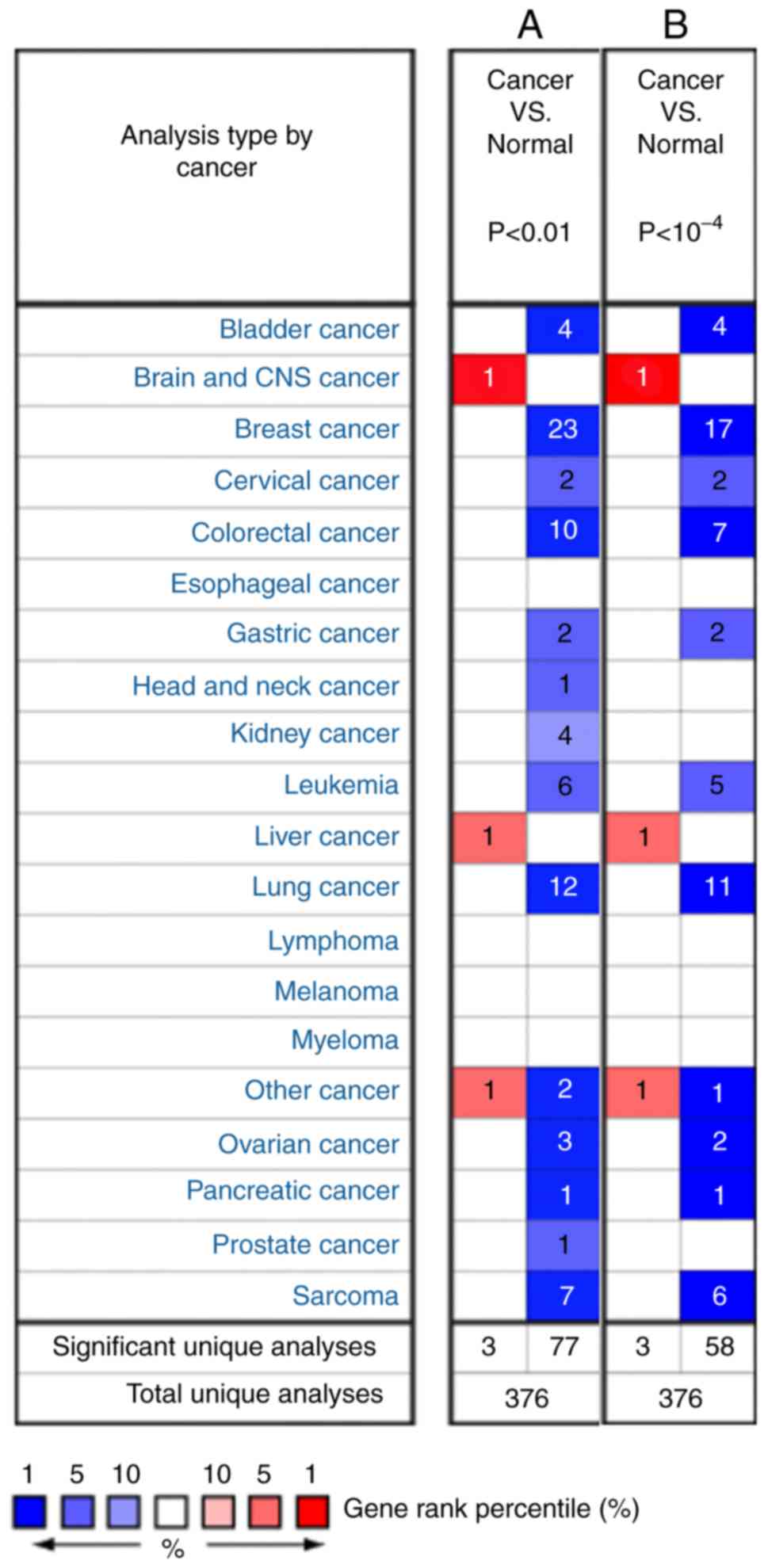
Consistent with previous studies, the analysis showed that the
expression of SDPR was downregulated in breast (1,11–13), kidney (11,14) and
prostate cancer (11,15), and SARC (4,26). In
addition, the present results identified that SDPR is
downregulated in bladder, lung, cervical, colorectal, gastric,
ovarian and pancreatic cancer (Fig.
1).
Furthermore, gene expression levels of SDPR
in cancer subtypes were estimated as a fold-change and gene rank
(Table I). The fold change was
from-44.083 (invasive ductal breast carcinoma) to-2.102 (cervical
squamous cell carcinoma). The gene rank was between 6 and 1% (in
the top %). For instance, the entire subtype dataset of breast
cancer showed 1% downregulated gene ranks. Downregulated gene
expression of SDPR was also observed in lung cancer (in top
3–1%), pancreatic cancer (in top 1%) and SARC (in top 4–1%).
 | Table I.SDPR expression in cancer. |
Table I.
SDPR expression in cancer.
