Introduction
Preeclampsia is a pregnancy-specific disease
associated with obstruction of uterine spiral arteries, excessive
activation of inflammatory immunity, endothelial cell damage,
genetic factors, metabolic dysfunction and nutritional deficiencies
(1–3). In total, ~4 million women are
diagnosed with preeclampsia annually worldwide, leading to over
70,000 maternal and 500,000 infant deaths (4). The prognosis of preeclampsia remains
elusive, particularly in late onset cases. SPARC plays a crucial
role in placenta development, and its downregulation under hypoxic
conditions potentially contributes to preeclampsia and -associated
intrauterine growth restriction (5). Similarly, maternal microRNA-125b
plasma levels in the first trimester have been shown to strongly
predict pre-eclampsia way before clinical manifestations (6). Women with a history of pre-eclampsia
have approximately double the risk of cardiovascular disease,
including cardiovascular-related death, coronary artery disease,
heart failure and stroke, persisting for up to 39 years of
follow-up (7). According to global
cancer statistics for 2020, breast cancer (BC) is the most common
malignant tumor worldwide, posing serious risks to women's physical
and mental health (8). The
epidemiological link between preeclampsia and BC was first
described in a 1983 case-control study by Polednak and Janerich,
which showed reduced BC risk before age 45 among women with
preeclampsia during their first pregnancy (9). Subsequent studies over the past 40
years have provided increasing evidence of this association.
Neutrophils, the most abundant immune cells, have
been identified to play a significant role in preeclampsia. In
preeclamptic patients, neutrophils are heavily activated,
increasing superoxide production, systemic inflammation and
vascular endothelial damage (10).
Activated neutrophils interact with platelets through signaling
pathways involving chemokines and adhesion factors, contributing to
preeclampsia development (11).
Platelet-activating factor released by platelets also plays a key
role in the condition's pathogenesis (12). Previous studies revealed that
neutrophils accelerate endothelial cell apoptosis through increased
NET expression, a critical step in vascular endothelial injury in
preeclampsia (13). In tumors,
neutrophils, known as tumor-associated neutrophils (TANs), are a
heterogeneous and integral component of the tumor microenvironment,
with dual roles in tumor promotion and prevention (14,15).
Fridlender et al (16) in
2009 introduced the N1 (antitumor) and N2 (protumor) nomenclature
for TANs. Neutrophils can mediate cytotoxicity against tumor cells
via reactive oxygen species (ROS) such as hydrogen peroxide and
superoxide, with ROS-mediated elimination depending on TRPM2, an
H2O2-dependent Ca2+ channel
(17). Neutrophils can also present
antigens to T cells, inducing IFN-γ production and adaptive immune
responses, or directly interact with T cells to lower activation
thresholds, aiding tumor cell clearance (18,19).
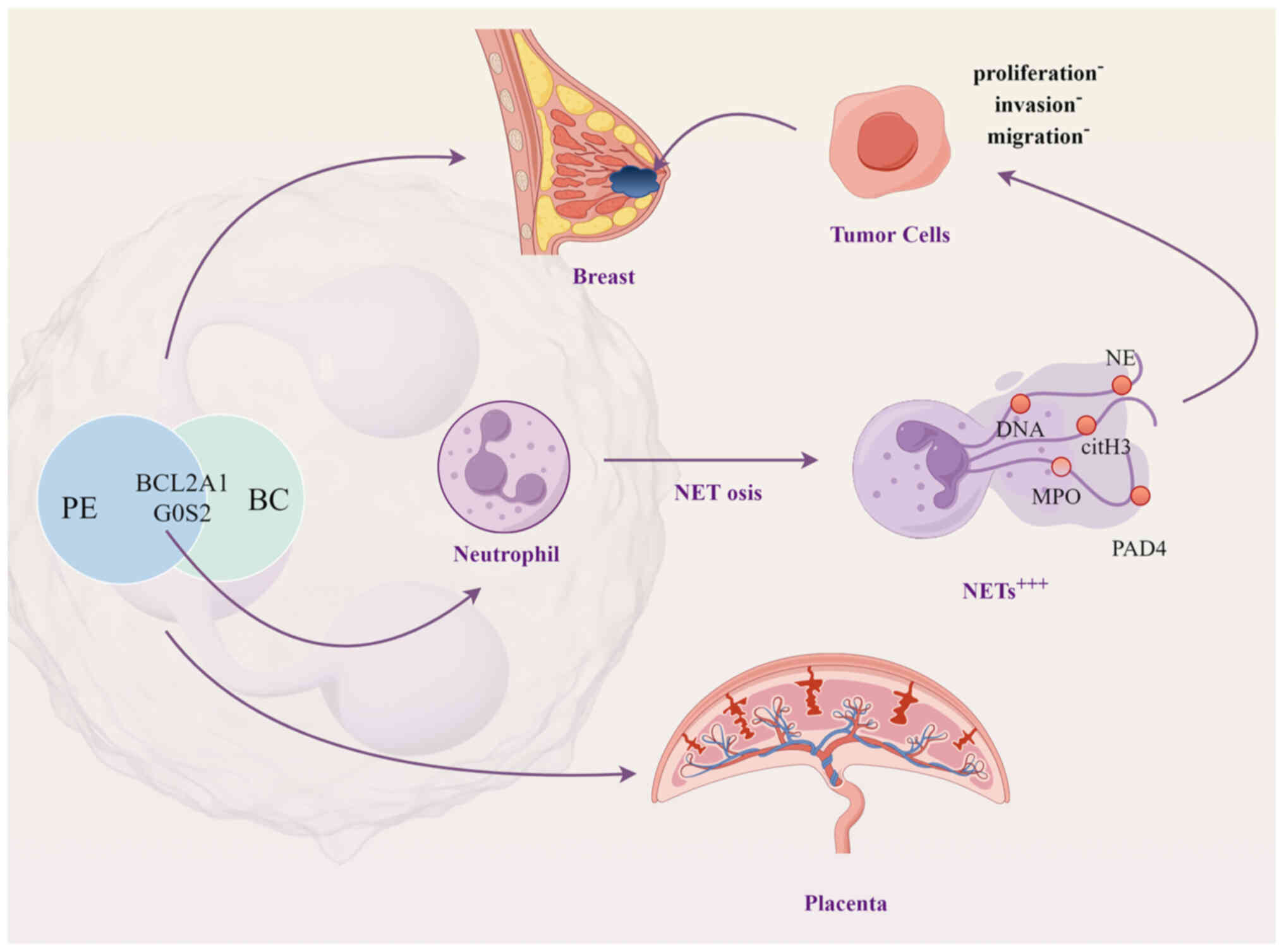
These findings suggest that NETs produced by neutrophils may
provide a crucial link between preeclampsia and reduced BC
risk.
In recent years, integrative genomic analyses have
advanced the understanding of shared pathogenesis between diseases.
The present study employed single-cell sequencing and
bioinformatics to investigate the relationship between neutrophils
and the reduced risk of BC in preeclamptic patients. Supported by
in vitro validation, the findings suggest that NETs mediate
a protective mechanism linking preeclampsia to a reduced risk of
BC. The pivotal roles of BCL2A1 and G0/G1 switch gene 2 (G0S2) in
regulating neutrophil activity and NET production underscore their
potential as therapeutic targets for cancer and inflammatory
diseases.
Materials and methods
Reagents and kits
List of reagents and kits used in the study are
provided along with company names and cat. nos. in Table SI.
Data sources
A scRNA-seq dataset (GSE173193) was obtained from
Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). This dataset
contained placental tissue samples of the gestational diabetes
group (n=2), preeclampsia group (n=2), advanced age group (n=2) and
normal control group (n=2). Normal control group (n=2) and
preeclampsia group (n=2) were used to characterize cellular
landscape of preeclampsia.
Single cell sequencing data analysis
processing
Quality control
Initially, the DropletUtils package (20) was utilized to assess the expression
levels of each cell and to filter out barcoded cells that showed no
expression. Next, cells were further filtered based on their unique
molecular identifier counts. Subsequently, the scatter package
(21) was employed to quantify gene
expression in the cells, leading to the exclusion of cells with
mitochondrial gene proportions >10% and ribosomal gene
proportions <10%. Finally, gene counts were obtained.
Data preprocessing and principal
component analysis (PCA)
First, the NormalizeData function from the Seurat
package (22) was employed to
standardize the expression matrix of the filtered samples. Next,
the FindVariableFeatures function was used to identify the first
2,000 genes with the most significant differences among cells.
Concentrating on these genes in subsequent analyses will enhance
the detection of biological signals in single-cell datasets.
Following this, the expression data were linearly scaled using the
ScaleData function from the Seurat package. Finally, PCA was
conducted using the RunPCA function within the Seurat package.
Cell cluster and annotation
First, principal components with high standard
deviations were selected. Next, cell clustering analysis was
conducted using the FindNeighbors and FindClusters functions from
the Seurat package. Then, Uniform Manifold Approximation and
Projection (UMAP) for dimensionality reduction was performed using
the RunUMAP function of the Seurat package. Cell types were
annotated based on the known marker genes.
Identification of marker genes
To identify differentially expressed genes (DEGs)
between each cluster and all other cells, the FindAllMarkers
function from the Seurat package was utilized. Novel marker genes
were determined based on the following criteria: (log2FC ≥0.1, a
minimum expression ratio of cell population=0.25, and P≤0.05),
resulting in the selection of the top 500 logFC markers (23). Cells were then labeled according to
the known marker genes, and a cluster display diagram was
created.
Cell-cell communication analysis
CellChatv1.1 (24)
was used to infer the communication between cells according to the
corresponding receptor ligand gene expression values of various
cells. And then, the receptor ligand pair network between cells was
obtained.
Pseudotime analysis
Pseudotime analysis was conducted using Monocle
(25). First, genes expressed in at
least 5% of the cells were selected. Next, the reduceDimension
function was applied to perform dimensionality reduction analysis,
followed by clustering of the cells using the clusterCells
function. The differentialGeneTest function was then used to
identify candidate genes that differed between the clusters with a
P<0.05. Dimensionality reduction analysis of the cells was
performed using the DDRTree method and the reduceDimension function
based on the candidate genes. Finally, the orderCells function was
utilized to arrange and visualize the cells along a
quasi-chronological trajectory.
DEG analysis and functional enrichment
analysis
DEGs were identified using the FindMarkers function
(test.use=MAST) in Seurat. A P<0.05 and a log2 fold change
>0.58 were established as the criteria for significant
differential expression. Gene Ontology (GO) enrichment and KEGG
pathway enrichment analyses of the DEGs were performed using the
DAVID database, (https://davidbioinformatics.nih.gov/) encompassing all
GO categories, including biological processes, cellular components
and molecular functions.
Identification of significant DEGs in
preeclampsia and BC
The ‘limma’ function from the R suite was employed
to identify DEGs from The Cancer Genome Atlas-BC and GSE24129
datasets. DEGs were classified as those exhibiting |log2 (fold
change)|>1 and P<0.01. The findings are presented using
volcano plots.
