Introduction
Esophageal cancer (ESCA) is a highly aggressive
malignancy and its incidence and mortality are increasing globally
(1). As ESCA is asymptomatic in
the early stage, most patient cases of ESCA are diagnosed in the
locally advanced stage or even in the distant metastasis stage
(2). Administering neoadjuvant
chemoradiotherapy as standard treatment for locally advanced ESCA
is beneficial (3). Due to the
aggressiveness, late diagnosis and treatment-refractory nature of
ESCA, most patients are not sensitive to the standard treatment.
Thus, the incidence of postoperative local recurrence or distant
metastases is high and the prognosis is poor. A previous study
showed that immunotherapy with chemoradiotherapy or chemotherapy as
neoadjuvant therapy can effectively improve anti-tumor effects in
the treatment of ESCA (4).
However, assessing the response to neoadjuvant therapy accurately
is difficult and the treatment of a number of patients diagnosed
with ESCA with immunotherapeutic combinations is unsatisfactory
(5). Therefore, novel early
diagnostic biomarkers and effective immunotherapeutic targets need
to be identified to improve the outcome of patients with ESCA.
In mammalian cells, histone 3 lysine 4 (H3K4)
methylation modifications are important for DNA methylation and
epigenetic inheritance. H3K4, as a histone which codes or as an
identifier of gene promoters, is closely associated with the cell
cycle, DNA replication, cell proliferation, tumorigenesis, invasion
metastasis and immune evasion in various malignant tumors (6). The agonist of H3K4 methylation
depends on its core components [WD-repeat protein-5,
retinoblastoma-binding protein-5, absent, small, homeotic
disks-2-like and dpy-30 homolog (DPY30)] and the inactivation of
DPY30 decreases the levels of H3K4 methylation (7,8).
DPY30 is involved in several physiological functions such as cell
proliferation, differentiation, apoptosis and senescence (9,10).
For example, DPY30 can promote the proliferation and
differentiation of hematopoietic stem cells by directly and
preferentially controlling the methylation of H3K4 and the
expression of a number of genes required for hematopoiesis
(11,12). Additionally, DPY30 deficiency can
induce apoptosis of neural stem cells (13). DPY30 also strongly influences
tumorigenesis in several types of cancers (14–16).
The knockdown or overexpression of DPY30 might directly regulate
H3K4 methylation levels and significantly affect the ability of
gastric cancer cells to proliferate, migrate and invade (6). DPY30 can also promote the expression
of vimentin by regulating the H3K4 methylation level of the
vimentin gene promoter, which promotes the epithelial-mesenchymal
transition (EMT) in epithelial ovarian cancer (15). High tissue levels of DPY30 can
induce EMT through the activation of the Wnt/β-catenin signaling
pathway during tumorigenesis and the development of cervical
squamous cell carcinoma (16).
However, the role of DPY30 in the development and carcinogenesis of
ESCA remains to be elucidated.
Therefore, the present study first determined the
relationship between the mRNA expression level of DPY30 and its
diagnostic and prognostic values in ESCA using bioinformatics.
Then, based on the collected clinical information and tissue
microarray of patients with ESCA, it evaluated the DPY30 protein
expression level and its diagnostic and prognostic value in ESCA.
Finally, it specifically analyzed the relationship between the
expression level of DPY30 and the tumor immune microenvironment to
identify a novel immunotherapeutic target for treating patients
with ESCA.
Materials and methods
ESCA data sources and mRNA expression
of DPY30
In July 2022, data on the gene expression of ESCA
tissues and normal esophageal tissues were obtained from The Cancer
Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx)
databases. Among them, 162 ESCA tissue samples and 11
tumor-adjacent normal esophageal epithelial tissue samples were
obtained from TCGA database and 653 normal esophageal tissue
samples were obtained from the GTEx database. The data on the
clinical characteristics and survival information of 173 patients
with ESCA were also downloaded from TCGA database. The data on ESCA
included transcripts per million (TPM) types and fragments per
kilobase per million (FPKM) types. The level of expression of the
DPY30 mRNA in ESCA tissues and normal esophageal tissues was
determined using the aforementioned data.
Patients and tissue microarray of
specimens
In total, 57 patients with ESCA underwent surgical
resection at Union Hospital of Huazhong University of Science and
Technology, Tongji Medical College (Wuhan, China) between 2013 and
2015. Paraffin specimens (57 pairs) from these patients were
obtained by surgical resection. The clinical features of the
patients are presented in Table
SI. All ESCA tissues and paired paracancerous tissues (diameter
of 1.5 mm; selected by a pathologist) from each paraffin specimen
were extracted and used to constructed a tissue microarray (TMA)
paraffin block. After checking the quality of the specimens, the
TMA was cut into sections (3 mm thick) for immunohistochemistry
analysis. The study was approved by the Institutional Ethics
Committee of Tongji Medical College, Huazhong University of Science
and Technology [approval number (2021) 0158]. Written informed
consent was obtained from all patients before they underwent
surgical procedures.
Immunohistochemistry staining and
assessment
Immunohistochemistry (IHC) staining was performed
for the TMA with 57 pairs of tissues by two professional
pathologists. The primary rabbit anti-DPY30 antibody (dilution
1:100; incubation at 37°C for 30 min; cat. no. CSB-PA861193LA01HU)
was purchased from Cusabio Technology LLC. Using the CaseViewer 2.4
software (3DHISTECH Ltd.) and HALO image analysis software
(v2.1.1637.26; Indica Labs), images were captured and analyzed.
Each specimen was first assigned a score based on the staining
intensity and the extent of stained cells (no staining=0, weak
staining=1, moderate staining=2 and strong staining=3) and (0–5%=0,
5–25%=1, 26–50%=2, 51–75%=3 and 76–100%=4). Based on the AI
pathwell v2 image analysis software (Servicebio) using the
principles of AI deep learning, the percentage of weak, moderate
and strong intensity, percentage of positive area, mean density and
area density was calculated (Table
SII). Then, two professional pathologists validated the results
of the immunohistochemical staining analysis and the results were
automatically calculated for each specimen based on the original
basic data and formulae. The final IHC scores were calculated using
the formula ∑(pi × i)=(percentage of weak intensity ×1) +
(percentage of moderate intensity ×2) + (percentage of strong
intensity ×3). Here, ‘pi’ represents the ratio of the positive
signal pixel area and ‘i’ represents the staining intensity; the
IHC scores (area) are data between 0–300 (17).
Identification of the diagnostic and
prognostic value of DPY30 in ESCA
By performing the Receiver Operating Curve (ROC)
analysis in TCGA database, the diagnostic value of DPY30 expression
in ESCA was assessed using the area under the curve (AUC).
Kaplan-Meier survival analysis was performed to determine the
relationship between DPY30 expression and overall survival (OS) in
patients with ESCA. The groups were formed based on the median
value of the expression of DPY30. Based on the clinical
characteristics of the patients with ESCA in TCGA database, the
relationship between clinicopathological characteristics and DPY30
mRNA expression levels was first determined to establish a
prognostic nomogram. Then, according to the collected
clinicopathological characteristics from 57 patients with ESCA, the
relationship between the clinicopathological characteristics and
DPY30 protein expression levels was confirmed.
Functional mechanisms and
protein-protein interaction (PPI) networks of DPY30 co-expressed
genes
Based on the sequencing data of tumor tissues from
patients with ESCA in TCGA, the DPY30-associated genes were
analyzed by Spearman's method. The co-expressed genes of DPY30 were
screened with correlation coefficients greater than 0.4 and
P-values less than 0.01. The genes were then divided into two
groups based on the median value of the expression level of DPY30.
The genes that were significantly correlated (positively and
negatively) with DPY30 were obtained by performing the Gene Set
Enrichment Analysis (GSEA). A protein-protein correlation network
was constructed based on the co-expressed genes using the STRING
database (http://string-db.org/) and Cytoscape
software (version 3.7.2; www.cytoscape.org). The Gene Ontology (GO)/Kyoto
Encyclopedia of Genes and Genomes (KEGG) function analysis was
conducted to determine the role of DPY30 in ESCA.