| A, Bladder
cancer |
|---|
|
|---|
| Author, year | Cancer subtype | P-value | Fold change | Gene rank (%) | Samples (n) | (Refs.) |
|---|
| Sanchez-Carbayo
et al, 2006 | Superficial bladder
cancer | <0.001 | −6.264 | 1 | 28 | (34) |
| Kim et al,
2010 | Superficial bladder
cancer | <0.001 | −3.076 | 3 | 126 | (35) |
| Sanchez-Carbayo
et al, 2006 | Infiltrating
bladder urothelial carcinoma | <0.001 | −3.517 | 2 | 81 | (34) |
| Kim et al,
2010 | Infiltrating
bladder urothelial carcinoma | <0.001 | −2.315 | 6 | 62 | (35) |
|
| B, Breast
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Curtis et
al, 2012 | Invasive lobular
breast carcinoma | <0.001 | −4.857 | 1 | 148 | (12) |
| TCGA Breast,
Current study | Invasive lobular
breast carcinoma | <0.001 | −10.124 | 1 | 36 | – |
| Curtis et
al, 2012 | Tubular breast
carcinoma | <0.001 | −5.474 | 1 | 67 | (12) |
| Curtis et
al, 2012 | Medullary breast
carcinoma | <0.001 | −8.162 | 1 | 32 | (12) |
| Curtis et
al, 2012 | Invasive ductal
breast carcinoma | <0.001 | −44.083 | 1 | 1,556 | (12) |
| TCGA Breast,
Current study | Invasive ductal
breast carcinoma | <0.001 | −25.347 | 1 | 389 | – |
| Curtis et
al, 2012 | Mucinous breast
carcinoma | <0.001 | −5.711 | 1 | 46 | (12) |
|
| C, Cervical
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Biewenga et
al, 2008 | Cervical squamous
cell carcinoma | <0.001 | −2.102 | 5 | 40 | (36) |
| Scotto et
al, 2008 | Cervical squamous
cell carcinoma | <0.001 | −2.123 | 3 | 32 | (37) |
|
| D, Colorectal
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Skrzypczak et
al, 2010 | Colorectal
adenocarcinoma | <0.001 | −3.48 | 4 | 45 | (38) |
| Ki et al,
2007 | Colon
adenocarcinoma | <0.001 | −2.413 | 1 | 50 | (39) |
|
| E, Gastric
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| D'Errico et
al, 2009 | Gastric intestinal
type adenocarcinoma | <0.001 | −2.126 | 3 | 26 | (40) |
| Cho et al,
2011 | Diffuse gastric
adenocarcinoma | <0.001 | −4.626 | 4 | 31 | (41) |
|
| F,
Leukemia |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Haferlach et
al, 2010 | Pro-B acute
lymphoblastic | <0.001 | −4.65 | 3 | 70 | (42) |
| Haferlach et
al, 2010 | Acute myeloid
leukemia | <0.001 | −2.442 | 4 | 542 | (42) |
| Haferlach et
al, 2010 | B-cell acute
lymphoblastic Leukemia | <0.001 | −4.513 | 4 | 147 | (42) |
| Haferlach et
al, 2010 | T-Cell acute
lymphoblastic leukemia | <0.001 | −4.366 | 4 | 174 | (42) |
| Haferlach et
al, 2010 | B-cell childhood
acute lymphoblastic Leukemia | <0.001 | −4.187 | 5 | 359 | (42) |
|
| G, Lung
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Garber et
al, 2001 | Lung
aenocarcinoma | <0.001 | −9.404 | 2 | 40 | (43) |
| Okayama et
al, 2012 | Lung
aenocarcinoma | <0.001 | −5.976 | 1 | 226 | (44) |
| Selamat et
al, 2012 | Lung
aenocarcinoma | <0.001 | −6.617 | 1 | 58 | (45) |
| Su et al,
2007 | Lung
aenocarcinoma | <0.001 | −3.803 | 2 | 27 | (46) |
| Landi et al,
2008 | Lung
aenocarcinoma | <0.001 | −4.783 | 2 | 58 | (47) |
| Hou et al,
2010 | Lung
aenocarcinoma | <0.001 | −7.204 | 3 | 45 | (48) |
| Garber et
al, 2001 | Squamous cell lung
carcinoma | <0.001 | −11.994 | 1 | 13 | (43) |
| Wachi et al,
2005 | Squamous cell lung
carcinoma | <0.001 | −5.833 | 1 | 5 | (49) |
| Hou et al,
2010 | Squamous cell lung
carcinoma | <0.001 | −8.845 | 2 | 27 | (48) |
| Garber et
al, 2001 | Large cell lung
carcinoma | <0.001 | −9.981 | 1 | 4 | (43) |
| Crabtree et
al, 2009 | Large cell lung
carcinoma | <0.001 | −10.929 | 3 | 19 | (50) |
|
| H, Other
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Yoshihara et
al, 2009 | Uterine Corpus
Leiomyoma | <0.001 | −2.418 | 1 | 50 | (51) |
|
| I, Ovarian
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| TCGA Ovarian,
Current study | Ovarian serous
adenocarcinoma | <0.001 | −38.254 | 1 | 37 | – |
| Pei et al,
2009 | Ovarian serous
cystadenocarcinoma | <0.001 | −3.519 | 4 | 586 | (52) |
|
| J, Pancreatic
cancer |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Barretina et
al, 2010 | Pancreatic
carcinoma | <0.001 | −2.616 | 1 | 36 | (26) |
|
| K,
Sarcoma |
|
| Author,
year | Cancer
subtype | P-value | Fold
change | Gene rank
(%) | Samples
(n) | (Refs.) |
|
| Barretina et
al, 2010 | Pleomorphic
myxofibrosarcoma | <0.001 | −8.447 | 1 | 3 | (26) |
| Barretina et
al, 2010 | Pleomorphic
liposarcoma | <0.001 | −4.486 | 1 | 23 | (26) |
| Barretina et
al, 2010 | Dedifferentiated
liposarcoma | <0.001 | −4.804 | 1 | 46 | (26) |
| Barretina et
al, 2010 |
Myxofibrosarcoma | <0.001 | −6.96 | 1 | 31 | (26) |
| Barretina et
al, 2010 | Leiomyosarcoma | <0.001 | −3.417 | 2 | 26 | (26) |
| Barretina et
al, 2010 | Myxoid/round cell
liposarcoma | <0.001 | −2.673 | 4 | 20 | (26) |
SDPR gene expression alteration and
survival of patients
The PrognoScan platform, which integrates published
cancer microarray datasets with clinical annotation (19), was used in the systematic
meta-analysis and to determine the prognostic value of SDPR
in multiple datasets. Survival analysis consists of two steps,
patient grouping and comparing the determined risks of these
groups. Since gene expression is continuous data, the PrognoScan
platform employed a minimum P-value approach for grouping patients
in the survival analysis, which determines the optimal cut-off
point in the continuous gene-expression measurement.