Construction and visualization of the
co-expression network
The DEGs were analyzed for co-expression networks
using the R package Weighted Gene Co-expression Network Analysis
(WGCNA) (26). WGCNA was employed
to construct a gene co-expression network based on the standardized
gene data in order to identify the optimal soft threshold. A
scale-free network was built using this best soft threshold, and
the genes were then clustered into functional modules, each
represented by different colors, with clustering and classification
performed using the dynamic tree cutting algorithm. Finally, PCA
was applied to describe the module eigengenes, which reflect a
unique expression profile for all the genes within each module. The
correlations between these eigengenes and clinical characteristics
were calculated to determine which modules were clinically
relevant.
Patient samples
In the present study, the inclusion criteria for
patients with BC included a preoperative diagnosis confirmed by
Mammotome biopsy and no prior history of radiation therapy or
chemotherapy. Tumor tissue samples, along with adjacent normal
tissues (control), were collected from 3 patients with BC, aged 40
to 70 years, with a median age of 55. The inclusion criteria for
patients with preeclampsia required a clear clinical diagnosis,
absence of other complications and a singleton cesarean delivery. A
total of 3 placenta samples were collected from both patients with
preeclampsia and control subjects, aged 20–40 years, with a median
age of 29 in the preeclampsia group and 32 in the control group.
Written informed consent was obtained from all study participants
for the use of tissue samples in scientific research. The present
study was conducted in accordance with the ethical guidelines and
was approved (approval no. 2022-E0118) by the Ethical Review
Committee of the First Affiliated Hospital of Guangxi Medical
University (Nanning, China).
Flow cytometric analysis
The cells obtained from freshly excised preeclampsia
and control tissues were washed and centrifuged (300 × g, 4°C, 5
min), after which a dye was added, and the cells were incubated in
the dark for 30 min before being promptly quantified using flow
cytometry. The detection reagents used were FITC-conjugated
anti-GR-1 (cat. no. 65140-1-Ig; Proteintech Group, Inc.) and
PE-Cy5-conjugated anti-F4/80 (cat. no. B281020; BioLegend, Inc.).
Fluorochrome labeling: GR-1: FITC (detected in FL1 channel) and
F4/80: PE-Cy5 (detected in FL3 channel). BD FACSVerse flow
cytometer (Becton Dickinson) was used, and data analysis was
conducted using FlowJo software (version 10; Tree Star, Inc.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from preeclampsia and BC
samples using TRIzol® reagent and cDNA was synthesized
with the TRUEscript H Minus M-MuLV Reverse Transcriptase. Enzymes
and reagents used included: Reverse transcriptase (TRUEscript H
Minus M-MuLV; cat. no. PC1703; Aidlab), RNase inhibitor (RNasin;
cat. no. RN3501; Aidlab) and nucleotides (dNTP Mixture; 10 mM each;
cat. no. PC2403; Aidlab). All primers were designed using Primer
Premier 5.0 software (https://primer-premier.software.informer.com/) and
were commercially synthesized by Sangon Biotech Co., Ltd. GAPDH was
used as an internal control. The cDNA was stored at −20°C until
needed. The reverse transcription protocol was performed as
follows: 42°C for 60 min followed by 70°C for 10 min. A two-step
RT-qPCR was conducted with SYBR Green Master Mix (cat. no. PC3302;
Aidlab) to assess the expression levels of BCL2A1 and G0S2.
Thermocycling conditions were as follows: Initial denaturation at
94°C for 10 min; followed 40 cycles of denaturation at 94°C for 20
sec, annealing at 55°C for 20 sec and extension at 72°C for 20 sec.
The primers used in the present study are listed in Table SII. GAPDH was used as housekeeping
control. Data were calculated using the 2−ΔΔCq method
(27).
Western blot analysis
Total proteins were extracted from cells lysed in
RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology).
Next, the concentration of proteins was detected by BCA analysis.
The extracted proteins were collected, denatured and separated
using 10% SDS-PAGE gels. The samples (30 µg) were loaded onto the
gels, and electrophoresis was conducted for 120 min. Following
this, the proteins were transferred to PVDF membranes and blocked
with 5% skimmed milk by incubation at 37°C for 2 h. After shaking
for 1 h, elution was performed. Next, the membranes were incubated
with primary antibodies (Table
SIII) at 4°C overnight with shaking. Next day, the PVDF
membrane was washed with TBST (0.1% Tween-20) and subjected to
secondary antibody (Table SIII)
incubation for 1 h at room temperature (RT). The images were
acquired using the Infrared electrochemical luminescence (cat. no.
ECL-0011; Beijing Dingguo Changsheng Biotechnology Co., Ltd.). The
gray value of the target bands was calculated using ImageJ software
2.0 (National institutes of Health). The experiment was repeated 3
times. The housekeeping gene β-actin was used as loading
control.
Immunofluorescence staining
After rehydration through graded ethanol (100→70%),
tissue sections were washed with PBS and underwent antigen
retrieval (cat. no. P0086; Beyotime Institute of Biotechnology) via
microwave heating in retrieval buffer at low power for 15 min.
Following PBS washes, sections were permeabilized with 0.1% Triton
X-100 for 10 min at RT and blocked with 5% BSA at 37°C for 1 h.
Primary antibodies (anti-BCL2A1 and anti-G0S2; diluted at
1:10,000-1:40,000) were incubated at 37°C for 2 h, followed by 3×5
min PBS washes. Fluorescent secondary antibodies, including
Cy3-labeled anti-rabbit (cat. no. SA00009-2) and FITC-labeled
anti-mouse (cat. no. SA00003-1; both from Proteintech Group, Inc.),
were incubated at 37°C for 1 h. After nuclear staining with Hoechst
(cat. no. P0133; Beyotime Institute of Biotechnology) at RT for 15
min and final PBS washes, slides were dehydrated in absolute
ethanol for 1 min, air-dried, and mounted with antifade medium
(cat. no. P0128M; Beyotime Institute of Biotechnology). Images were
acquired using an inverted confocal microscope (IX71; Olympus
Corporation).
Cell culture and transfection
Cells were used to identify neutrophils, which were
subsequently cultured in RPMI-1640 (cat. no. 12633020; Gibco;
Thermo Fisher Scientific, Inc.) medium supplemented with 10% FBS +
1% penicillin-streptomycin at 37°C and 5% CO2. Cells
with favorable proliferation status were transfected with 50 nM
small interfering RNA (siRNA). Cells were seeded in six-well plates
at a density of 5×105 cells per well and cultured until
reaching 80% confluency. Transfection was then performed using
Lipofectamine 3000 (Thermo Fisher Scientific, Inc.), followed by 6
h incubation at 37°C with 5% CO2 before medium change
Neutrophils were divided into 5 groups: Control, vector, BCL2A1-OE,
G0S2-OE and BCL2A1-OE + G0S2-OE. Neutrophils and MCF-7 cells
(5×105 cells/ml) were separately seeded in a two-chamber
culture system. Using a Transwell model, MCF-7 cells were plated in
the lower chamber with culture medium in the upper chamber.
Neutrophils (1×106 cells/ml) were then seeded in the
upper compartment of the Transwell insert and co-cultured with the
lower-chamber breast cancer cells for 24 h under standard
incubation conditions. MCF-7 cells (cat. no. HTB-22) were purchased
from the American Type Culture Collection.
Cell Counting Kit-8 (CCK-8)
assays
Assays were conducted using a CCK-8 reagent. The
cells were plated in 96-well plates at a density of
1×104 cells per well. Next, 10 µl of CCK-8 was added to
each well in a light-protected environment. The cells were then
incubated at 37°C in a 5% CO2 atmosphere for 1.5 h.
Finally, absorbance was measured at 450 nm using a ThermoMax
microplate reader.
Transwell migration assay
For the cell migration assay, 200 µl of cell
suspension (1×105 cells) was added to the upper
compartment of a Transwell chamber with an 8-µm pore size and a
24-well insert. In each well, 50 µl of serum-free medium
supplemented with 10 g/l BSA was mixed with the co-cultured cells
in the upper chamber. The lower chambers contained 10% FBS. Cell
migration ability was assessed by counting the number of cells that
migrated to the lower chamber of the Transwell using a fluorescence
microscope (IX51; Olympus Corporation).
Cell scratch assay
A total of ~5×105 cells were placed into
a six-well plate, with three replicate wells designated for each
group. Cells were maintained in serum-free medium throughout the
assay. Once the cells adhered to the surface in a single layer, a
200-µl pipette tip was employed to create vertical scratches in the
six-well plate. The cells were then washed three times with PBS,
and the suspension was discarded before incubating the cells in a
chamber with 5% CO2 at 37°C. Images were captured under
a microscope at 0 and 48 h, and the experiment was repeated three
times.
Statistical analysis
All statistical analyses were conducted using
GraphPad Prism 8 (version 8.0; Dotmatics) and R software (version
4.2) (https://cran.r-project.org/bin/windows/base/old/4.2.0/).
The data are presented as the mean ± standard deviations (SDs).
Statistical analyses were performed using unpaired two-tailed
t-test for two groups or one-way ANOVA for more than two groups,
followed by Tukey's post hoc test for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Single-cell RNA sequencing (scRNA-seq)
reveals the cell composition of preeclamptic and control
placentas
To examine cell-type-specific changes in
preeclampsia at the single-cell level, scRNA-seq) was performed on
the GSE173193 dataset, which included two control patients and two
preeclamptic patients. After conducting quality control and
normalization, the first 2,000 highly variable genes within the
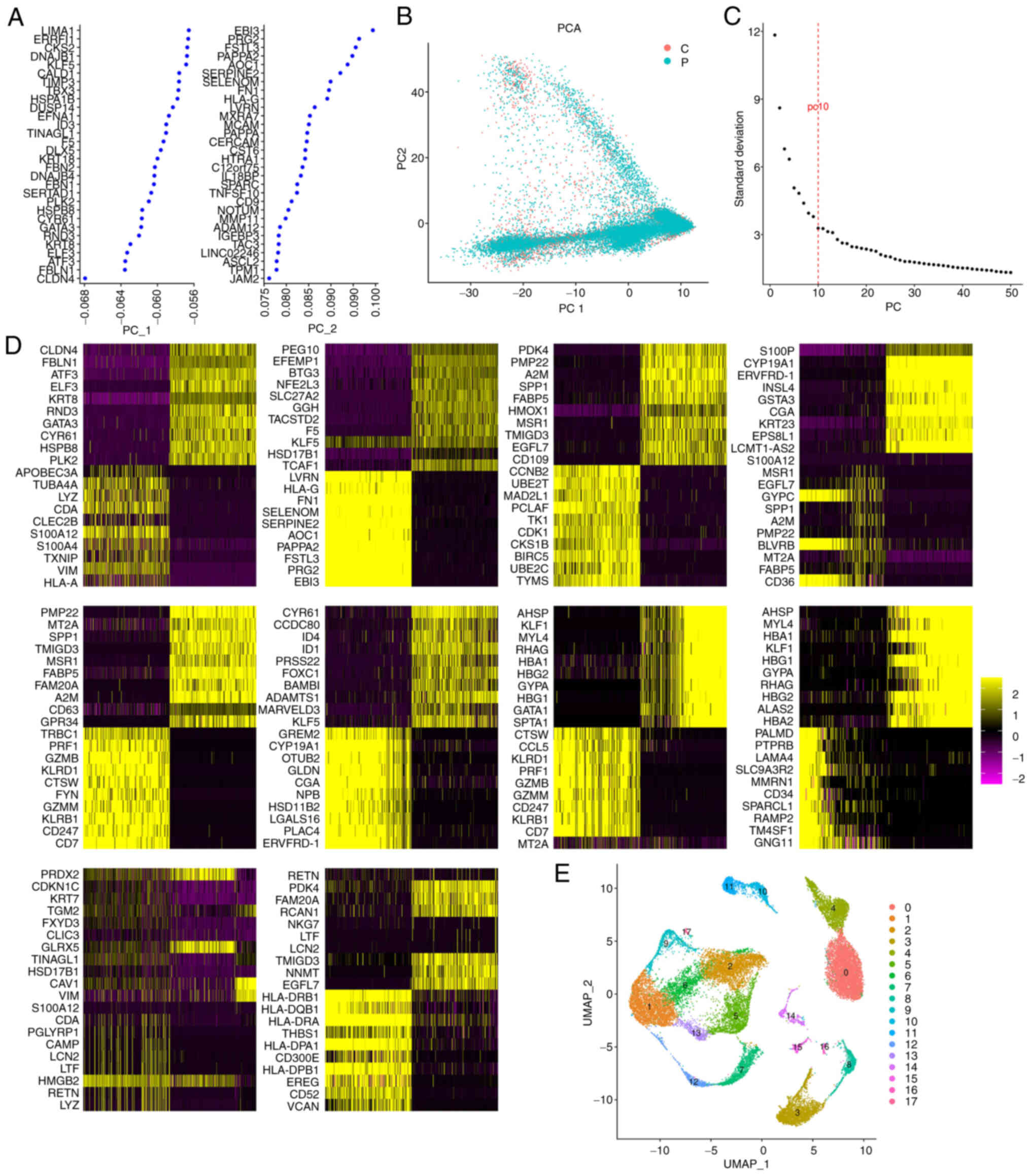
cells were identified (Fig. 1A).