Correlation of DPY30 expression with
the tumor microenvironment of ESCA
ESCA tissues from TCGA database were evaluated by
performing immune scoring via the Estimation of Stromal and Immune
cells in Malignant Tumor tissues using the Expression data
(ESTIMATE) and ssGSEA algorithm (18,19).
The relationship between the DPY30 expression level and ESCA
infiltrating immune cells was investigated using Spearman's
correlation.
Statistical analysis
Statistical analysis was performed using SPSS v22.0
statistical software (IBM Corp.). The significant differences
between groups were determined by performing unpaired and paired
Student's t-tests. The association between DPY30 expression levels
with the clinicopathological features of patients with ESCA was
evaluated by performing a one-way ANOVA. If the association was
significant, it was evaluated by Scheffe post hoc test (20). All quantitative results were
expressed as the mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
DPY30 mRNA and protein are
overexpressed in ESCA
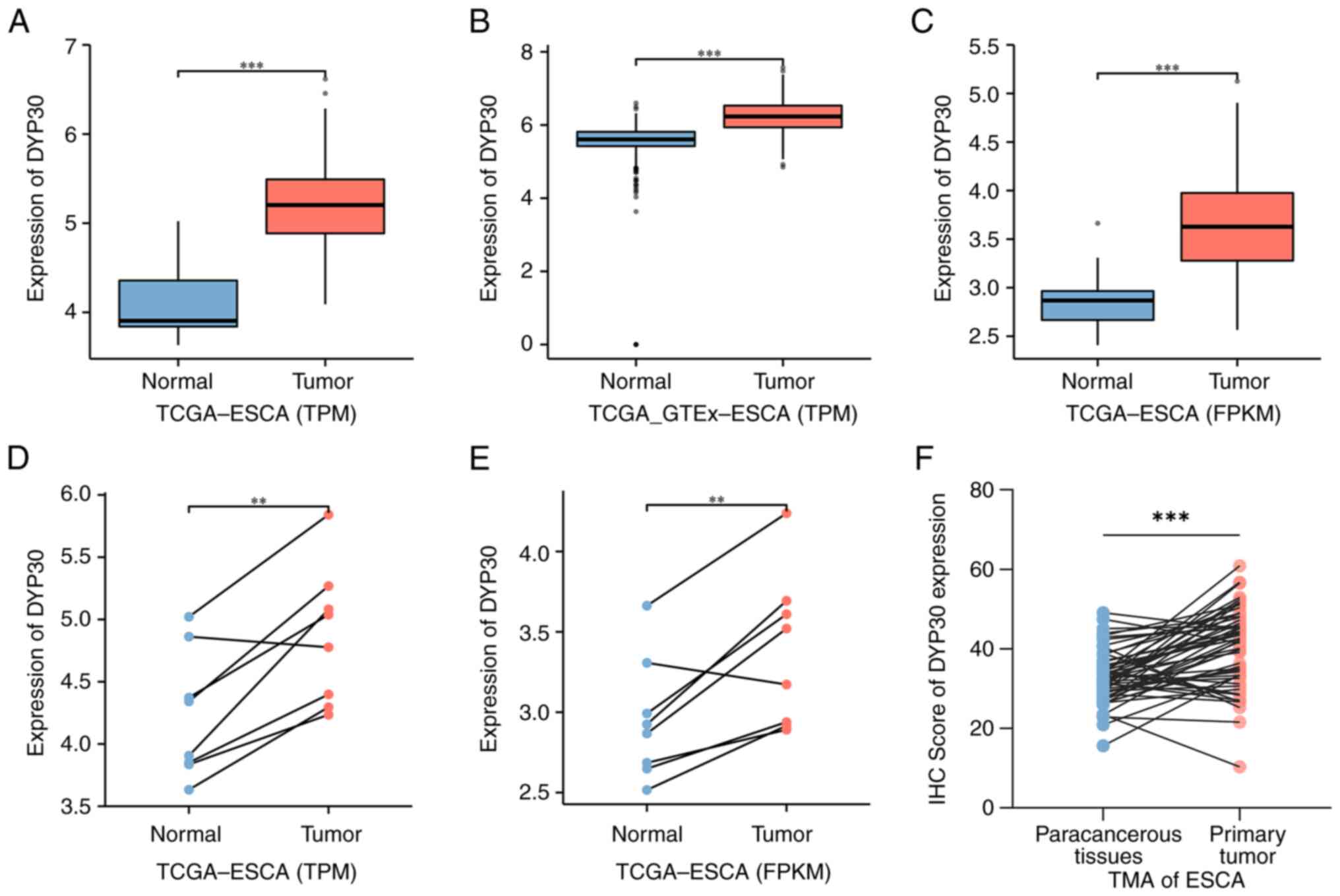
Based on TCGA and GTEx databases, the results
demonstrated that the expression of DPY30 mRNA was significantly
higher in unpaired ESCA tissues compared with normal esophageal
tissues (Fig. 1A-C). For
one-to-one matching, the expression of DPY30 mRNA in paired ESCA
tissues was significantly higher compared with normal esophageal
tissues (Fig. 1D and E).
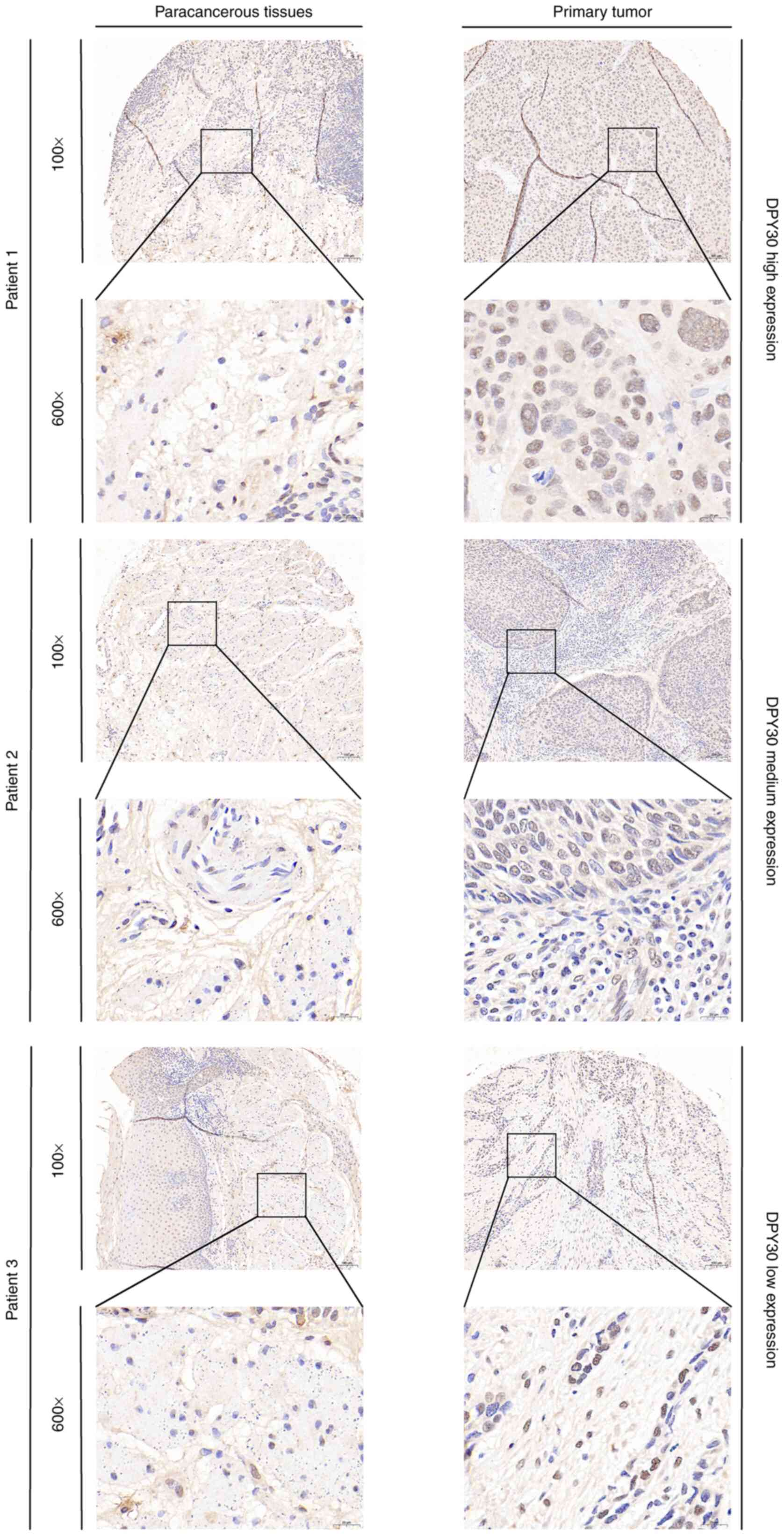
Additionally, the results of the immunohistochemistry analysis
demonstrated that the protein of DPY30 was significantly
overexpressed in ESCA tissues compared with that in paracancerous
tissues (Figs. 1F and S1). For the DPY30 protein expression in
the clinical TMA from 57 paired ESCA (ESCA) tissues, the
representative immunohistochemical images of the high, medium and
low protein expression of DPY30 are shown in Fig. 2. Thus, DPY30 was overexpressed in
ESCA patients and might influence the diagnosis and prognosis of
ESCA.