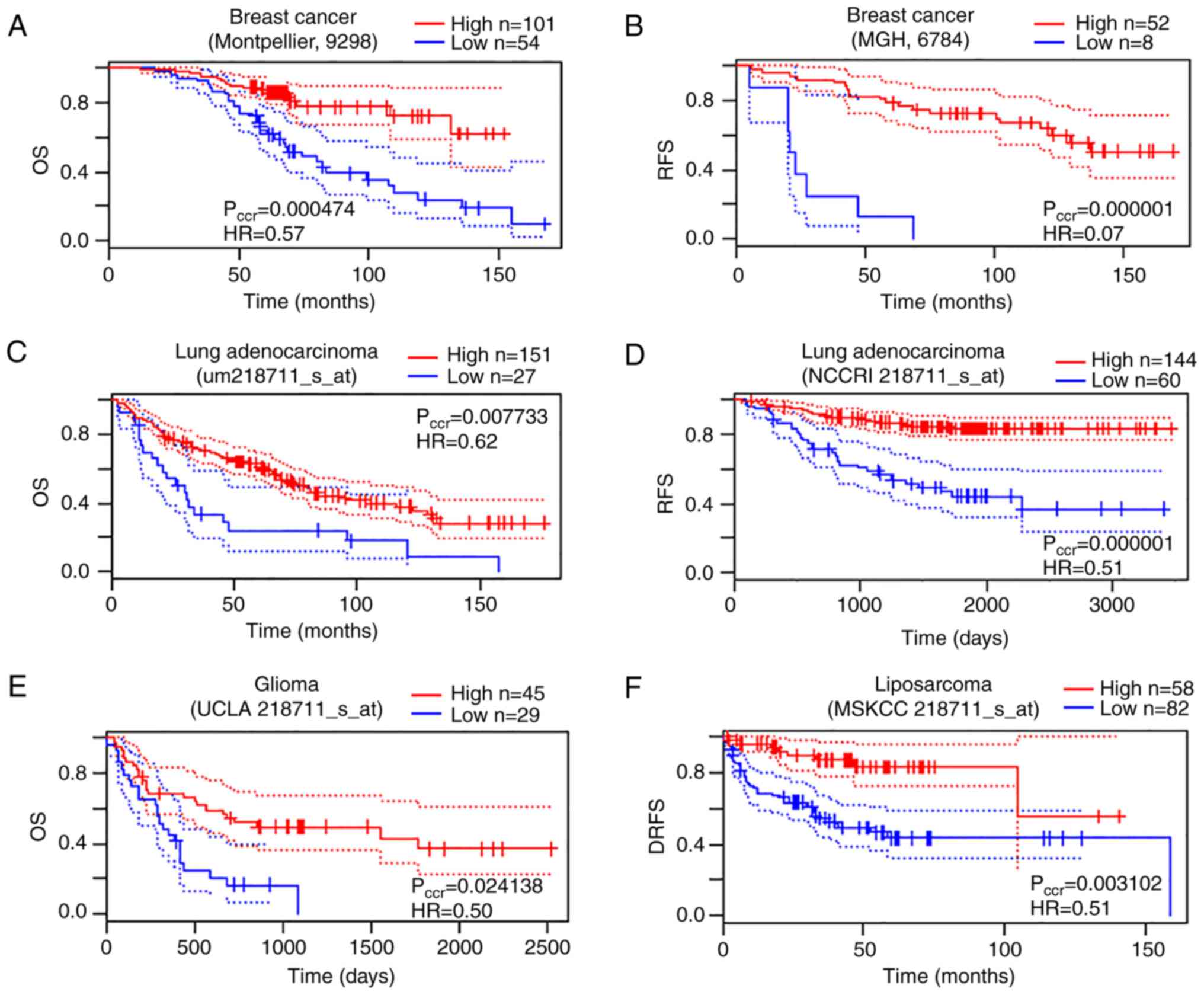
In the present study, PrognoScan indicated a
significant association between microarray SDPR expression
and cancer prognosis in several tests: Brain (1/8), breast (10/40),
lung (6/19) and soft tissue (1/1). In all these 18 tests, low
SDPR expression was associated with poor survival (data not
shown), suggesting its protective function in cancer malignancy.
Decreased SDPR expression was significantly associated with
decreased overall survival (OS) and relapse-free survival (RFS) of
patients with breast cancer (Fig. 2A and
B), consistent with a previous study (1). In addition, SDPR downregulation
was significantly associated with OS and RFS in patients with lung
cancer adenocarcinoma (Fig. 2C and
D), OS in patients with glioma (Fig.
2E) and distant-recurrence free survival (DRFS) in liposarcoma
(Fig. 2F).
SDPR is hypermethylated in specific
types of cancer
In a previous next-generation sequencing study,
SDPR was not identified as a frequent mutational gene target
(16) and it was also observed that
SDPR was epigenetically silenced in breast cancer cell lines
(1). Therefore, the present study
hypothesized that DNA methylation alteration, which is an important
epigenetic regulator for transcription, may be one of the
mechanisms for SDPR gene expression differences. In order to
reveal the underlying molecular mechanisms responsible for
SDPR downregulation, MethHC was used to identify
differential DNA methylation data between tumor and non-tumor
tissues, which included 18 human types of cancer in >6,000
samples. MethHC is a database that systematically integrates DNA
methylation and mRNA expression data from TCGA. The methylation
profile of SDPR across 18 tumors is presented in Fig. 3. In this profile, significantly
SDPR gene methylation differences were observed in most
types of cancer (17/18). Consistent with previously published
experimental data, SDPR was significantly hypermethylated in
breast invasive carcinoma compared with normal breast tissues
(1). In addition, SDPR was
observed to be significantly hypermethylated in bladder urothelial
carcinoma, cervical squamous cell carcinoma, head and neck squamous
cell carcinoma, LUAD, lung squamous cell carcinoma (LUSC),
pancreatic adenocarcinoma, prostate adenocarcinoma, SARC, skin
cutaneous melanoma, stomach adenocarcinoma and uterine corpus
endometrial carcinoma (P<0.005). Statistical differences were
also found in colon adenocarcinoma, rectum adenocarcinoma and
thyroid carcinoma (P<0.05). These results suggested that DNA
methylation may be responsible for SDPR downregulation in
these types of cancer. As shown in Fig.
1, SDPR was downregulated in kidney cancer. However,
significant SDPR hypomethylation was found in both kidney
renal clear cell carcinoma (KIRC) and kidney renal papillary cell
carcinoma (KIRP) (P<0.005), suggesting the existence of other
regulatory pathways.