Dimensionality reduction was then performed using PCA to analyze
the linearly scaled scRNA data, focusing on the top two principal
components for further investigation. The PCA results indicated a
distinct separation between preeclamptic and control placental
cells (Fig. 1B). Based on the elbow
point criterion, the optimal number of principal components was
determined to be 10 (Fig. 1C).
Heatmaps illustrating the top 20 marker genes for each principal
component are shown (Fig. 1D).
Using the UMAP method, the placental cells were clustered into 18
groups (Fig. 1E). The top ten
marker genes of each cell cluster are presented in Figs. S1 (group 0–8) and S2 (group 9–17).
Identification of cell types and their
marker genes in placental cells
Next it was aimed to identify various cell types
within placental cells from both normal individuals and patients
with preeclampsia. Using known marker genes, 12 distinct cell types
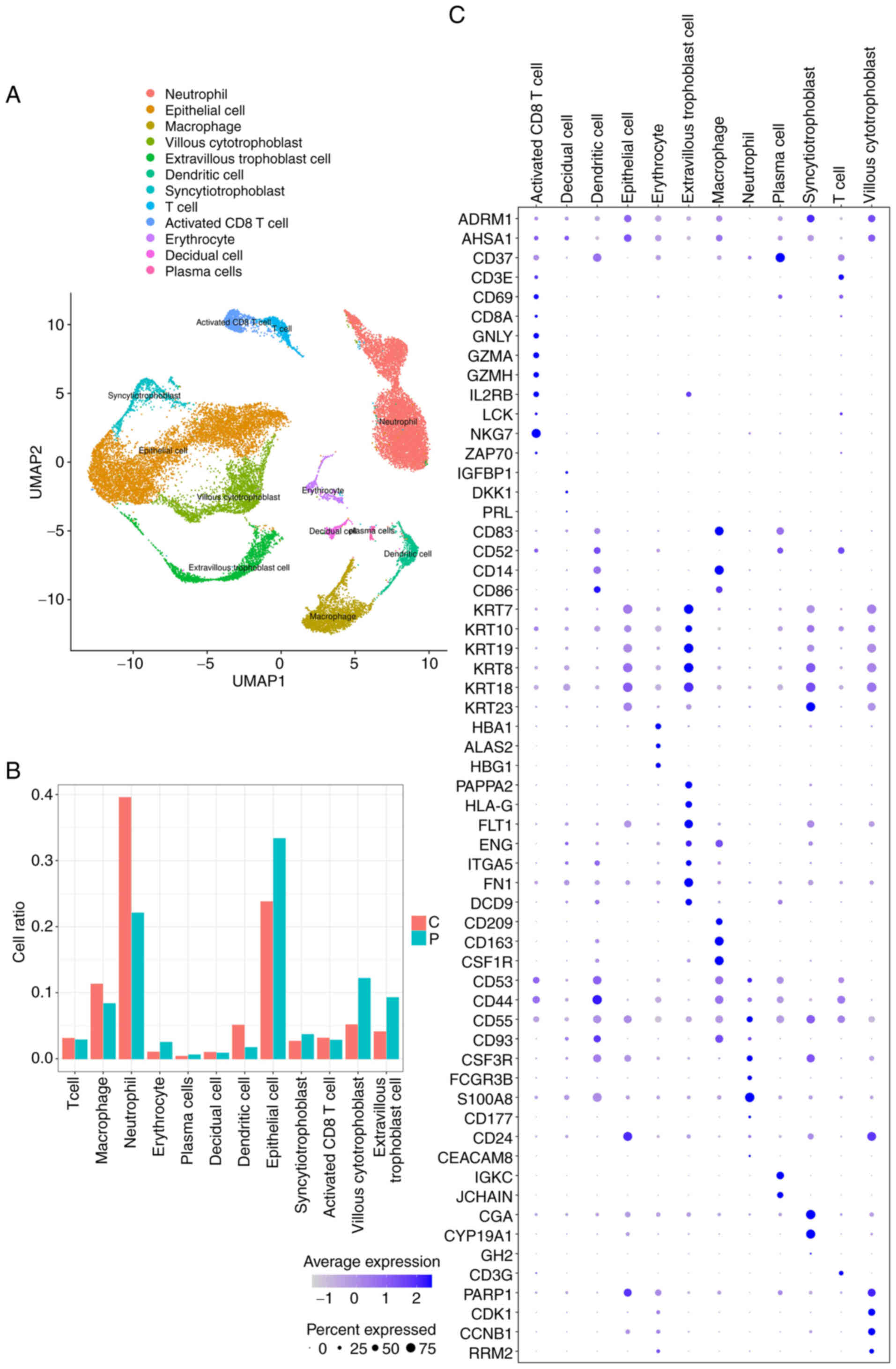
were annotated (Fig. 2A). The
relative abundance of each cell type is illustrated in Fig. 2B. Notably, differences in the ratios
of placental cells were observed between preeclampsia patients and
controls. By applying a |logFC|≥0.1, a minimum expression ratio of
the cell population of 0.25, and a P≤0.05, novel marker genes were
identified for each cell type. The top ten marker genes for each
cell type were visualized as follows: Neutrophils (ALOX5AP, BCL2A1,
CAMP, CXCL8, G0S2, IFITM2, RETN, S100A12, S100A8 and S100A9);
villous cytotrophoblast cells (CCNB1, CDK1, HIST1H4C, HMGB1, PTTG1,
STMN1, TUBA1B, TUBB, TYMS and UBE2C); extravillous trophoblast
cells (AOC1, EBI3, FN1, FSTL3, HPGD, NOTUM, PAPPA2, PRG2, SERPINE2
and TAC3); and syncytio-trophoblast cells (CGA, CYP19A1, ERVFRD-1,
GADD45G, GDF15, HOPX, INSL4, KISS1, KRT23 and CSH1) (Fig. S3). Additionally, the expression
levels of the known marker genes used for cell type annotation were
analyzed (Fig. 2C).
Cell-cell interactions based on
ligand-receptor interactions and reconstruction of the temporal
dynamics of trophoblasts
The placenta is formed through a complex process
that requires the collaborative efforts of various cell lineages.
Intercellular communications among different cell types govern the
proper functions of metazoans and heavily depend on the
interactions between secreted ligands and cell-surface receptors.
Based on marker genes, ligand-receptor interactions were
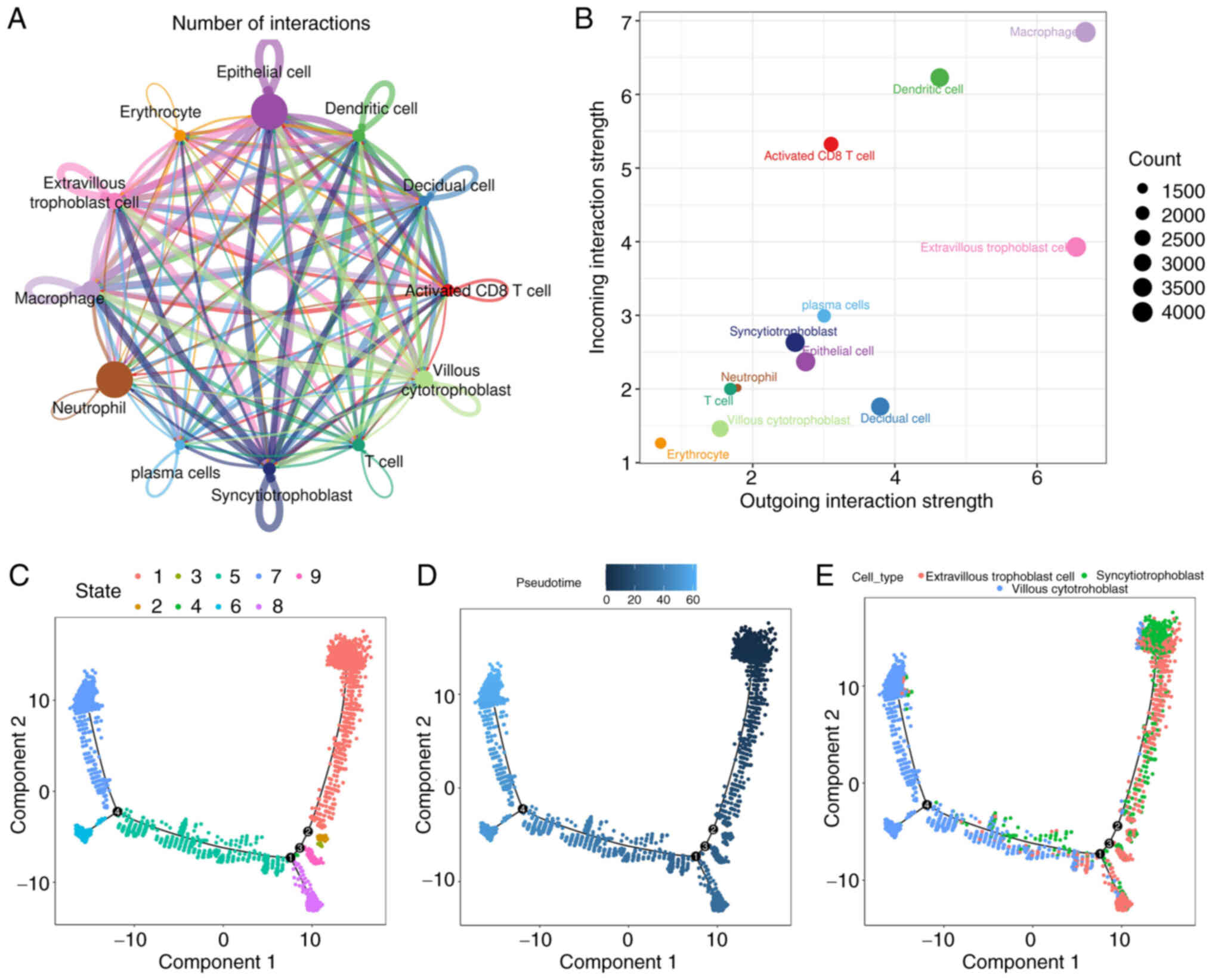
identified. It was demonstrated that each cell type possesses a
significant number of receptor ligands (Fig. 3A and B). Among these interactions,
those occurring between macrophages and other cells show the
highest number and intensity in the placentas of patients with
preeclampsia. Additionally, there are numerous interactions between
neutrophils and other cells that are characterized by high
intensity as well. Overall, the present findings suggest that
complex intercellular communication occurs within the placental
microenvironment, with distinct changes in this communication.