DPY30 has diagnostic value in
ESCA
The diagnostic value of DPY30 in ESCA was determined
based on the ROC curves. The TPM type of TCGA database analysis
demonstrated that the area under the ROC curve (AUC) value of DPY30
was 0.938 in ESCA (Fig. 3A). By
analyzing the TPM type of TCGA and GTEx databases, the AUC value of
DPY30 was calculated to be 0.858 in ESCA (Fig. 3B). By analyzing the FPKM type of
TCGA database, the AUC value of DPY30 was calculated to be 0.909 in
ESCA (Fig. 3C). These results
suggested that DPY30 plays an important role in the diagnosis of
ESCA.
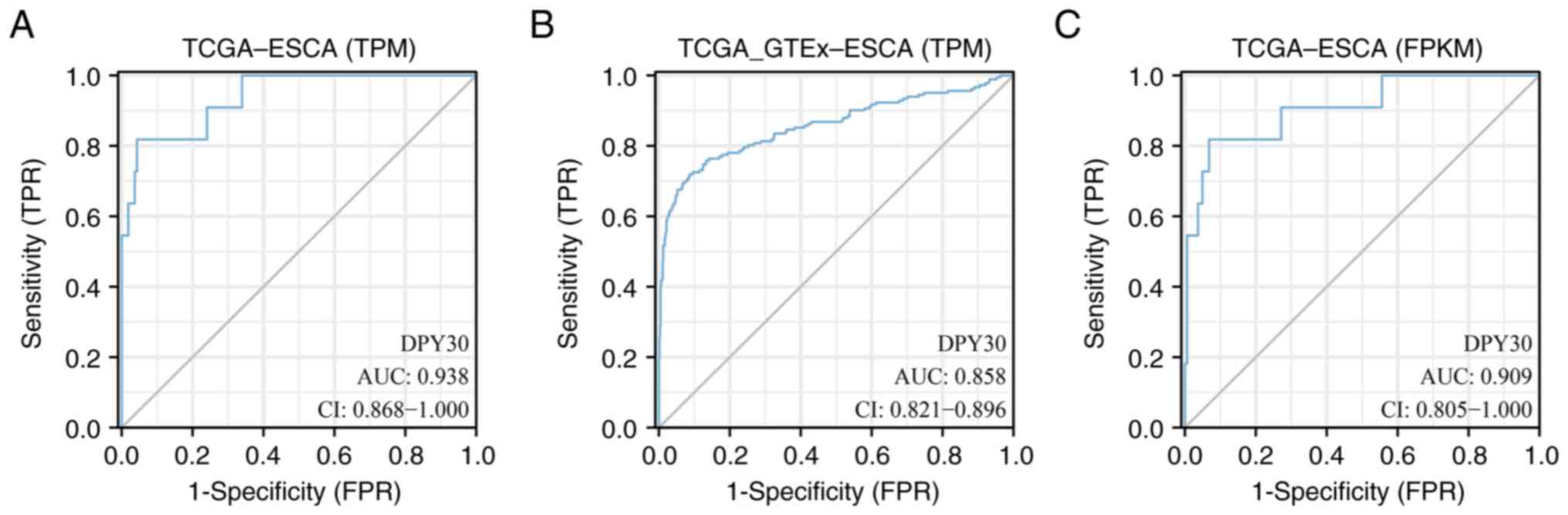 | Figure 3.ROC curve was used to evaluate the
diagnostic value of DPY30 in ESCA. (A) The ROC curve was used to
analyze the TPM type of TCGA database in ESCA. (B) The ROC curve
was used to analyze the TPM type of TCGA and GTEx databases in
ESCA. (C) The ROC curve was used to analyze the FPKM type of TCGA
database in ESCA. ROC, receiver operating characteristic; DPY30,
dpy-30 homolog; ESCA, esophageal cancer; TPM, transcripts per
million; TCGA, the Cancer Genome Atlas; GTEx, Genotype-Tissue
Expression; FPKM, fragments per kilobase per million; AUC, the area
under the curve; CI, confidence interval; TPR, true positive rate;
FPR, false positive rate. |
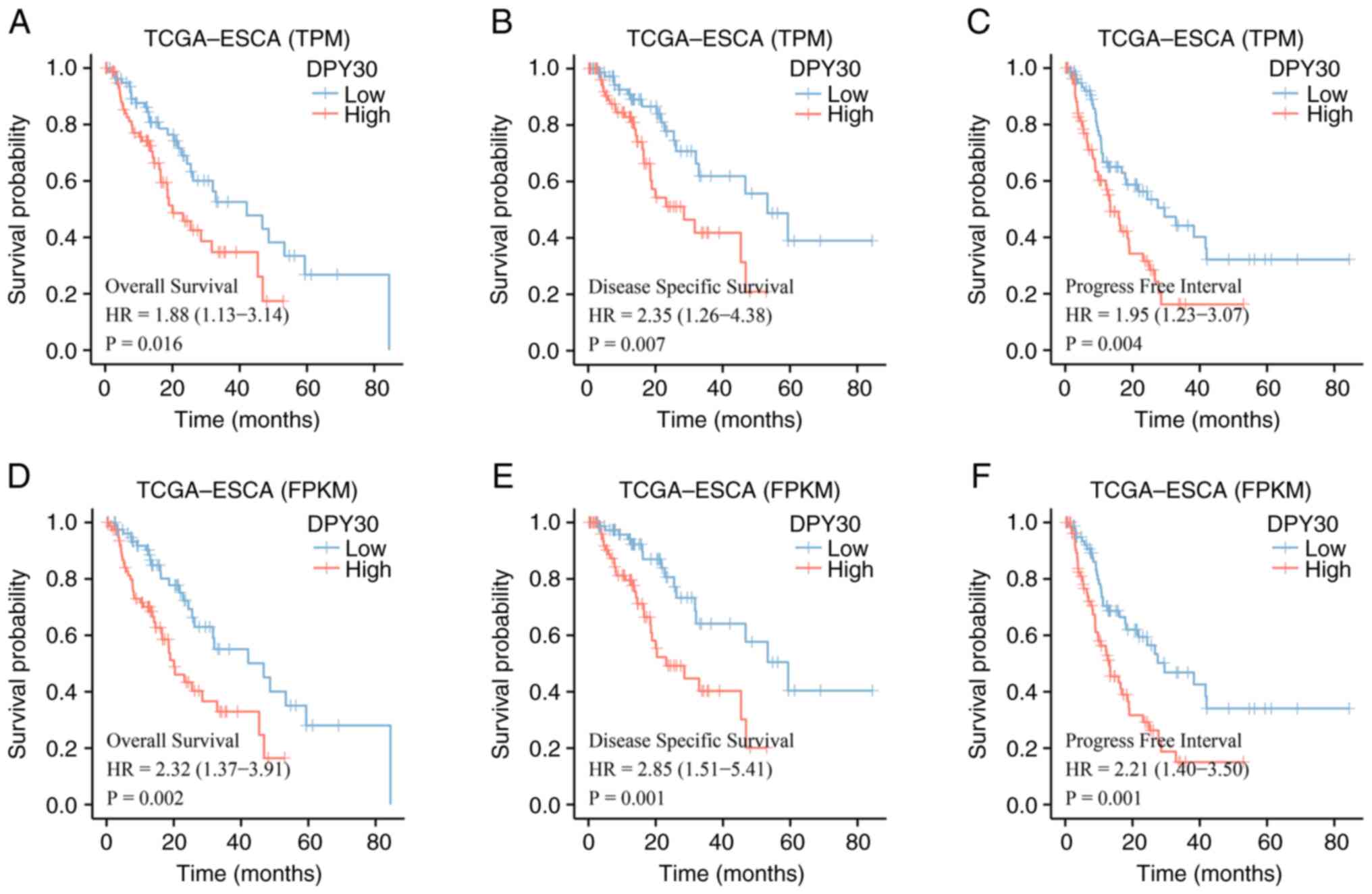
High expression of DPY30 is related to
a poor prognosis of ESCA
The prognostic value of DPY30 was determined using
different types of TCGA databases. Based on the TPM and FPKM types
of TCGA databases, the results of the analysis revealed a
significant correlation between the high expression of DPY30 and a
poor prognosis in ESCA (Fig. 4).