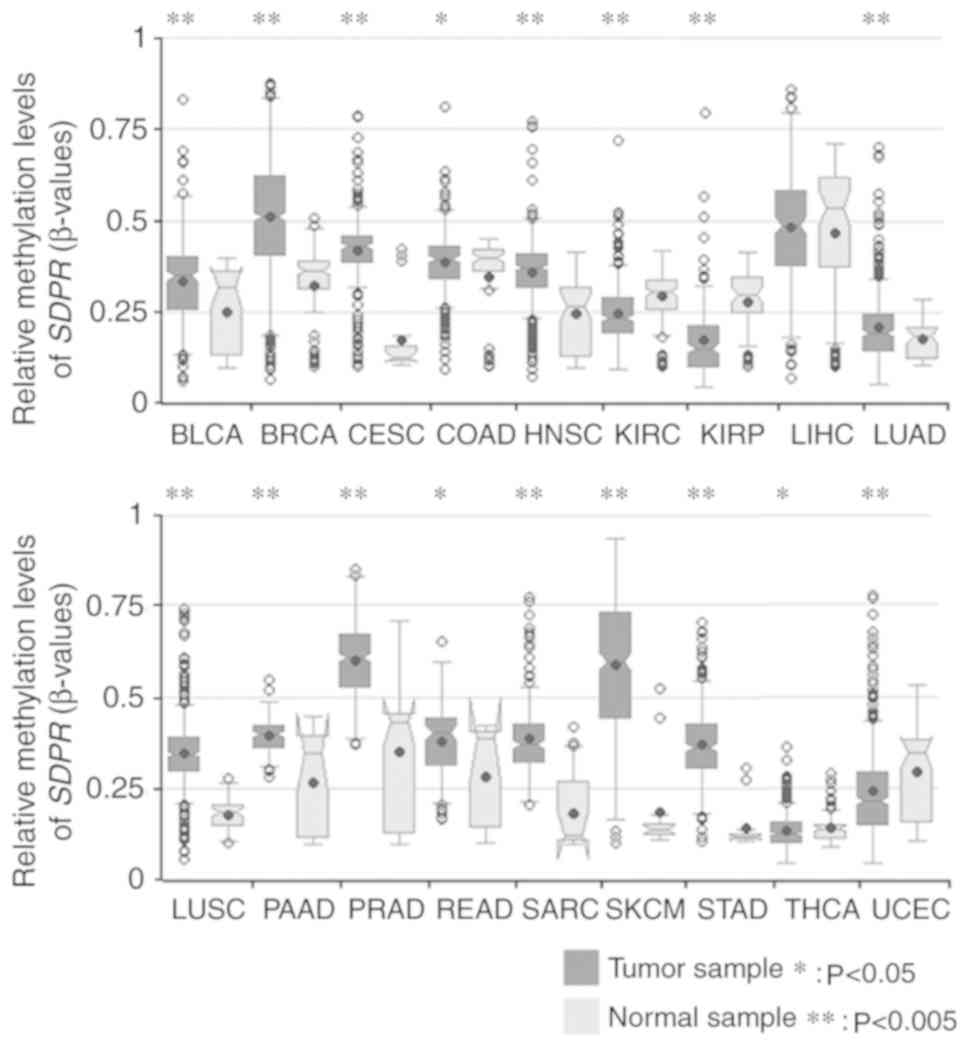 | Figure 3.Methylation profiles of SDPR
across tumors. The differential methylation statuses of each
transcript of SDPR in different tumor types are presented as
boxplots. Tumor samples are in dark grey and are compared with
normal samples in light grey. The shapes of notched boxplots
indicated the characteristics of the data in each sub-dataset.
P-values were adjusted using the Benjamini-Hochberg procedure.
*P<0.05, **P<0.0005 vs. normal sample. SDPR, serum
deprivation response; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical squamous cell carcinoma;
COAD, colon adenocarcinoma; HNSC, head and neck squamous cell
carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM,
skin cutaneous adenocarcinoma; STAD, stomach adenocarcinoma; THCA,
thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma. |
Gene methylation of SDPR is associated
with patient survival in LUAD and SARC
As downregulation of SDPR gene expression was
observed in lung cancer and SARC, SDPR was indicated to be
hypermethylated compared with normal tissues, and downregulation of
SDPR expression was significantly associated with poor
patient survival in LUAD and SARC, MethSurv was used to identify
whether hypermethylation of SDPR was associated with patient
survival in LUAD or SARC. In addition, since the differential
methylation levels at the CpG island are tissue-specific, the DNA
methylation patterns were analyzed, taking into consideration the
differential methylation levels in different gene subregions
(Fig. 4A). MethSurv was the first
database that indicated an association with overall survival and
DNA methylation patterns, in which the methylation levels are from
TCGA methylation profile using the HM450k array. Based on the UCSC
database, the CpG sites were grouped into gene subregions:
‘TSS200’, ‘TSS1500’, ‘1st exon’, ‘5′UTR’, ‘body’ and ‘3′UTR’
(23). As shown in Fig. 4B and C, clustering visualization in
the form of heat maps evaluated the association of methylation
levels with the available patient characteristics and gene
subregions in LUAD and SARC. According to the heatmaps, methylation
sites cg18843739, cg10082589 and cg17809945 in the ‘TSS1500’
subregion showed differential methylation levels in patients with
LUAD (461 samples). Similar to patients with SARC, the differently
methylated sites were cg07488576 in the ‘1st Exon’ subregion and
cg18843739 in the ‘TSS1500’ subregion (261 samples).