Furthermore, to explore the evolutionary processes of trophoblasts,
the present study utilized the Monocle tool to uncover
pseudo-temporal ordering due to the similarity of cell clusters
with developmental lineages. The trends of pseudotime-dependent
genes along the pseudo-timeline were categorized into nine cell
clusters of trophoblasts, each exhibiting diverse expression
dynamics. The analysis indicated a progressive development of
trophoblasts from Cluster 7 to Cluster 9 (Fig. 3C-E); syncytio-trophoblast and
extravillous trophoblast cells were positioned at the beginning of
the differentiation process, while villous cytotrophoblast cells
were found at the end. This suggests that extravillous trophoblast
cells or syncytio-trophoblast cells may differentiate into villous
cytotrophoblast cells during the development of preeclamptic
placentas, suggesting that different trophoblast subtypes may
fulfill distinct biological functions.
Biological processes associated with
marker genes of trophoblast and neutrophil subsets
To further investigate the functional status and
potential regulatory factors associated with trophoblast and
neutrophil subsets in preeclamptic placentas, GO and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the
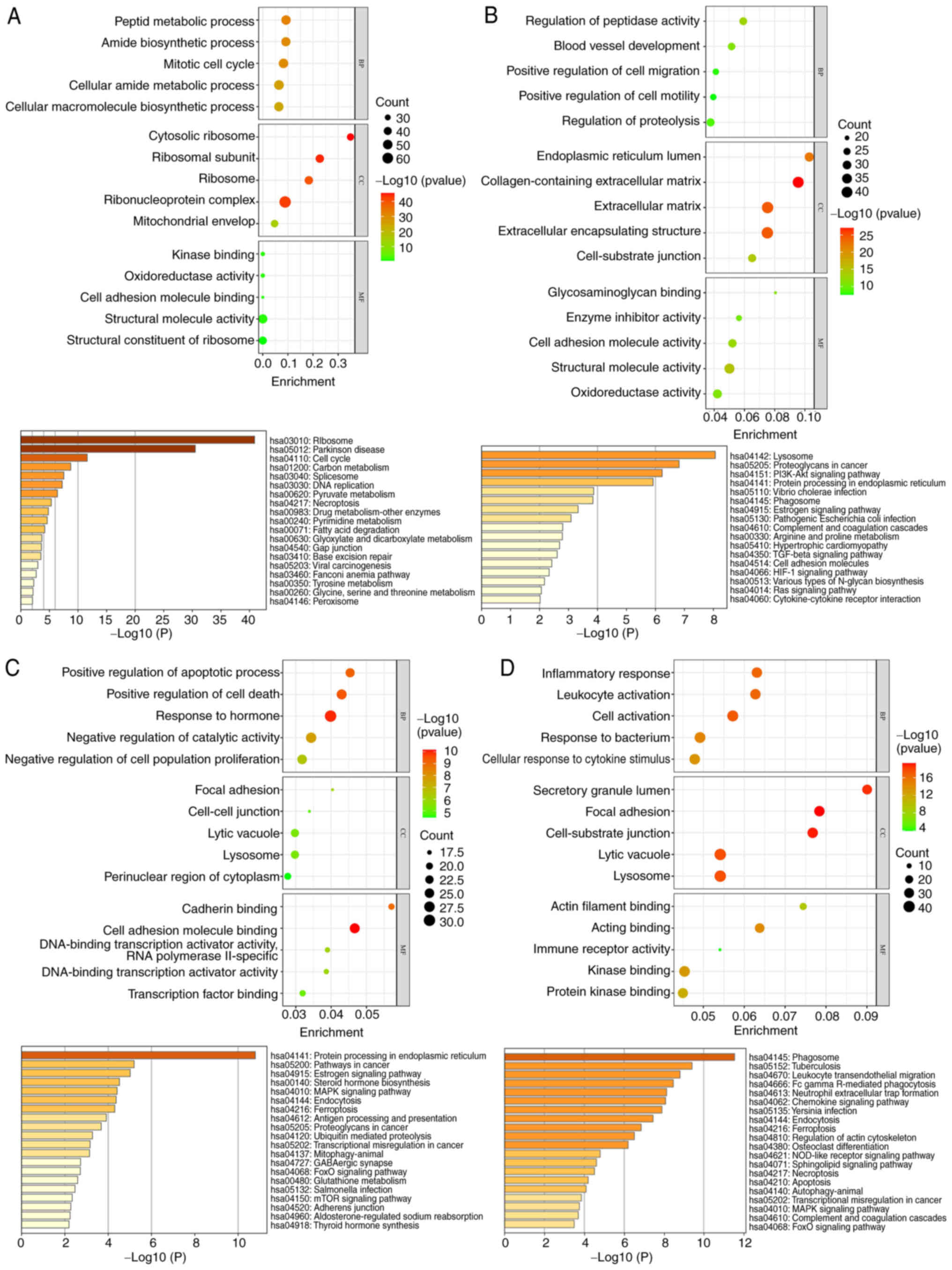
DEGs identified in the cell populations (Fig. 4A-D) were performed in the present
study. The results indicated that in the annotated villous
cytotrophoblast cell populations, the biological functions were
primarily enriched in the regulation of cell metabolism,
biosynthesis, mitosis and cell cycle processes. This aligns with
the findings of Zhou et al (28). In the annotated extravillous
trophoblast cell population, biological functions were mainly
related to proteolysis, cell migration and movement, angiogenesis
and regulation. For the annotated syncytio-trophoblast cell
population, genes linked to biological functions were enriched in
the positive regulation of cell apoptosis and death, as well as
hormone regulation. In the annotated neutrophil population,
biological functions predominantly involved cell activation,
responses to cytokine stimulation, cell migration, movement, the
actin filament process and the inflammatory response. Regarding
cell components, the genes were mainly enriched in lysosomes and
secretory granules. In terms of molecular function, the genes
showed enrichment primarily in kinase and protein kinase binding,
actin binding, and immune receptor activity. The KEGG pathway
enrichment analysis revealed significant pathways such as
phagosomes, pulmonary tuberculosis, leukocyte migration across the
endothelium, Fcγ receptor-mediated phagocytosis and the formation
of neutrophil extracellular traps.
BCL2A1 and G0S2 genes are
significantly increased in neutrophils from preeclamptic placental
tissue
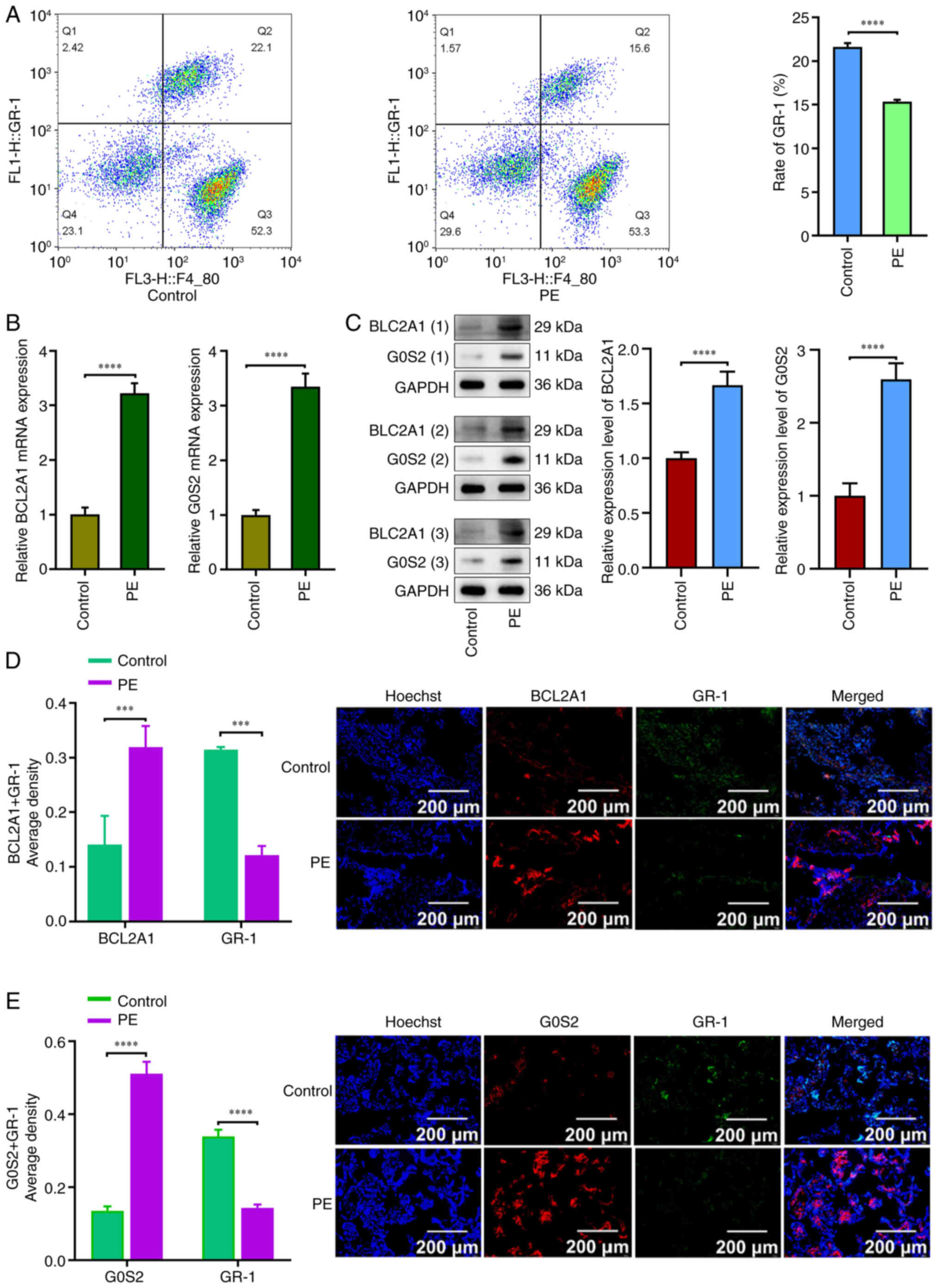
The flow cytometric results indicated that GR-1 was
downregulated in the preeclampsia group compared with the normal
group (P<0.05; Fig. 5A). RT-qPCR
results revealed that BCL2A1 and G0S2 were upregulated in the
preeclampsia group relative to the normal group (P<0.05;
Fig. 5B). Western blot analysis
revealed that the expression of BCL2A1 and G0S2 was increased in
the preeclampsia group compared with the control group (P<0.05;
Fig. 5C). Immunofluorescence
results demonstrated that the expression levels of BCL2A1 and G0S2
were significantly higher in the preeclampsia group than in the
control group, while GR-1 expression was significantly lower in the
preeclampsia group (P<0.05; Fig. 5D
and E).