High expression of DPY30, predicted worse OS, disease-specific
survival (DSS) and progression-free survival (PFS) based on
different data types in patients with ESCA. The results of the
prognostic analysis of patients with ESCA demonstrated that the
high expression of DPY30 predicted poor overall survival.
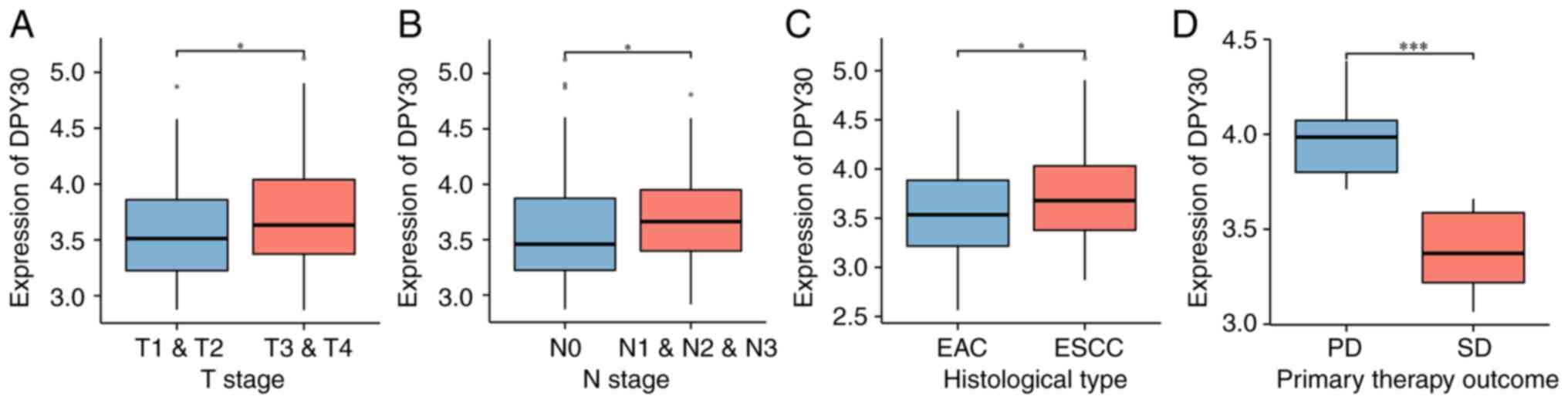
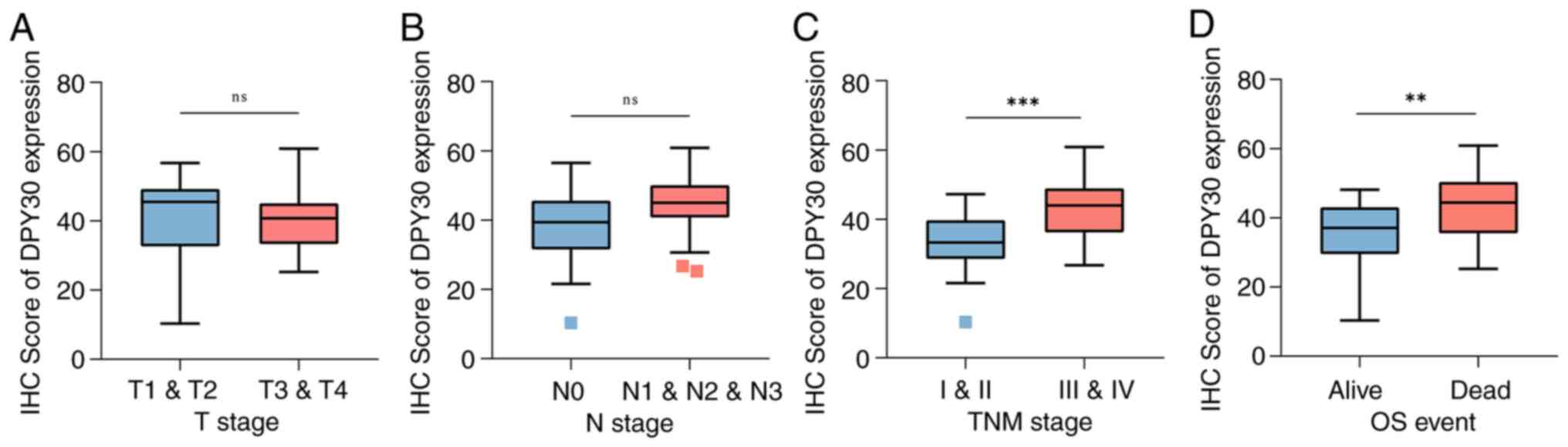
DPY30 overexpression is associated
with clinicopathological features and therapeutic efficacy in
patients with locally advanced ESCA
The correlation between the expression level of the
DPY30 mRNA and the clinicopathological features of patients with
ESCA was analyzed from TCGA database. The results demonstrated that
DPY30 overexpression was associated with the advanced pathologic T
stage and advanced pathologic N stage (Fig. 5A and B). The expression levels of
DPY30 were higher in esophageal squamous carcinoma than that in
esophageal adenocarcinoma (Fig.
5C). More notably, the overexpression of DPY30 suggested a
worse outcome for the neoadjuvant treatment of ESCA (Fig. 5D). The results of the analysis of
the clinical characteristics and the expression level of the DPY30
protein in 57 patients with ESCA (Table I) demonstrated that DPY30
overexpression was associated with the advanced pathologic TNM
stage and OS (Fig. 6).
 | Table I.Clinical characteristics and dpy-30
homolog protein expression of patients with esophageal cancer. |
Table I.
Clinical characteristics and dpy-30
homolog protein expression of patients with esophageal cancer.