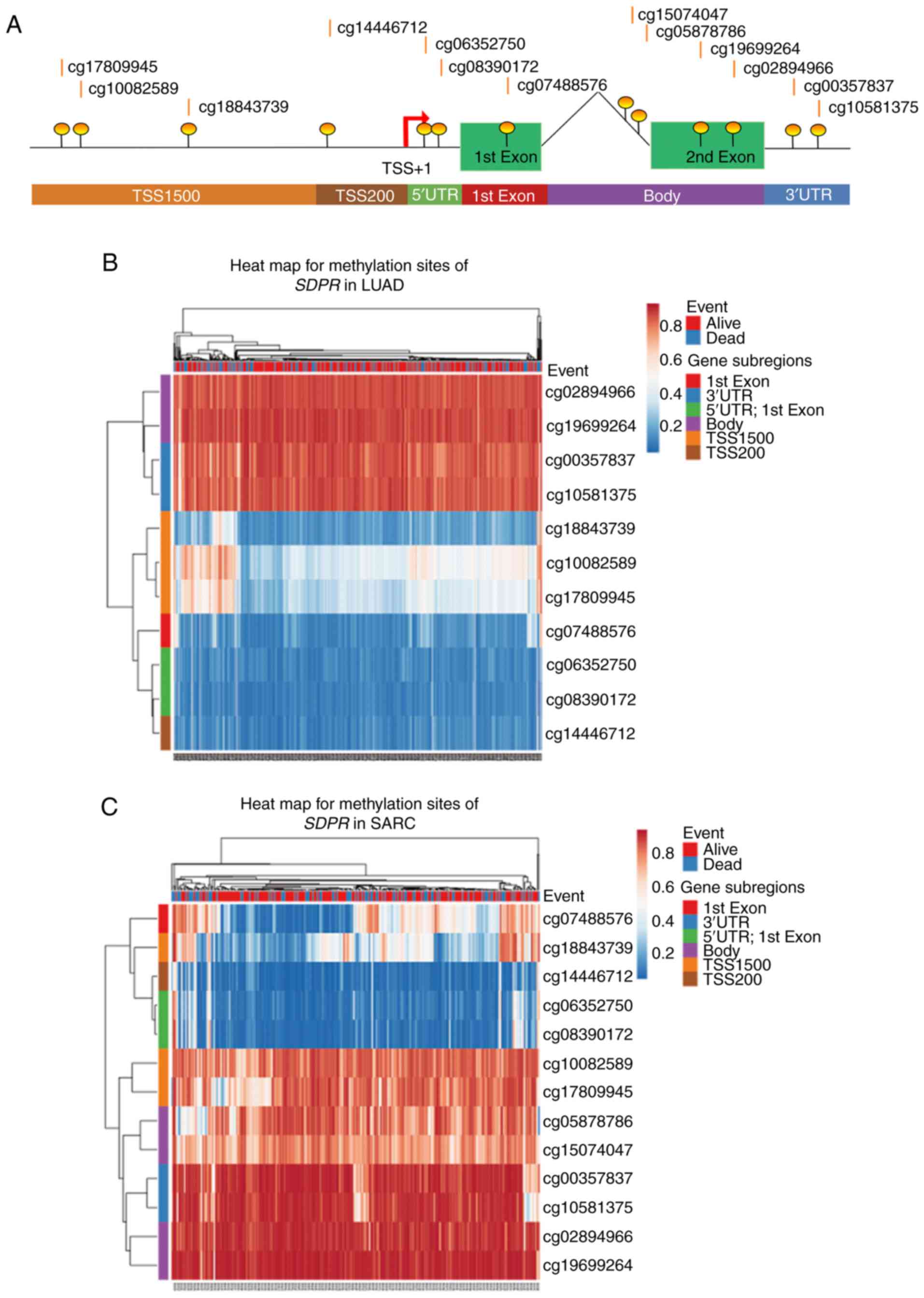 | Figure 4.Gene methylation of SDPR is
associated with patient survival in LUAD and SARC. (A) Diagram
showing the relative location from the TSS for each CpG site of
SDPR in Methsurv. Heat map depicting clustering analysis of the CpG
methylation levels within the SDPR gene in (B) LUAD and (C)
SARC. Methylation levels (0 represents fully unmethylated and 1
represents fully methylated) are shown as a continuous variable
from blue to red color. Kaplan-Meier plot showed survival curve in
higher (red) and lower (blue) methylation groups in methylation
sites (D) cg17809945 and (E) cg10082859 in LUAD, and (F) cg07488576
and (G) cg18843739 in SARC. Violin plots showing the methylated
levels of (H) cg17809945 and (I) cg10082859 among stage I, II, III
and IV LUAD samples. The interquartile range and median methylation
levels are shown in each violin plot as boxplots. SDPR,
serum deprivation response; LUAD, lung adenocarcinoma; SARC,
sarcoma; TSS, transcription start site; TSS1500, covering-200
to-1500 nucleotides upstream of TSS; TSS200, TSS to-200 nucleotides
upstream of TSS; UTR, untranslated region; HR, hazard ratio; LR,
log-likelihood ratio. |
MethSurv was used to identify which methylation
sites were significantly associated with patient survival. By
analyzing all 11 methylation sites in LUAD hypermethylation, it was
indicated that two sites in LUAD (cg17809945 and cg10082589,
located in the ‘TSS1500’ subregion of SDPR) were
significantly associated with poor overall survival (Fig. 4D and E). In SARC, 2 out of 13
methylation sites (cg07488576 in the ‘1st Exon’ subregion and
cg18843739 in the ‘TSS1500’ subregion) were identified to be
associated with poor survival (Fig. 4F
and G). The results were consistent with the heatmap presented
in Fig. 4B and C. In this analysis,
CpG (cg10082589; HR=1.887; 95% CI: 1.371–2.596; LR test
P-value=0.001) was identified as the optimal survival methylation
marker for LUAD (Fig. 4E) and CpG
(cg07488576; HR=2.406; 95% CI: 1.608–3.599; LR test
P-value=0.00001) was identified as the optimal survival methylation
marker for SARC (Fig. 4F). In
addition, when LUAD samples were grouped according to clinical
stage, the methylation levels of cg17809945 (Fig. 4H) and cg10082589 (Fig. 4I) were indicated to be higher in late
stages, suggesting that they may be involved in LUAD progression.
However, no association between methylation levels of CpG sites and
clinical stages was observed in SARC (data not shown).
Prediction of transcription factors
regulating SDPR expression in LUAD
Since epigenetic patterns may not be the only reason
for gene expression alternations in cancer, transcriptional
regulation by deregulated transcription factors was investigated.