Overexpression of the BCL2A1 and G0S2
genes in neutrophils promotes the production of NETs
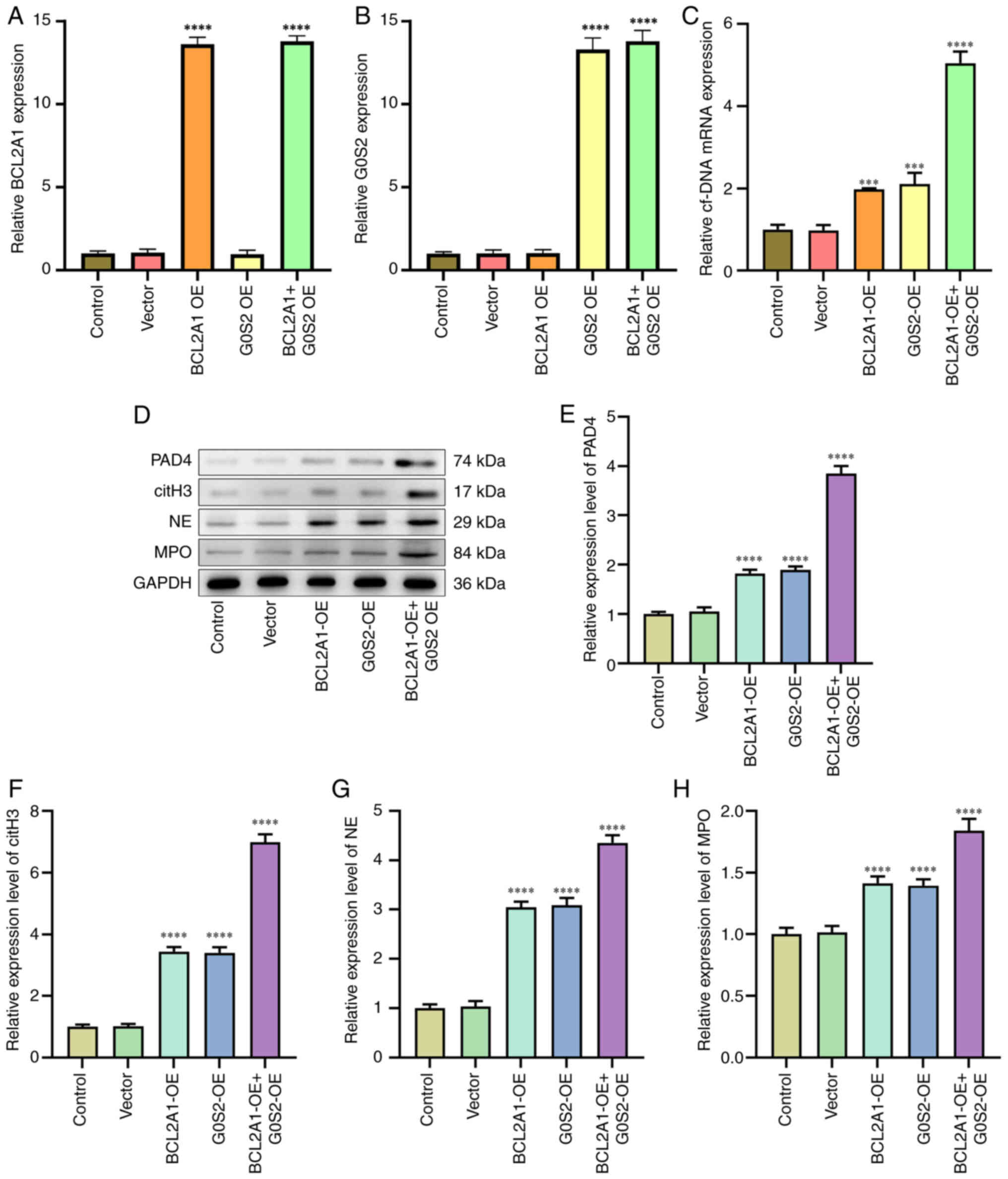
The RT-qPCR results indicated that circulating free
DNA expression was upregulated in the groups overexpressing BCL2A1
and G0S2 compared with the blank group (P<0.05; Fig. 6A-C). Western blot analysis showed
that the expression of PAD4, citH3, NE and MPO was increased in the
BCL2A1-overexpressing and G0S2-overexpressing groups relative to
the blank group (P<0.05; Fig.
6D-H).
Clinical relationship between BCL2A1
and G0S2 genes and BC
The combined analysis of the GEO (GSE24129) and
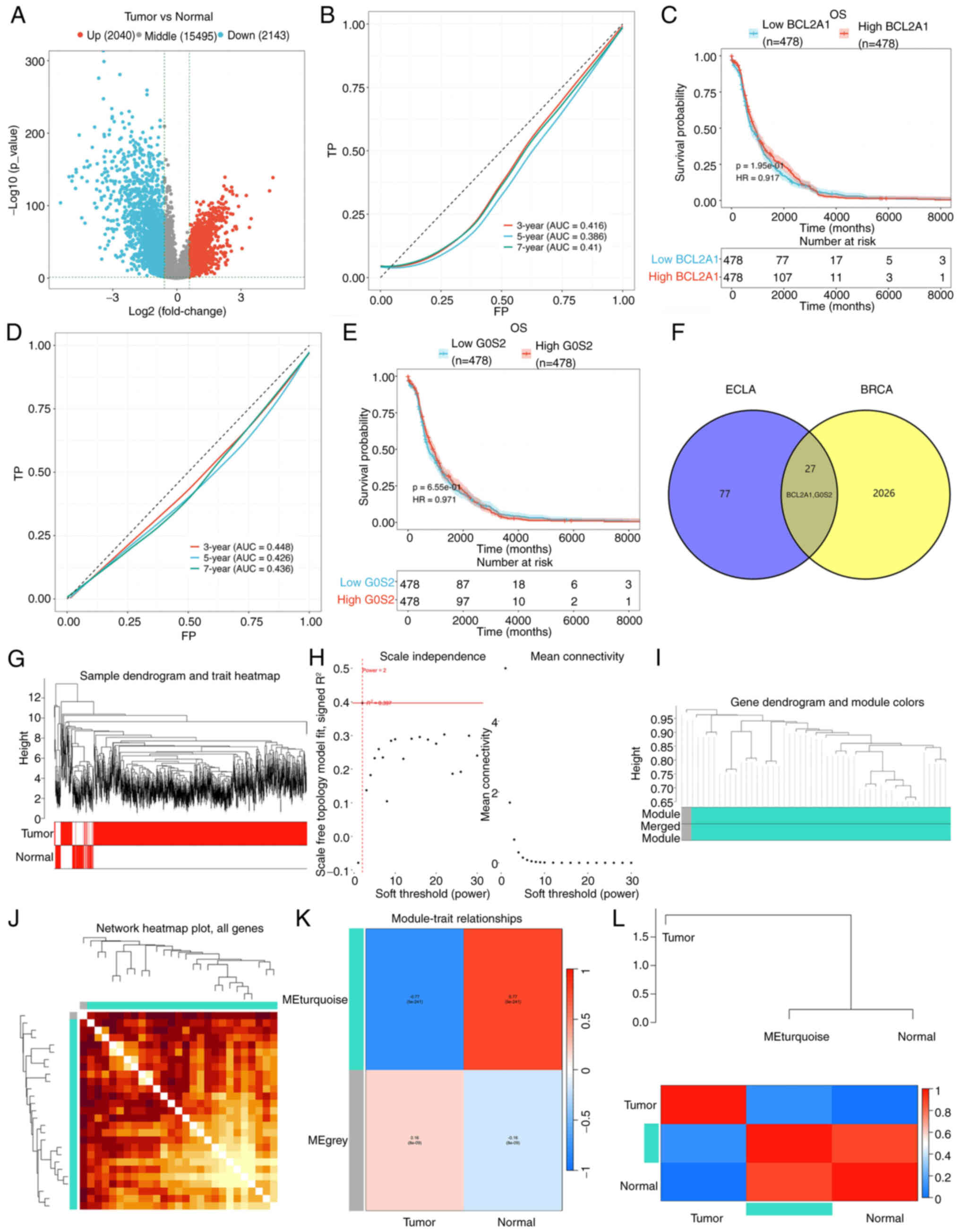
TCGA-BC datasets identified 4,183 DEGs, comprising 2,040
upregulated genes and 2,143 downregulated genes (Fig. 7A). The present study specifically
focused on the upregulated genes BCL2A1 and G0S2. Using the TCGA-BC
database, patients were categorized into high- and low-BCL2A1
groups. To assess the diagnostic accuracy of the prognostic risk
model, the areas under the time-dependent ROC curves (AUCs) were
calculated. The AUCs for the risk model in predicting 3-, 5-, and
7-year survival were 0.416, 0.386, and 0.410, respectively
(Fig. 7B). Additionally, patients
with high BCL2A1 expression did not show significantly different
overall survival (OS) compared with those with low BCL2A1
expression (Fig. 7C). In the
subgroup analyses based on G0S2 expression, the AUC of the risk
model for predicting 3-, 5-, and 7-year survival was 0.448, 0.426,
and 0.436, respectively (Fig. 7D).
Patients with high G0S2 expression also did not exhibit
significantly different OS when compared with those with low G0S2
expression (Fig. 7E). These results
indicated that the models lack strong predictive power.
Consequently, further analysis of the DEGs between patients with
preeclampsia and BC was performed using Venn diagrams, which
revealed 27 overlapping upregulated DEGs, including BCL2A1 and G0S2
(Fig. 7F).
Co-expression modules for 27 DEGs
To further investigate these 27 DEGs, WGCNA was
conducted to uncover the interactions and coregulatory mechanisms
among them. A cluster tree of samples was constructed using a
scale-free network and topological overlap based on dynamic hybrid
cutting (Fig. 7G). The optimal soft
threshold was determined according to the fitting index and the
average degree of network connection, in line with the scale-free
topology criterion (Fig. 7H). Based
on this optimal soft threshold, the gene modules were categorized
into two distinct modules, and a module cluster graph was created
(Fig. 7I). A correlation analysis
was performed between the modules and clinical features, presenting
the results in a heatmap (Fig. 7J).