| Characteristic | High
expression | Low expression | P-value |
|---|
| n | 33 | 24 |
|
| Sex, n (%) |
|
| 0.261 |
|
Female | 3 (5.3) | 5 (8.8) |
|
|
Male | 30 (52.6) | 19 (33.3) |
|
| Pathological type,
n (%) |
|
| 0.119 |
|
Esophageal adenosquamous or
adenocarcinoma | 2 (3.5) | 5 (8.8) |
|
|
Esophageal squamous cell
carcinoma | 31 (54.4) | 19 (33.3) |
|
| T stage, n (%) |
|
| 0.193 |
| T1
& T2 | 15 (26.3) | 6 (10.5) |
|
| T3
& T4 | 18 (31.6) | 18 (31.6) |
|
| N stage, n (%) |
|
| 0.016 |
| N0 | 16 (28.1) | 20 (35.1) |
|
| N1
& N2 & N3 | 17 (29.8) | 4 (7.0) |
|
| Pathologic TNM
stage, n (%) |
|
| <0.001 |
| Stage I
& II | 2 (3.5) | 13 (22.8) |
|
| Stage
III & IV | 31 (54.4) | 11 (19.3) |
|
| Differentiation
grade, n (%) |
|
| 0.855 |
| High
differentiation | 8 (14.0) | 7 (12.3) |
|
| Low
differentiation | 5 (8.8) | 2 (3.5) |
|
| Medium
differentiation | 20 (35.1) | 15 (26.3) |
|
| Survival state, n
(%) |
|
| 0.020 |
|
Succumbed | 25 (43.9) | 10 (17.5) |
|
|
Survived | 8 (14.0) | 14 (24.6) |
|
| Age, mean ± SD | 61.24±8.53 | 61.46±6.9 | 0.919 |
| Time (day), mean ±
SD | 769.58±321.56 |
1,170.83±428.16 | <0.001 |
|
Immunohistochemistry score, median
(interquartile range) | 45.69 (43.53,
50.39) | 31.91 (28.01,
34.84) | <0.001 |
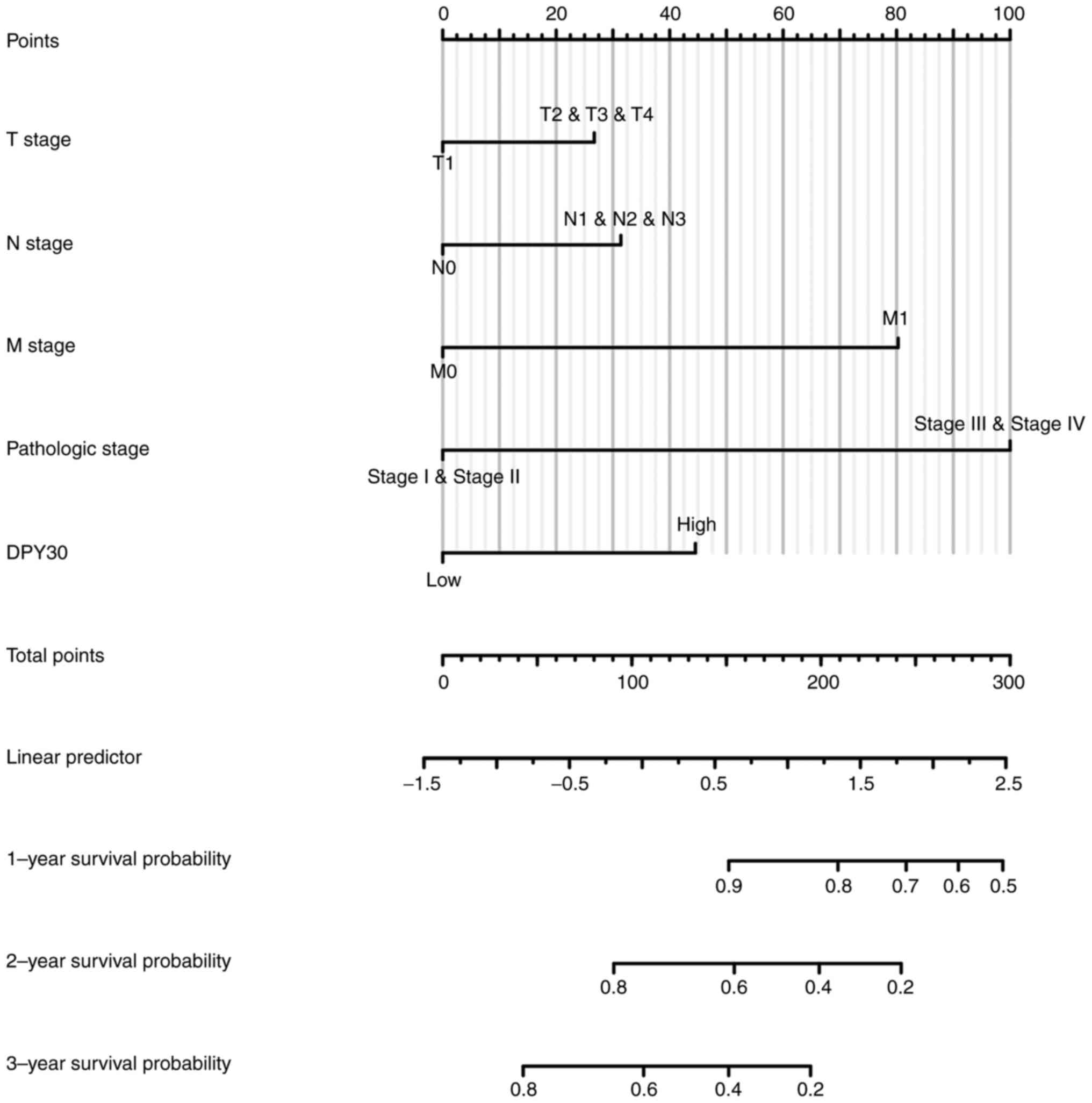
DPY30 overexpression is a risk factor
for patients with ESCA
To find the prognostic risk factor for patients with
ESCA in TCGA database, univariate Cox regression analysis was
performed to show that pathologic N stage, pathologic M stage,
pathological stage, treatment outcome and DPY30 mRNA expression
level might be the risk factors for OS (Table II). Based on univariate Cox
regression was performed to analyze the clinical characteristics of
57 patients with ESCA. The results demonstrated that the
overexpression of DPY30 was associated with the advanced pathologic
N stage, pathologic TNM stage and the expression level of the DPY30
protein (Table III). The results
of the multivariate Cox regression analysis did not show
independent risk factors for ESCA in patients with ESCA. An
accurate prognostic nomogram based on the clinicopathological
characteristics and DPY30 expression level in ESCA was constructed
(Fig. 7).
 | Table II.Risk factors predicting poor overall
survival of patients with esophageal cancer in The Cancer Genome
Atlas databases |
Table II.
Risk factors predicting poor overall
survival of patients with esophageal cancer in The Cancer Genome
Atlas databases
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Total (n) | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| T stage | 145 |
|
|
|
|
| T1
& T2 | 64 | Reference |
|
|
|
| T3
& T4 | 81 | 1.312
(0.756-2.277) | 0.334 |
|
|
| N stage | 144 |
|
|
|
|
| N0 | 66 | Reference |
|
|
|
| N1
& N2 & N3 | 78 | 2.970
(1.606-5.493) |
<0.001a | 10.618
(1.913-58.945) | 0.007a |
| M stage | 129 |
|
|
|
|
| M0 | 121 | Reference |
|
|
|
| M1 | 8 | 5.075
(2.312-11.136) |
<0.001a | 43.317
(4.395-426.952) | 0.001a |
| Pathologic
stage | 142 | 57 |
|
|
|
| Stage I
& Stage II | 85 | Reference |
|
|
|
| Stage
III & Stage IV | 57 | 3.223
(1.807-5.747) |
<0.001a | 0.427
(0.098-1.860) | 0.257 |
| Age | 162 |
|
|
|
|
|
<60 | 83 | Reference |
|
|
|
|
>60 | 79 | 0.831
(0.506-1.365) | 0.466 |
|
|
| Histological
type | 162 |
|
|
|
|
|
Adenocarcinoma | 80 | Reference |
|
|
|
|
Squamous cell carcinoma | 82 | 0.875
(0.526-1.455) | 0.607 |
|
|
| Primary therapy
outcome | 94 |
|
|
|
|
| PD | 10 | Reference |
|
|
|
| SD
& PR & CR | 84 | 0.239
(0.095-0.602) | 0.002a | 1.734
(0.309-9.732) | 0.532 |
| Dpy-30 homolog
mRNA |
|
|
|
|
|
| expression | 162 |
|
|
|
|
| Low
expression | 81 | Reference |
|
|
|
| High
expression | 81 | 2.316
(1.373-3.907) | 0.002a | 1.186
(0.310-4.538) | 0.803 |
 | Table III.Risk factors predicting poor overall
survival of 57 patients with ESCA. |
Table III.