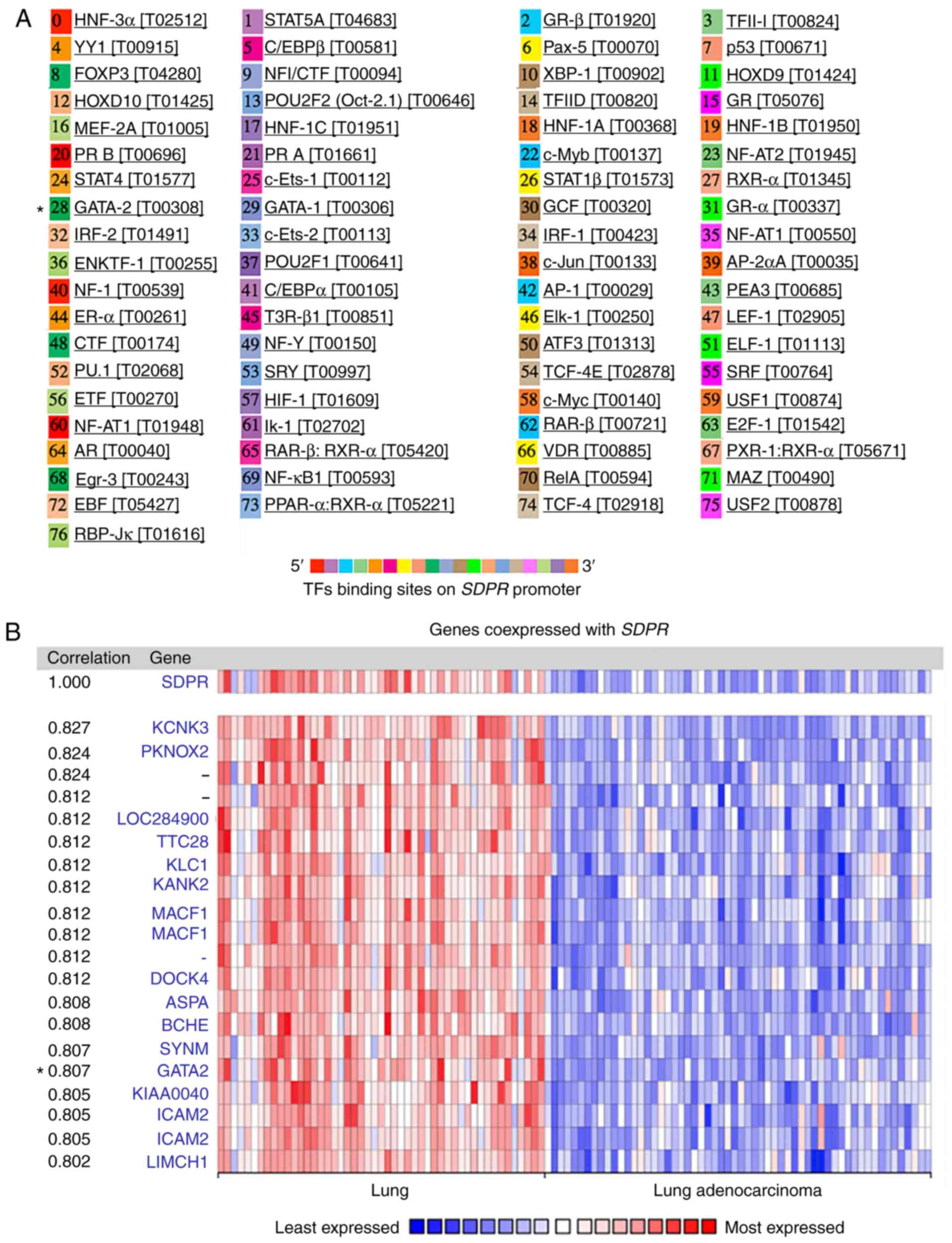
The UCSC database was used to identify the promoter sequences of
SDPR and PROMO was subsequently used at the ALGGEN server
showing potential transcriptional factors on the promoter (Fig. 5A). Meanwhile, genes that were
co-expressed with SDPR were analyzed using the Oncomine
database, which were subsequently grouped into normal lung tissue
and LUAD (Fig. 5B). By comparing the
potential transcriptional factors with genes significantly
co-expressed with SDPR (node correlation value >0.5),
GATA binding protein 2 (GATA2; node correlation value=0.807)
was identified as a potential transcription factor. GATA2 is a
member of the GATA family, serves as a transcriptional activator
during development and carcinogenesis, and was indicated to be
epigenetically repressed in both human and mouse lung tumors
(27). This result suggested that
GATA2 may be a potential transcription factor regulating
SDPR gene expression in LUAD.
Discussion
SDPR has been previously reported to show
characteristic gene signatures in specific types of cancer,
including breast (1,2,11–13), thyroid (3,28), oral
(5) and kidney (11,14)
cancer, and SARC (4,26). In breast cancer, SDPR was
identified as a novel tumor suppressor, which was significantly
associated with patient survival (1,2). However,
a systemic profile of SDPR alterations or analysis of its
relevance in clinical outcomes in different types of cancer has yet
to be performed, to the best of our knowledge. In the present
study, SDPR downregulation was observed in bladder, breast,
lung, kidney, cervical, colorectal, gastric, ovarian and pancreatic
cancer. Consistent with previous studies (1,2),
SDPR downregulation was significantly associated with
patient survival in breast cancer. Furthermore, the analysis also
indicated that decreased expression of SDPR was
significantly associated with poor OS and RFS in patients with
adenocarcinoma of lung cancer, OS in patients with glioma and DRFS
in liposarcoma.
In breast cancer, SDPR, which is partially
silenced by DNA methylation, has been elucidated to execute an
anti-metastatic function by promoting apoptosis (1), and depletion of SDPR-induced EMT
through TGF-β signaling activation, according to previously
published experimental studies (1,2). To
clarify why SDPR expression was altered in other types of
cancer, the gene methylation level of SDPR was subsequently
profiled in 18 types of cancer and was compared with normal
tissues. It was indicated to be significantly altered in 17 out of
18 types of cancer.
If DNA methylation of SDPR is involved in its
downregulation in LUAD or SARC, it might be associated with patient
survival. As not all of the methylation sites are responsible for
gene expression, the most significant methylation sites require
further investigation. Therefore, the present study analyzed the
DNA methylation patterns in LUAD and SARC, considering the
differential methylation levels in different gene subregions of
SDPR. To the best of our knowledge, the present study for
the first time, indicated that CpG (cg10082589) serves as the
optimal survival methylation marker for LUAD, and CpG (cg07488576)
serves as the optimal survival methylation marker for SARC.
Furthermore, when LUAD samples were grouped according to clinical
stage, the methylation level of CpG (cg10082589) and CpG
(cg1780995) were higher in late stages, suggesting that they may be
involved in LUAD progression.