The analysis revealed that the turquoise module exhibited the
strongest correlation with tumors (R=0.77, P=5e-241; Fig. 7K). The genes within this module were
then analyzed and the core genes closely associated with
preeclampsia and BC (BCL2A1 and G0S2) were further identified
(Fig. 7L).
Overexpression of the BCL2A1 and G0S2
genes in neutrophils inhibits malignant progression of BC
cells
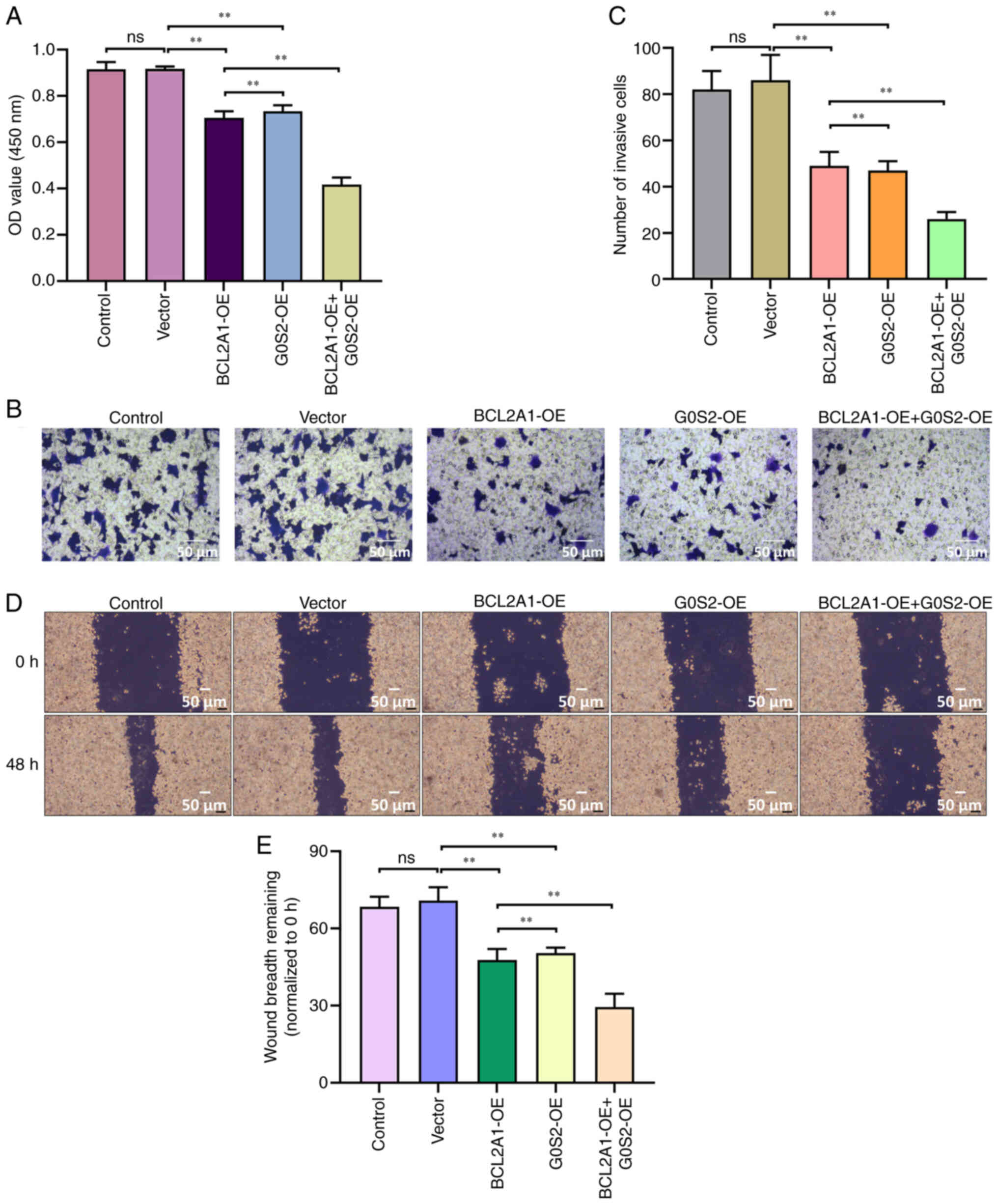
To validate this hypothesis, neutrophils that were
overexpressing BCL2A1 and G0S2 were co-cultured with MCF-7 cells
indirectly, followed by functional analysis. The results from the
CCK-8 assay indicated that the combination of BCL2A1-OE and G0S2-OE
led to a reduced proliferation capacity in MCF-7 cells (P<0.05;
Fig. 8A). Transwell experiments
demonstrated that the BCL2A1-OE + G0S2-OE group showed a
significant reduction in the invasive ability of MCF-7 cells
(P<0.05; Fig. 8B and C).
Additionally, this group also displayed the lowest level of
migration (P<0.05; Fig. 8D and
E). These findings suggest that the co-culture of BC cells with
neutrophils overexpressing BCL2A1 and G0S2 inhibits the
proliferation, invasion and migration of BC cells.
BCL2A1 and G0S2 gene expression levels
were significantly elevated in neutrophils from BC tissue, and
there was also an increase in NETs
To confirm the present findings, the expression
levels of BCL2A1, G0S2, cf.-DNA, PAD4, citH3, NE and MPO in BC
neutrophils were assessed using RT-qPCR and western blotting.
Additionally, immunofluorescence was employed to further validate
the expression of the BCL2A1 and G0S2 genes in neutrophils.
Notably, the RT-qPCR results indicated that the mRNA expression
levels of BCL2A1 and G0S2 were significantly elevated in BC
neutrophils compared with those in the control group (P<0.05;
Fig. 9A). Furthermore, western blot
analysis confirmed that the levels of BCL2A1, G0S2, PAD4, citH3, NE
and MPO were upregulated in the BC neutrophil group compared with
the blank group (P<0.05; Fig.
9B). The immunofluorescence results demonstrated that the
expression of BCL2A1 and G0S2 was significantly higher in the BC
tissue than in the control group, while GR-1 expression was
significantly lower in the BC tissue (P<0.05; Fig. 9C and D). Collectively, these data
suggest that the BCL2A1 and G0S2 genes play essential roles in
regulating neutrophil survival in BC tissues.
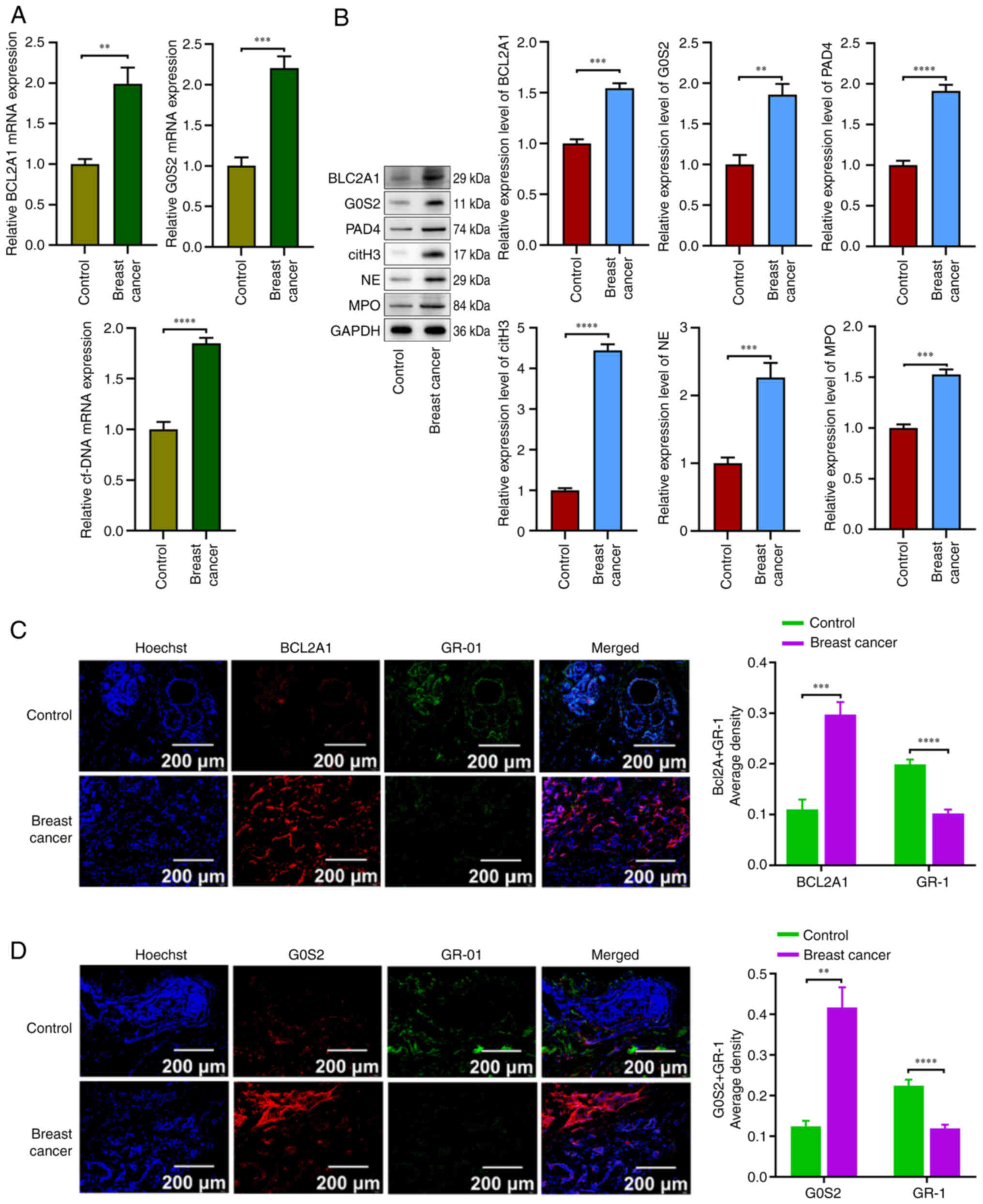 | Figure 9.BCL2A1 and G0S2 gene expression
levels were significantly elevated in neutrophils from breast
cancer tissue, along with increase in production of neutrophil
extracellular traps. (A) The mRNA expression level of BCL2A1, G0S2
and cf-DNA in breast cancer tissue compared with control. (B) The
protein expression level of BCL2A1, G0S2, PAD4, citH3, NE and MPO
in breast cancer tissue compared with control. (C and D) Expression
and localization of neutrophils, and BCL2A1 and G0S2 in breast
cancer cells at ×10 magnification. **P<0.01, ***P<0.001 and
****P<0.0001. All experiments were conducted in triplicate. MPO,
myeloperoxidase; G0S2, G0/G1 switch gene 2. |
Discussion
A number of epidemiological studies have shown that
preeclampsia can reduce the risk of BC through different pathways
(29–32). However, the specific underlying
mechanism is not yet clear. This is the first study in which
multi-omics analysis, clinical validation and cell culture
experiments have been combined to reveal the specific mechanisms by
which preeclampsia affects the incidence of BC. First, single-cell
sequencing data provided a clear understanding of the
cell-type-specific transcriptome alterations that occur in
preeclampsia placental tissue. The biological processes of 27,724
cells, including trophoblasts and neutrophils, were identified in
12 different cell types. These findings indicate that the
biological activities of these cells are mostly related to the cell
cycle, cell migration, angiogenesis, hormone release and the
inflammatory response. This stage of disease progression is crucial
to our understanding of this disease. Therefore, the authors
focused on the genes whose expression increased in neutrophils,
BCL2A1 and G0S2, to further investigate the relationship between
preeclampsia and BC.