Risk factors predicting poor overall
survival of 57 patients with ESCA.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Total (n) | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age | 57 | 1.002
(0.959-1.047) | 0.936 |
|
|
| Sex | 57 |
|
|
|
|
|
Male | 49 | Reference |
|
|
|
|
Female | 8 | 0.925
(0.359-2.387) | 0.872 |
|
|
| T stage | 57 |
|
|
|
|
| T3
& T4 | 36 | Reference |
|
|
|
| T1
& T2 | 21 | 1.086
(0.552-2.137) | 0.812 |
|
|
| N stage | 57 |
|
|
|
|
| N0 | 36 | Reference |
|
|
|
| N1
& N2 & N3 | 21 | 3.430
(1.740-6.763) | <0.001a | 2.741
(1.371-5.481) | 0.004a |
| Grade | 57 |
|
|
|
|
| Medium
differentiation | 35 | Reference |
|
|
|
| High
differentiation | 15 | 0.687
(0.294-1.602) | 0.385 |
|
|
| Low
differentiation | 7 | 1.042
(0.396-2.742) | 0.934 |
|
|
| Pathological
stage | 57 |
|
|
|
|
| Stage
III & IV | 42 | Reference |
|
|
|
| Stage I
& II | 15 | 0.227
(0.080-0.646) | 0.005a | 0.359
(0.111-1.157) | 0.086 |
| Dpy-30 homolog
protein |
|
|
|
|
|
| expression | 57 |
|
|
|
|
| High
expression | 33 | Reference |
|
|
|
| Low
expression | 24 | 0.354
(0.169-0.740) | 0.006a | 0.864
(0.354-2.105) | 0.747 |
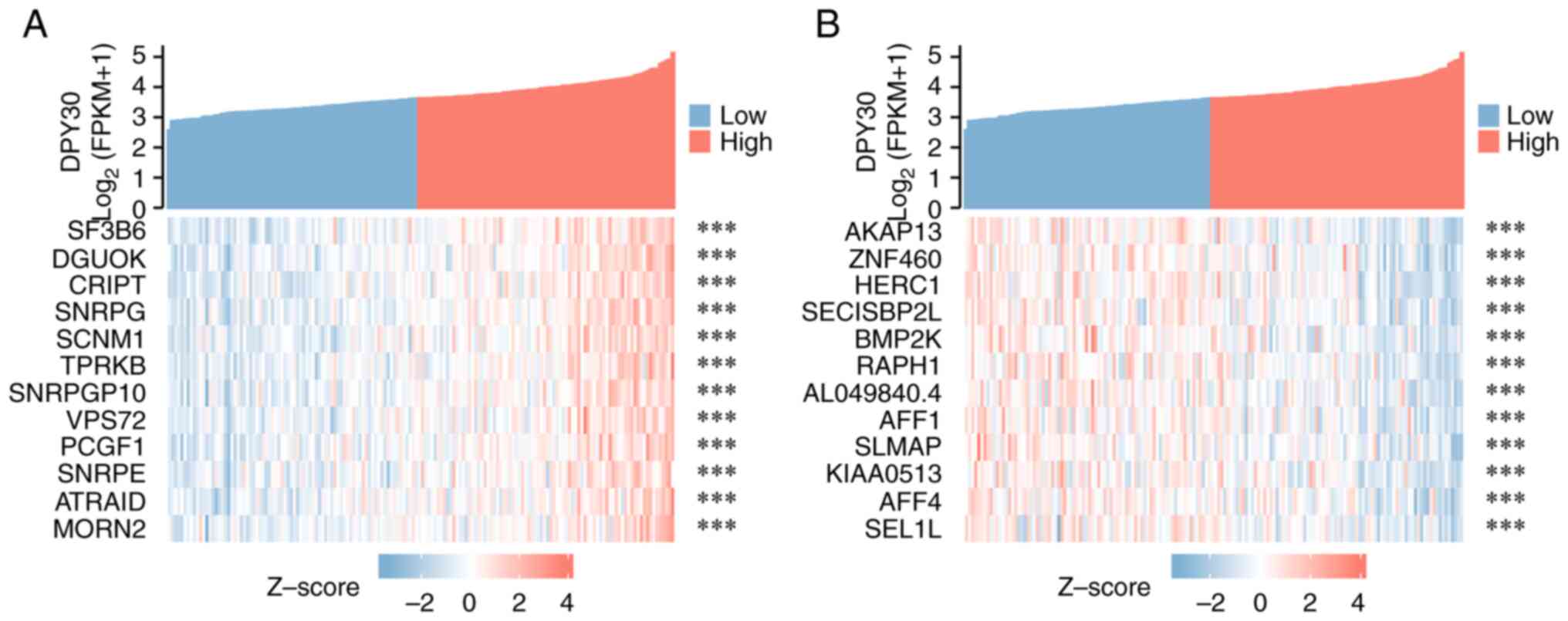
Functions and signaling mechanisms of
DPY30 co-expressed genes
Based on the correlation analysis of the
DPY30-related genes, 669 genes were found to be co-expressed with
DPY30 (Table SIII). The top 12
genes with positive and negative correlations with DPY30 in ESCA
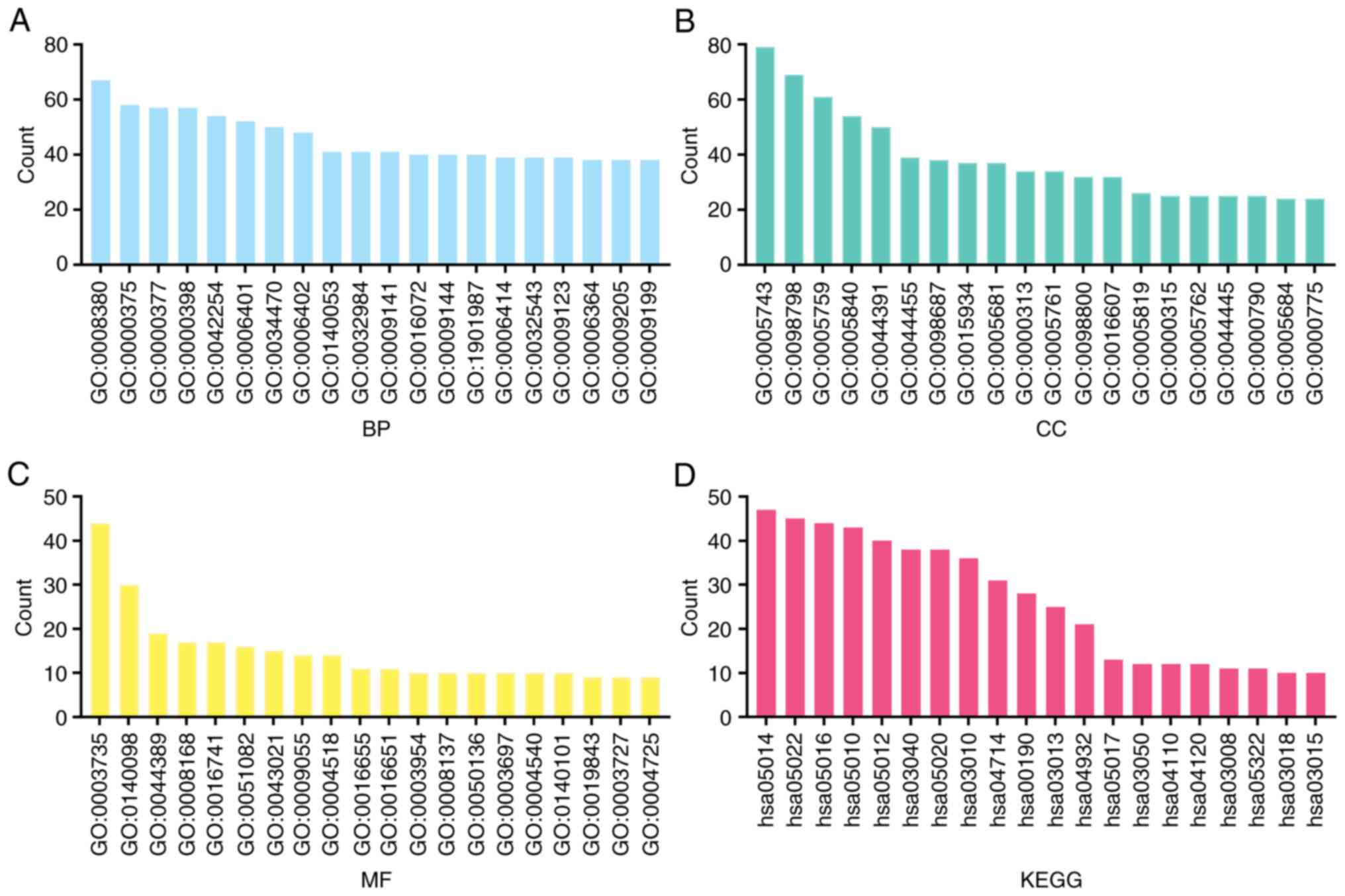
are shown in Figs. 8 and S2. Based on GO analysis, it was found
that DPY30 and its co-expressed genes were involved in RNA and DNA
modification, regulation of mitochondria and lysosome function and
interference of the cell cycle (Fig.