In addition, by analyzing both the co-expressed
genes with SDPR and the putative transcription factor
binding sites on SDPR, GATA2 was identified as a potential
transcription factor for SDPR transcription in LUAD.
Transcriptional factor GATA2 regulates genes critical for embryonic
development, self-renewal maintenance (29), functionality of blood-forming
(30) and lymphatic vessel valve
development (31). GATA2 has
been reported to be frequently epigenetically repressed in both
human and mouse lung tumors, and aberrant GATA2 methylation
occurred early during lung carcinogenesis (27). GATA2 may serve a role in the
downregulation of SDPR in LUAD.
Analysis of the associations between SDPR
expression and DNA methylation with patient survival in LUSC was
additionally performed. However, neither the OS nor disease-free
survival of patients had been observed to be significantly
associated with SDPR expression alternations, according to
the Prognoscan database. Furthermore, similar to LUAD, the same 13
CpG sites grouped in gene subregions based on the UCSC database
were analyzed using MethSurv. However, the results indicated no
significant association between DNA methylation data and patient
survival (data not shown). Since gene expression patterns differ in
the subtypes of lung cancer, this may provide novel insight for the
examination of potential mechanisms of SDPR function in
LUAD.
SDPR was also downregulated in kidney cancer.
Significant hypomethylation was found in both KIRC and KIRP,
suggesting other regulatory pathways. By comparing methylation
patterns with patient survival in KIRC and KIRP, the present study
indicated that hypomethylation in the ‘TSS200’ and ‘TSS1500’
subregions of SDPR was significantly associated with longer
survival time, while hypomethylation in the gene ‘body’ and the
‘3′UTR’ subregions was associated with poor OS (data not shown).
The function of DNA methylation status seems to vary in context;
this may be due to hypermethylation in the promoter region inducing
the downregulation of gene expression, while hypermethylation in
the gene body may not block and may even stimulate transcription
elongation, and the gene body methylation may have an impact on
splicing (32). In addition, long
non-coding RNA (lncRNA) SDPR-antisense (SDPR-AS) has been verified
to be co-expressed with SDPR, and elevated lncRNA SDPR-AS
increases the OS in renal cell carcinoma, suggesting the
possibility that lncRNAs may serve a regulatory role in the
SDPR pathway (33).
The specific methylation CpG sites, which are
significantly associated with patient survival, require further
study in order to verify the present results, such as
pyrosequencing and reverse transcription-quantitative PCR. The
potential transcription factor GATA2 should also be experimentally
investigated to determine whether it is responsible in SDPR
transactivation. Despite taking age and sex into consideration as
covariates in survival analysis based on the Methsurv database, due
to the lack of available clinical data, the survival analysis based
on PrognoScan is univariable. Since several factors, such as
co-morbidities, performance status and treatments, may potentially
affect the prognosis of patients with cancer, it is important to
consider all potential relevant specific features for specific
cancer types in future clinical investigations of SDPR.
In summary, the present study suggested that the
role of SDPR as a tumor suppressor may have broader clinical
relevance beyond breast cancer. The present study on SDPR
may help to examine its underlying molecular mechanism in cancer
progression, reveal novel perspectives for prognostic evaluation in
specific cancer, and provide insight for further research in the
field.
Acknowledgements
Not applicable.
Funding
The present study was supported by Foundation of
Sichuan Educational Committee, People's Republic of China (grant
nos. 17ZA0164 and 18CZ0010) and Foundation of Chengdu University of
Traditional Chinese Medicine, China (grant nos. ZRQN1660 and
CGZH1709).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and YL designed the present study. YW analyzed
the gene expressing data, patient survival data and the potential
transcriptional factors of SDPR. ZS analyzed the
co-expressing genes with SDPR. YL and PL analyzed the gene
methylation data. YW wrote the manuscript and YL revised it. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ozturk S, Papageorgis P, Wong CK, Lambert
AW, Abdolmaleky HM, Thiagalingam A, Cohen HT and Thiagalingam S:
SDPR functions as a metastasis suppressor in breast cancer by
promoting apoptosis. Proc Natl Acad Sci USA. 113:638–643. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tian Y, Yu Y, Hou LK, Chi JR, Mao JF, Xia
L, Wang X, Wang P and Cao XC: Serum deprivation response inhibits
breast cancer progression by blocking transforming growth factor-β
signaling. Cancer Sci. 107:274–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Poma AM, Giannini R, Piaggi P, Ugolini C,
Materazzi G, Miccoli P, Vitti P and Basolo F: A six-gene panel to
label follicular adenoma, low- and high-risk follicular thyroid
carcinoma. Endocr Connect. 7:124–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Codenotti S, Vezzoli M, Poliani PL,
Cominelli M, Monti E and Fanzani A: Cavin-2 is a specific marker
for detection of well-differentiated liposarcoma. Biochem Biophys
Res Commun. 493:660–665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Unozawa M, Kasamatsu A, Higo M, Fukumoto
C, Koyama T, Sakazume T, Nakashima D, Ogawara K, Yokoe H, Shiiba M,
et al: Cavin-2 in oral cancer: A potential predictor for tumor
progression. Mol Carcinog. 55:1037–1047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen CG, Bright NA, Howard G and Nichols
BJ: SDPR induces membrane curvature and functions in the formation
of caveolae. Nat Cell Biol. 11:807–814. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nassar ZD and Parat MO: Cavin family: New
players in the biology of caveolae. Int Rev Cell Mol Biol.
320:235–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gustincich S, Vatta P, Goruppi S, Wolf M,
Saccone S, Della Valle G, Baggiolini M and Schneider C: The human
serum deprivation response gene (SDPR) maps to 2q32-q33 and codes
for a phosphatidylserine-binding protein. Genomics. 57:120–129.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baig A, Bao X, Wolf M and Haslam RJ: The
platelet protein kinase C substrate pleckstrin binds directly to
SDPR protein. Platelets. 20:446–457. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gupta R, Toufaily C and Annabi B: Caveolin
and cavin family members: Dual roles in cancer. Biochimie.