Based on information in the TCGA database, the
associations between the differential mRNA expression in BC,
patients and the BCL2A1 and G0S2 genes, and the survival rate of
patients with BC were examined. Similarly, the expression of the
BCL2A1 and G0S2 genes was elevated in BC. The genes upregulated in
BC from the TCGA database were combined with the preeclampsia
dataset from the GEO database by Venn diagram analysis to identify
27 overlapping genes, including BCL2A1 and G0S2, to study the
relationship between preeclampsia and BC. According to our
findings, the BCL2A1 and G0S2 genes are associated with both
preeclampsia and BC, and they may be crucial in the onset and
progression of these two diseases. Using WGCNA analysis, it was
also discovered that BCL2A1 and G0S2 were both important genes. As
a result, BCL2A1 and G0S2 may be crucial in the disorders
preeclampsia and BC.
Like other Bcl-2 family members, BCL2A1 is crucial
for controlling apoptosis. It has been revealed that BCL2A1 also
plays additional critical roles in vascular endothelial cells. In
activated endothelial cells, TNF-α was found to be a gene caused by
cytokine treatment (33).
Preeclampsia is a significant pregnancy complication characterized
by hypertension, proteinuria, endothelial dysfunction and an
immune-inflammatory response (1).
In the present study, it was discovered that neutrophils from the
placental tissue of preeclampsia patients had considerably greater
BCL2A1 expression. BCL2A1 is specifically involved in the
pathogenesis of preeclampsia. BCL2A1 is a tightly regulated target
gene of nuclear factor κB (NF-κB) that plays a crucial role in cell
survival (34). Additionally,
BCL2A1 is primarily expressed in the hematopoietic system, where it
enhances the survival and inflammatory responses of specific
subsets of leukocytes. According to previous studies, BCL2A1 is
overexpressed in various cancer types, including ovarian cancer
(35), BC (36), colon cancer (37) and prostate cancer (38). Furthermore, as an NF-κB target gene,
BCL2A1 also plays a significant role in inflammation (34). Inflammation is a key aspect of the
innate immune response, with NOD-like receptors participating in
inflammasome formation under the influence of pattern recognition
receptors (PRRs). One of the primary signaling pathways activated
by PRRs involves the activation of NF-κB and the upregulation of
proinflammatory genes (39).
Consequently, the formation of inflammasomes may also trigger the
expression of BCL2A1, thereby supporting the survival of
proinflammatory cells during the immune response (34). The present findings suggest that the
overexpression of BCL2A1 in neutrophils could promote apoptosis and
inhibit the proliferation of BC cells, providing a plausible
explanation for this observation.
G0S2 was initially discovered to be involved in the
transition of the cell cycle from the G0 to the G1 phase induced by
lectin (40). The G0S2 gene plays
an important role in cell cycle regulation and apoptosis (41). Studies have shown that G0S2 can
affect cell apoptosis and survival by inhibiting lipolysis and
fatty acid oxidation under certain conditions (42,43).
Furthermore, the expression of G0S2 may be associated with the
function of vascular endothelial cells and vascular pathology. For
example, G0S2 can ameliorate oxidative low-density
lipoprotein-induced vascular endothelial cell damage by regulating
mitochondrial apoptosis (44).
Placental abnormalities and metabolic disorders are important
features of preeclampsia, and adipose metabolism plays a role in
these conditions (45). Moreover,
adipose tissue also plays an important regulatory role in the
progression of breast tumors, as adipose tissue can provide
nutrients and adipokines for proliferating tumor cells. Studies
have shown that fatty acid metabolism also plays an important role
in various aspects of tumor cell proliferation, transformation and
migration (46). It has been
reported that siRNA-mediated knockdown of G0S2 leads to reduced
proliferation, migration and invasion of BC cells, suggesting that
G0S2 is a major factor that promotes the survival and metastasis of
BC cells (47). Notably, the
present findings revealed that the overexpression of G0S2 in
neutrophils can reduce the proliferation, invasion and migration
capacity of BC cells. This suggests that the invasion and migration
of BC cells are influenced by fatty acid metabolism, where the
overexpression of G0S2 can inhibit fatty acid oxidation, thereby
disrupting the invasive and migratory abilities of BC cells.
Furthermore, G0S2 can localize to the endoplasmic
reticulum and mitochondria (48,49).
G0S2 can interact with Bcl-2, preventing the formation of the
Bcl-2/Bax heterodimeric complex by controlling mitochondrial
membrane permeability and cytochrome release, thereby modulating
its antiapoptotic activity in human cancer cells (49). Thus, these results suggest that the
synergistic effect of BCL2A1 and G0S2 in neutrophils can inhibit
the growth and metastasis of BC cells.
Neutrophils are the first line of defense in the
immune system, and they function by phagocytosis and degranulation.
Recent research has unveiled the existence of a distinctive variant
of neutrophil death, known as neutrophil necrosis. This process
actively contributes to the extermination of pathogens through the
extracellular release of NETs (50). NETs are intricate DNA networks
filled with antimicrobial peptides that are diligently discharged
by neutrophils in response to diverse stimuli. NETs are not only
essential for neutrophil innate immune responses but also play a
role in autoimmune diseases such as systemic lupus erythematosus,
rheumatoid arthritis (51) and
psoriasis (52). Additionally, they
are implicated in non-infectious conditions, including
coagulopathies (53), thrombosis
(54), diabetes (55) and atherosclerosis (56). Notably, NETs play dual roles in
tumors, serving both as facilitators and inhibitors of tumor
progression. NETs composed of myeloperoxidase, proteases and
histones can eliminate tumors, and impede tumor proliferation and
metastasis. However, they also have the potential to degrade the
extracellular matrix, promoting the escape and metastasis of cancer
cells (57). In the present study,
an increase was observed in the release of NETs in BC. In
vitro cell experiments indicated that the BCL2A1 and G0S2 genes
regulate the generation of NETs, leading to the inhibition of
proliferation, migration and invasion in BC cells. Consequently,
NETs can potentially impede the malignant progression of BC by
suppressing the biological functions of these cells.
The present study has several limitations. First,
the small public dataset used could skew the outcomes of the
investigation. Second, the present study confirmed a lower risk of
BC in preeclamptic women, which is consistent with the results
reported in the majority of related studies (29–32);
however, there are also conflicting results. A positive correlation
was reported in one study (58),
and it was also reported in two additional studies (59,60).
Finally, additional research is still needed to confirm the
co-expression of BCL2A1 and G0S2 as a reliable indicator of
preeclampsia and BC. In conclusion, the present study proposed that
neutrophils may be co-pathogenic factors between preeclampsia and
BC, and further elucidated that BC risk was reduced in patients
with preeclampsia due to the regulatory role of the BCL2A1 and G0S2
genes in neutrophil-mediated NET production (Fig. 10).
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangxi Natural Science
Foundation Program (grant nos. 2023GXNSFAA026037 and
2024GXNSFAA010368), the Guangxi medical and health appropriate
technology development and application project (grant nos. S2022080
and S2022062), the Excellent medical talents training program of
the First Affiliated Hospital of Guangxi Medical University and the
Project on Enhancement of Basic Research Ability of Young and
Middle-aged Teachers in Guangxi Universities and Colleges (grant
no. 2024KY0110).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LX and JinL performed data analysis and prepared
figures. LX, JinL, JiaL, MW and XL performed experiments. LX wrote
the manuscript. JieL and YZ designed and supervised the study, and
revised the manuscript. LX and JL confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all study
participants for the use of tissue samples in scientific research.
The present study was conducted in accordance with the ethical
guidelines and was approved (approval no. 2022-E0118) by the
Ethical Review Committee of the First Affiliated Hospital of
Guangxi Medical University (Nanning, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
DEGs
|
differentially expressed genes
|
|
G0S2
|
G0/G1 switch gene 2
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
NETs
|
neutrophil extracellular traps
|
|
NF-κB
|
nuclear factor κB
|
|
PCA
|
principal component analysis
|
|
PRRs
|
pattern recognition receptors
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
ROS
|
reactive oxygen species
|
|
scRNA-seq
|
single-cell RNA sequencing
|
|
TANs
|
tumor-associated neutrophils
|
|
UMAP
|
Uniform Manifold Approximation and
Projection
|
|
WGCNA
|
weighted gene co-expression network
analysis
|
References
|
1
|
Rana S, Lemoine E, Granger JP and
Karumanchi SA: Preeclampsia: Pathophysiology, challenges, and
perspectives. Circ Res. 124:1094–1112. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Sayed AAF: Preeclampsia: A review of
the pathogenesis and possible management strategies based on its
pathophysiological derangements. Taiwan J Obstet Gynecol.
56:593–598. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piani F, Agnoletti D, Baracchi A,
Scarduelli S, Verde C, Tossetta G, Montaguti E, Simonazzi G, Degli
Esposti D and Borghi C; HDP Bologna Study Group, : Serum uric acid
to creatinine ratio and risk of preeclampsia and adverse pregnancy
outcomes. J Hypertens. 41:1333–1338. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dimitriadis E, Rolnik DL, Zhou W,
Estrada-Gutierrez G, Koga K, Francisco RPV, Whitehead C, Hyett J,
da Silva Costa F, Nicolaides K and Menkhorst E: Pre-eclampsia. Nat
Rev Dis Primers. 9:82023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tossetta G, Fantone S, Giannubilo SR,
Marinelli Busilacchi E, Ciavattini A, Castellucci M, Di Simone N,
Mattioli-Belmonte M and Marzioni D: Pre-eclampsia onset and SPARC:
A possible involvement in placenta development. J Cell Physiol.
234:6091–6098. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Licini C, Avellini C, Picchiassi E, Mensà
E, Fantone S, Ramini D, Tersigni C, Tossetta G, Castellucci C and
Tarquini F: Pre-eclampsia predictive ability of maternal miR-125b:
A clinical and experimental study. Transl Res. 228:13–27. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Inversetti A, Pivato CA, Cristodoro M,
Latini AC, Condorelli G and Di Simone N: Update on long-term
cardiovascular risk after pre-eclampsia: A systematic review and
meta-analysis. Eur Heart J Qual Care Clin Outcomes. 10:4–13. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gilbert JS, Bauer AJ, Gilbert SA and Banek
CT: The opposing roles of anti-angiogenic factors in cancer and
preeclampsia. Front Biosci (Elite Ed). 4:2652–2669. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miller D, Motomura K, Galaz J, Gershater
M, Lee ED, Romero R and Gomez-Lopez N: Cellular immune responses in
the pathophysiology of preeclampsia. J Leukoc Biol. 111:237–260.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sreeramkumar V, Adrover JM, Ballesteros I,
Cuartero MI, Rossaint J, Bilbao I, Nácher M, Pitaval C, Radovanovic
I, Fukui Y, et al: Neutrophils scan for activated platelets to
initiate inflammation. Science. 346:1234–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chawla N, Shah H, Huynh K, Braun A,
Wollocko H and Shah NC: The role of Platelet-activating factor and
magnesium in obstetrics and gynecology: Is there crosstalk between
pre-Eclampsia, clinical hypertension, and HELLP syndrome?