9A-C). The results of the KEGG analysis demonstrated that these
genes were associated with RNA transport and degradation,
spliceosome, ribosome, oxidative phosphorylation, the cell cycle
and other mechanisms (Fig. 9D and
Table SIV). To visualize the
relationship of the DPY30 co-expressed genes, the PPI network was
constructed (Fig. 10).
The relationship between DPY30
expression and immune infiltrating cells in ESCA
Based on the diagnostic and prognostic value of
DPY30 in ESCA, the ESTIMATE algorithm was used to further determine
the value of DPY30 in the tumor microenvironment of ESCA. Based on
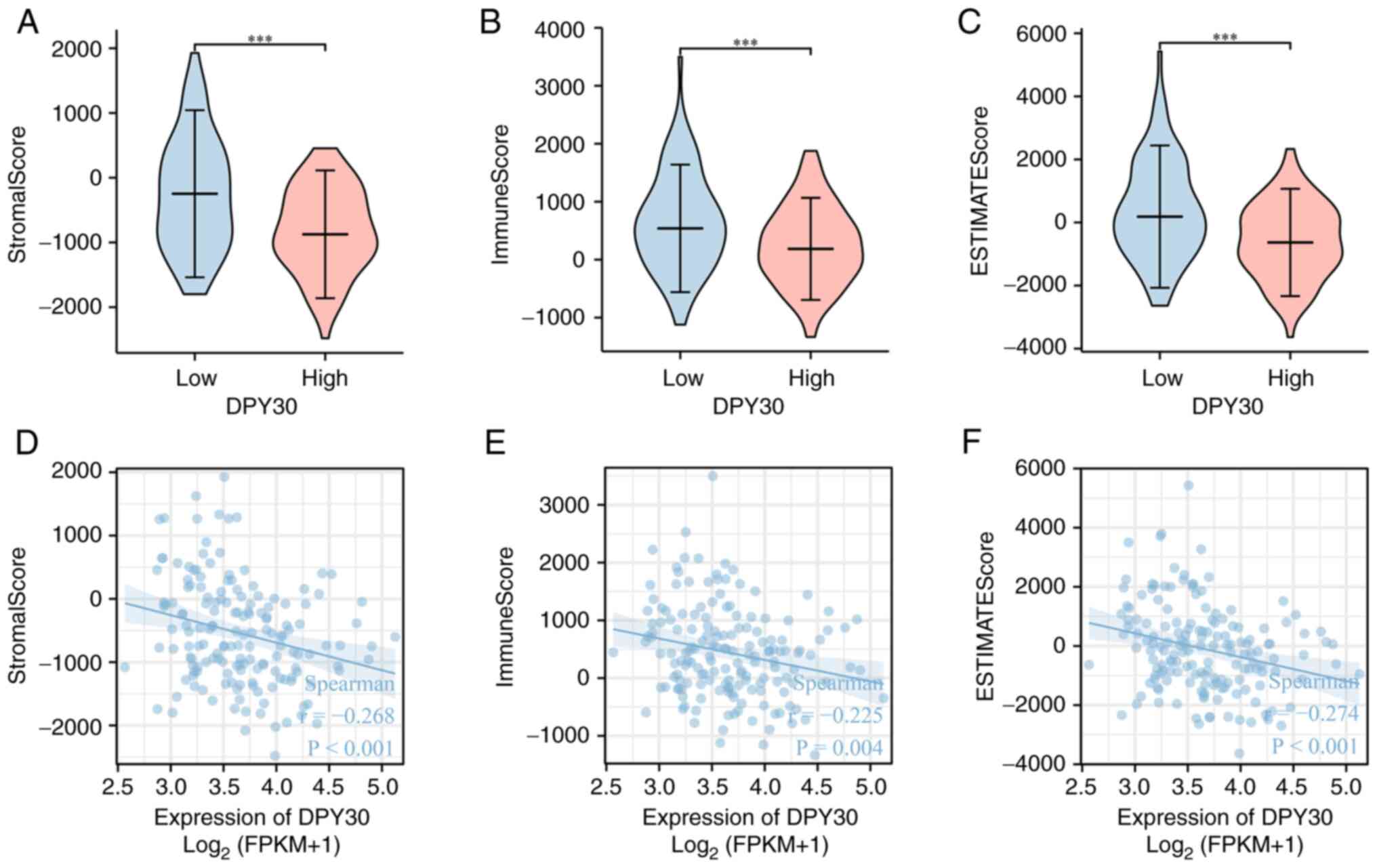
the result of the Stromal Score, Immune Score and ESTIMATE Score,
the high expression of DPY30 was found to be related to low immune
scores in the tumor microenvironment of ESCA (Fig. 11A-C). DPY30 expression was
negatively correlated with the overall immune infiltration, stromal
content and the combined score of both (Fig. 11D and E). The analysis of the
correlation between DPY30 expression and immune cells in ESCA
demonstrated that DPY30 expression was significantly correlated
with the number of types of immune cells (Table SV; Figs. S3 and S4). Among them, mast cells, effector
memory T cells, eosinophils, neutrophils, central memory T cells,
dendritic cells, T cells, T follicular helper cells and B cells
demonstrated a significant negative correlation with the expression
of DPY30 (Fig. 12).
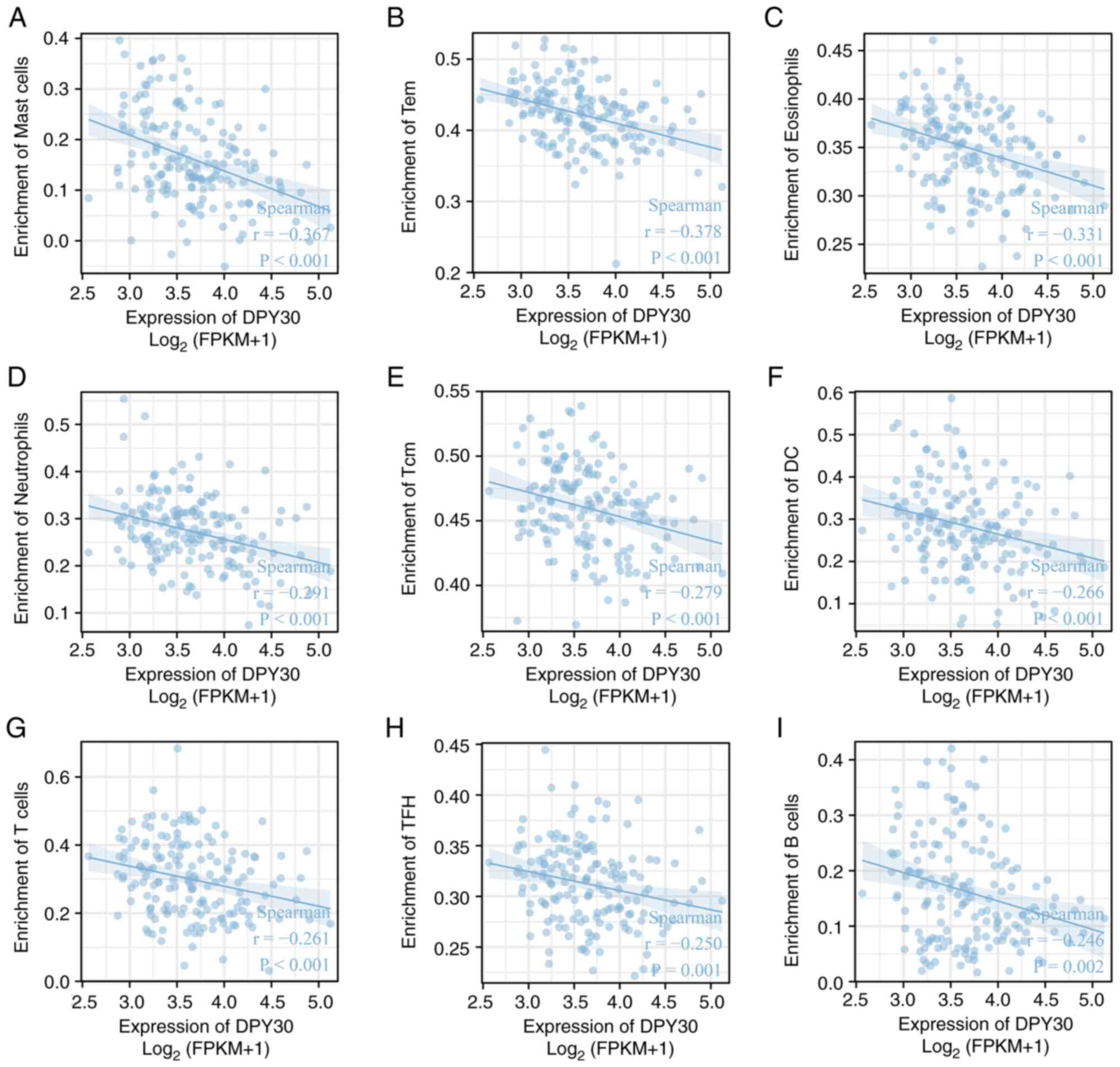 | Figure 12.Correlation between DPY30 expression
and immune cells in the tumor microenvironment of ESCA. The
correlation between DPY30 expression and (A) mast cells, (B) Tem
cells, (C) eosinophils, (D) neutrophils, (E) Tcm cells, (F) DC
cells, (G) T cells, (H) TFH cells and (I) B cells. DPY30, dpy-30
homolog; ESCA, esophageal cancer; FPKM, fragments per kilobase per
million; Tem, effector memory T cell; Tcm, central memory T cell;
DC, dendritic cells; TFH, T follicular helper cells. |
Discussion
ESCA is a major health problem worldwide. Due to the
lack of early symptoms and early diagnostic biomarkers, a number of
patients are diagnosed with locally advanced or even advanced ESCA.