107:188–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Jia Z, Shen Y, Ichikawa H, Jarvik J,
Nagele RG and Goldberg GS: Coordinate suppression of Sdpr and Fhl1
expression in tumors of the breast, kidney, and prostate. Cancer
Sci. 99:1326–1333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai L, Deng X, Li Q, Wang M, An W, Deli A,
Gao Z, Xie Y, Dai Y and Cong YS: Down-regulation of the cavin
family proteins in breast cancer. J Cell Biochem. 113:322–328.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gianazza E, Chinello C, Mainini V,
Cazzaniga M, Squeo V, Albo G, Signorini S, Di Pierro SS, Ferrero S,
Nicolardi S, et al: Alterations of the serum peptidome in renal
cell carcinoma discriminating benign and malignant kidney tumors. J
Proteomics. 76:125–140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altintas DM, Allioli N, Decaussin M, de
Bernard S, Ruffion A, Samarut J and Vlaeminck-Guillem V:
Differentially expressed androgen-regulated genes in
androgen-sensitive tissues reveal potential biomarkers of early
prostate cancer. PLoS One. 8:e662782013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
19
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miller R and Siegmund D: Maximally
selected chi square statistics. Biometrics. 38:1011–1016. 1982.
View Article : Google Scholar
|
|
21
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res 43 (Database
Issue). D856–D861. 2015. View Article : Google Scholar
|
|
22
|
Broad Institute TCGA Genome Data Analysis
Center (2016), . Firehose stddata__2016_01_28 run. Broad Institute
of MIT and Harvard. Doi: 10.7908/C11G0KM9.
|
|
23
|
Modhukur V, Iljasenko T, Metsalu T, Lokk
K, Laisk-Podar T and Vilo J: MethSurv: A web tool to perform
multivariable survival analysis using DNA methylation data.
Epigenomics. 10:277–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using principal
component analysis and heatmap. Nucleic Acids Res 43W. W566–W570.
2015. View Article : Google Scholar
|
|
25
|
Farré D, Roset R, Huerta M, Adsuara JE,
Roselló L, Albà MM and Messeguer X: Identification of patterns in
biological sequences at the ALGGEN server: PROMO and MALGEN.
Nucleic Acids Res. 31:3651–3653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barretina J, Taylor BS, Banerji S, Ramos
AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho
A, et al: Subtype-specific genomic alterations define new targets
for soft-tissue sarcoma therapy. Nat Genet. 42:715–721. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tessema M, Yingling CM, Snider AM, Do K,
Juri DE, Picchi MA, Zhang X, Liu Y, Leng S, Tellez CS and Belinsky
SA: GATA2 is epigenetically repressed in human and mouse lung
tumors and is not requisite for survival of KRAS mutant lung
cancer. J Thorac Oncol. 9:784–793. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Borup R, Rossing M, Henao R, Yamamoto Y,
Krogdahl A, Godballe C, Winther O, Kiss K, Christensen L, Høgdall
E, et al: Molecular signatures of thyroid follicular neoplasia.
Endocr Relat Cancer. 17:691–708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krendl C, Shaposhnikov D, Rishko V, Ori C,
Ziegenhain C, Sass S, Simon L, Müller NS, Straub T, Brooks KE, et
al: GATA2/3-TFAP2A/C transcription factor network couples human
pluripotent stem cell differentiation to trophectoderm with
repression of pluripotency. Proc Natl Acad Sci USA.
114:E9579–E9588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
de Pater E, Kaimakis P, Vink CS, Yokomizo
T, Yamada-Inagawa T, van der Linden R, Kartalaei PS, Camper SA,
Speck N and Dzierzak E: Gata2 is required for HSC generation and
survival. J Exp Med. 210:2843–2850. 2013. View Article : Google Scholar
|
|
31
|
Kazenwadel J, Betterman KL, Chong CE,
Stokes PH, Lee YK, Secker GA, Agalarov Y, Demir CS, Lawrence DM,
Sutton DL, et al: GATA2 is required for lymphatic vessel valve
development and maintenance. J Clin Invest. 125:2979–2994. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ni W, Song E, Gong M, Li Y, Yao J and An
R: Downregulation of lncRNA SDPR-AS is associated with poor
prognosis in renal cell carcinoma. Onco Targets Ther. 10:3039–3047.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sanchez-Carbayo M, Socci ND, Lozano J,
Saint F and Cordon-Cardo C: Defining molecular profiles of poor
outcome in patients with invasive bladder cancer using
oligonucleotide microarrays. J Clin Oncol. 24:778–789. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Biewenga P, Buist MR, Moerland PD, Ver
Loren van Themaat E, van Kampen AH, ten Kate FJ and Baas F: Gene
expression in early stage cervical cancer. Gynecol Oncol.
108:520–526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii): e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ki DH, Jeung HC, Park CH, Kang SH, Lee GY,
Lee WS, Kim NK, Chung HC and Rha SY: Whole genome analysis for
liver metastasis gene signatures in colorectal cancer. Int J
Cancer. 121:2005–2012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469. 2009.
View Article : Google Scholar
|
|
41
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Haferlach T, Kohlmann A, Wieczorek L,
Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK,
Mills KI, et al: Clinical utility of microarray-based gene
expression profiling in the diagnosis and subclassification of
leukemia: Report from the international microarray innovations in
leukemia study group. J Clin Oncol. 28:2529–2537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ,
Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH and Huang CY:
Selection of DDX5 as a novel internal control for Q-RT-PCR from
microarray data using a block bootstrap re-sampling scheme. BMC
Genomics. 8:1402007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wachi S, Yoneda K and Wu R:
Interactome-transcriptome analysis reveals the high centrality of
genes differentially expressed in lung cancer tissues.
Bioinformatics. 21:4205–4208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Crabtree JS, Jelinsky SA, Harris HA, Choe
SE, Cotreau MM, Kimberland ML, Wilson E, Saraf KA, Liu W,
McCampbell AS, et al: Comparison of human and rat uterine
leiomyomata: Identification of a dysregulated mammalian target of
rapamycin pathway. Cancer Res. 69:6171–6178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yoshihara K, Tajima A, Komata D, Yamamoto
T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K,
et al: Gene expression profiling of advanced-stage serous ovarian
cancers distinguishes novel subclasses and implicates ZEB2 in tumor
progression and prognosis. Cancer Sci. 100:1421–1428. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|