Biomedicines. 11:13432023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gupta AK, Hasler P, Holzgreve W and Hahn
S: Neutrophil NETs: A novel contributor to Preeclampsia-associated
placental hypoxia? Semin Immunopathol. 29:163–167. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McFarlane AJ, Fercoq F, Coffelt SB and
Carlin LM: Neutrophil dynamics in the tumor microenvironment. J
Clin Invest. 131:e1437592021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li MO, Wolf N, Raulet DH, Akkari L, Pittet
MJ, Rodriguez PC, Kaplan RN, Munitz A, Zhang Z, Cheng S and
Bhardwaj N: Innate immune cells in the tumor microenvironment.
Cancer Cell. 39:725–729. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fridlender ZG, Sun J, Kim S, Kapoor V,
Cheng G, Ling L, Worthen GS and Albelda SM: Polarization of
tumor-associated neutrophil phenotype by TGF-beta: ‘N1’ versus ‘N2’
TAN. Cancer Cell. 16:183–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Granot Z, Henke E, Comen EA, King TA,
Norton L and Benezra R: Tumor entrained neutrophils inhibit seeding
in the premetastatic lung. Cancer Cell. 20:300–314. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giese MA, Hind LE and Huttenlocher A:
Neutrophil plasticity in the tumor microenvironment. Blood.
133:2159–2167. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hirschhorn D, Budhu S, Kraehenbuehl L,
Gigoux M, Schröder D, Chow A, Ricca JM, Gasmi B, De Henau O,
Mangarin LMB, et al: T cell immunotherapies engage neutrophils to
eliminate tumor antigen escape variants. Cell. 186:1432–1447.e17.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lun ATL, Riesenfeld S, Andrews T, Dao TP,
Gomes T; participants in the 1st Human Cell Atlas Jamboree, ;
Marioni JC: EmptyDrops: Distinguishing cells from empty droplets in
Droplet-based Single-cell RNA sequencing data. Genome Biol.
20:632019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McCarthy DJ, Campbell KR, Lun AT and Wills
QF: Scater: Pre-processing, quality control, normalization and
visualization of single-cell RNA-seq data in R. Bioinformatics.
33:1179–1186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao Y, Hao S, Andersen-Nissen E, Mauck WM
III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al:
Integrated analysis of multimodal single-cell data. Cell.
184:3573–3587.e29. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Puthumana J, Thiessen-Philbrook H, Xu L,
Coca SG, Garg AX, Himmelfarb J, Bhatraju PK, Ikizler TA, Siew ED,
Ware LB, et al: Biomarkers of inflammation and repair in kidney
disease progression. J Clin Invest. 131:e1399272021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin S, Guerrero-Juarez CF, Zhang L, Chang
I, Ramos R, Kuan CH, Myung P, Plikus MV and Nie Q: Inference and
analysis of cell-cell communication using CellChat. Nat Commun.
12:10882021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trapnell C, Cacchiarelli D, Grimsby J,
Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS and
Rinn JL: The dynamics and regulators of cell fate decisions are
revealed by pseudotemporal ordering of single cells. Nat
Biotechnol. 32:381–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou W, Wang H, Yang Y, Guo F, Yu B and Su
Z: Trophoblast cell subtypes and dysfunction in the placenta of
individuals with preeclampsia revealed by SingleCell RNA
sequencing. Mol Cells. 45:317–328. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Powell M, Fuller S, Gunderson E and Benz
C: A common IGF1R gene variant predicts later life breast cancer
risk in women with preeclampsia. Breast Cancer Res Treat.
197:149–159. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nichols HB, House MG, Yarosh R, Mitra S,
Goldberg M, Bertrand KA, Eliassen AH, Giles GG, Jones ME, Milne RL,
et al: Hypertensive conditions of pregnancy, preterm birth, and
premenopausal breast cancer risk: A premenopausal breast cancer
collaborative group analysis. Breast Cancer Res Treat. 199:323–334.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Opdahl S, Romundstad PR, Alsaker MDK and
Vatten LJ: Hypertensive diseases in pregnancy and breast cancer
risk. Br J Cancer. 107:176–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vatten LJ, Romundstad PR, Trichopoulos D
and Skjaerven R: Pre-eclampsia in pregnancy and subsequent risk for
breast cancer. Br J Cancer. 87:971–973. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karsan A, Yee E, Kaushansky K and Harlan
JM: Cloning of human Bcl-2 homologue: Inflammatory cytokines induce
human A1 in cultured endothelial cells. Blood. 87:3089–3096. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vogler M: BCL2A1: The underdog in the BCL2
family. Cell Death Differ. 19:67–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang R, Yung MMH, He F, Jiao P, Chan KKL,
Ngan HYS and Chan DW: The Stress-inducible BCL2A1 is required for
ovarian cancer metastatic progression in the peritoneal
microenvironment. Cancers (Basel). 13:45772021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murthy SRK, Cheng X, Zhuang T, Ly L, Jones
O, Basadonna G, Keidar M and Canady J: BCL2A1 regulates Canady
Helios Cold Plasma-induced cell death in triple-negative breast
cancer. Sci Rep. 12:40382022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yue T, Liu X, Zuo S, Zhu J, Li J, Liu Y,
Chen S and Wang P: BCL2A1 and CCL18 are predictive biomarkers of
cisplatin chemotherapy and immunotherapy in colon cancer patients.
Front Cell Dev Biol. 9:7992782021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pucci P, Venalainen E, Alborelli I,
Quagliata L, Hawkes C, Mather R, Romero I, Rigas SH, Wang Y and
Crea F: LncRNA HORAS5 promotes taxane resistance in
castration-resistant prostate cancer via a BCL2A1-dependent
mechanism. Epigenomics. 12:1123–1138. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takeuchi O and Akira S: Pattern
recognition receptors and inflammation. Cell. 140:805–820. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Russell L and Forsdyke DR: A human
putative lymphocyte G0/G1 switch gene containing a CpG-rich island
encodes a small basic protein with the potential to be
phosphorylated. DNA Cell Biol. 10:581–591. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heckmann BL, Zhang X, Xie X and Liu J: The
G0/G1 switch gene 2 (G0S2): Regulating metabolism and beyond.
Biochim Biophys Acta. 1831:276–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang X, Heckmann BL, Campbell LE and Liu
J: G0S2: A small giant controller of lipolysis and adipose-liver
fatty acid flux. Biochim Biophys Acta Mol Cell Biol Lipids.
1862:1146–1154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang X, Lu X and Lombès M, Yang X, Lu X
and Lombès M: The G(0)/G(1) switch gene 2 regulates adipose
lipolysis through association with adipose triglyceride lipase.
Cell Metab. 11:194–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liang Z, Diao W, Jiang Y and Zhang Y: G0S2
ameliorates oxidized low-density lipoprotein-induced vascular
endothelial cell injury by regulating mitochondrial apoptosis. Ann
Transl Med. 10:13832022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kaaja R, Tikkanen MJ, Viinikka L and
Ylikorkala O: Serum lipoproteins, insulin, and urinary prostanoid
metabolites in normal and hypertensive pregnant women. Obstet
Gynecol. 85:353–356. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu X, Deng F, Li Y, Daniels G, Du X, Ren
Q, Wang J, Wang LH, Yang Y, Zhang V, et al: ACSL4 promotes prostate
cancer growth, invasion and hormonal resistance. Oncotarget.
6:44849–44863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cho E, Kwon YJ, Ye DJ, Baek HS, Kwon TU,
Choi HK and Chun YJ: G0/G1 switch 2 induces cell survival and
metastasis through Integrin-mediated signal transduction in human
invasive breast cancer cells. Biomol Ther (Seoul). 27:591–602.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zandbergen F, Mandard S, Escher P, Tan NS,
Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S,
et al: The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem
J. 392:313–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Welch C, Santra MK, El-Assaad W, Zhu X,
Huber WE, Keys RA, Teodoro JG and Green MR: Identification of a
protein, G0S2, that lacks Bcl-2 homology domains and interacts with
and antagonizes Bcl-2. Cancer Res. 69:6782–6789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Islam MM and Takeyama N: Role of
neutrophil extracellular traps in health and disease
pathophysiology: Recent insights and advances. Int J Mol Sci.
24:158052023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fresneda Alarcon M, McLaren Z and Wright
HL: Neutrophils in the pathogenesis of rheumatoid arthritis and
systemic lupus erythematosus: Same Foe Different M.O. Front
Immunol. 12:6496932021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shao S, Fang H, Dang E, Xue K, Zhang J, Li
B, Qiao H, Cao T, Zhuang Y, Shen S, et al: Neutrophil extracellular
traps promote inflammatory responses in psoriasis via activating
epidermal TLR4/IL-36R crosstalk. Front Immunol. 10:7462019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jin J, Wang F, Tian J, Zhao X, Dong J,
Wang N, Liu Z, Zhao H, Li W, Mang G and Hu S: Neutrophil
extracellular traps contribute to coagulopathy after traumatic
brain injury. JCI Insight. 8:e1411102023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Van Bruggen S and Martinod K: The coming
of age of neutrophil extracellular traps in thrombosis: Where are
we now and where are we headed? Immunol Rev. 314:376–398. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang S, Wang S, Chen L, Wang Z, Chen J, Ni
Q, Guo X, Zhang L and Xue G: Neutrophil extracellular traps delay
diabetic wound healing by inducing Endothelial-to-Mesenchymal
transition via the hippo pathway. Int J Biol Sci. 19:347–361. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sano M, Maejima Y, Nakagama S,
Shiheido-Watanabe Y, Tamura N, Hirao K, Isobe M and Sasano T:
Neutrophil extracellular traps-mediated Beclin-1 suppression
aggravates atherosclerosis by inhibiting macrophage autophagy.
Front Cell Deve Biol. 10:8761472022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Masucci MT, Minopoli M, Del Vecchio S and
Carriero MV: The emerging role of neutrophil extracellular traps
(NETs) in tumor progression and metastasis. Front Immunol.
11:17492020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Calderon-Margalit R, Friedlander Y, Yanetz
R, Deutsch L, Perrin MC, Kleinhaus K, Tiram E, Harlap S and Paltiel
O: Preeclampsia and subsequent risk of cancer: Update from the
Jerusalem Perinatal Study. Am J Obstet Gynecol. 200:63.e1–5. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Brasky TM, Li Y, Jaworowicz DJ Jr,
Potischman N, Ambrosone CB, Hutson AD, Nie J, Shields PG, Trevisan
M, Rudra CB, et al: Pregnancy-related characteristics and breast
cancer risk. Cancer Causes Control. 24:1675–1685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nechuta S, Paneth N and Velie EM:
Pregnancy characteristics and maternal breast cancer risk: A review
of the epidemiologic literature. Cancer Causes Control. 21:967–989.
2010. View Article : Google Scholar : PubMed/NCBI
|