Although various treatment strategies have been developed for ESCA,
the five-year survival rate is only 15–25% (21,22).
A number of studies have investigated ways to improve the prognosis
of ESCA by identifying new diagnostic markers and therapeutic
targets. DPY30 is highly expressed in hepatocellular liver cancer
tissues and plays a key role in the coordination of glucose
homeostasis and the modification of different types of histones
(23). The mRNA and protein levels
of DPY30 are significantly overexpressed in cholangiocarcinoma and
are independent prognostic factors in cholangiocarcinoma patients
(23). The knockdown of DPY30 can
significantly inhibit proliferation, induce cell cycle arrest in
the G2/M phase and decrease glycolysis in vitro
(24). The present study indicated
that DPY30 might have similar oncogenic effects in ESCA and may be
a potential therapeutic target for ESCA. DPY30 is highly expressed
in ESCA tissues and has an important diagnostic value. The
overexpression of DPY30 is significantly associated with a number
of clinicopathological features and poor prognosis in patients with
ESCA. Therefore, DPY30 might be used as a potential biomarker and
have a diagnostic, prognostic and therapeutic value in ESCA.
DPY30 and its homologs are mainly localized in the
nucleus and the encoded proteins that directly control the cell
cycle regulators. They play an important role in cell proliferation
and differentiation (25).
Additionally, DPY30 also serves as the core subunit of the
methyltransferase complex that catalyzes histone H3K4 methylation
(26). Histone methylation plays a
key role in gene regulation, cell differentiation, DNA
recombination and damage repair. Aberrant histone methylation can
cause mutations, translocations, or overexpression of a number of
important genes that usually lead to the development of diseases
such as cancer (27). Thus, the
overexpression of DPY30 might play a key role in the development of
tumorigenesis by increasing histone H3K4 methylation level. DPY30
can directly promote the expression of the MYC gene and also
regulate the efficient binding of the MYC oncoprotein to a number
of other genes (28,29). The DPY30/WNT/β-catenin signaling
mechanism can induce the EMT of cells in cervical cancer and
epithelial ovarian cancer (15,16).
However, these mechanisms need to be further investigated in ESCA
cell models.
DPY30 can significantly influence immune cell
infiltration in the tumor microenvironment of ESCA. It is also
significantly negatively correlated with multiple immune cells in
the tumor microenvironment. EMT is an important mechanism of ESCA
growth and metastasis. It also plays an important role in the
formation of the tumor immune microenvironment (30). DPY30 might promote tumor
progression and significantly affect tumor immunogenic cells by
inducing EMT. Thus, its expression level might predict the effect
of immunotherapy and it is a potential therapeutic target. The
peptides that can specifically inhibit DPY30 activity can
significantly inhibit the growth of hematological cancer cells
(29). Several molecules of the
methyltransferase complex are targets for drug intervention and
small molecule inhibitors of potential antitumor agents (31). Therefore, DPY30 is a potential
therapeutic target for ESCA.
The present study determined the diagnostic and
prognostic value of DPY30 in ESCA by bioinformatic analysis and
tissue microarray. However, it is also necessary to collect more
specimens and to use an antibody with strong specificity to
validate the results in further research. Also, the signaling
mechanism of DPY30 in tumorigenesis and the development of ESCA
need to be examined in cell experiments. It is important that the
present study first aimed to explore the relationship between DPY30
and tumor microenvironment of ESCA by bioinformatics. However, the
final regulatory network between DPY30 expression and the tumor
immune microenvironment needs to be elucidated by performing
specific experiments. Thus, future extensive studies should
concentrate on supplementing in vivo and in vitro
experiments and performing multiplex immunohistochemistry to
determine the role of DPY30 in tumor microenvironment.
To summarize, the present study found that DPY30 was
significantly overexpressed in ESCA tissues and associated with
poor prognosis in patients with ESCA. DPY30 might also be used as
an early diagnostic and prognostic indicator of ESCA. It is also
associated with the tumor immune microenvironment and is expected
to be a new target for the treatment of patients with ESCA.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81572277 and
82072593), and the Department of Science and Technology of Hubei
Province (grant no. 2020BCB027)
Availability of data and materials
The results shown here are fully or partly based on
the data generated by TCGA Research Network: https://www.cancer.gov/tcga. All other data generated
or analyzed during this study are included in this published
article.
Authors' contributions
PM, HX, YL and QH contributed to the study design
and provided important information. QG, PM, HX, WM and MW performed
data analysis. PM and HX confirm the authenticity of all the raw
data. PM and HX wrote the main text in the manuscript. All authors
revised and edited the manuscript and gave their final approval for
the version to be published.
Ethical approval and consent to
participate
The present study was performed with the approval of
the Institutional Ethics Committee of Tongji Medical College,
Huazhong University of Science and Technology [approval number
(2021) 0158]. All aspects of the study complied with the
Declaration of Helsinki. All participants provided written informed
consent before undergoing surgical treatment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ESCA
|
esophageal cancer
|
|
DPY30
|
dpy-30 homolog
|
|
TMA
|
tissue microarray
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Gouw DJJM, Klarenbeek BR, Driessen M,
Bouwense SAW, van Workum F, Fütterer JJ, Rovers MM, Ten Broek RPG
and Rosman C: Detecting pathological complete response in
esophageal cancer after neoadjuvant therapy based on imaging
techniques: A diagnostic systematic review and meta-analysis. J
Thorac Oncol. 14:1156–1171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leng XF, Daiko H, Han YT and Mao YS:
Optimal preoperative neoadjuvant therapy for resectable locally
advanced esophageal squamous cell carcinoma. Ann N Y Acad Sci.
1482:213–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bolger JC, Donohoe CL, Lowery M and
Reynolds JV: Advances in the curative management of oesophageal
cancer. Br J Cancer. 126:706–717. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar A, Kumari N, Nallabelli N and Prasad
R: Pathogenic and therapeutic role of H3K4 family of methylases and
demethylases in cancers. Indian J Clin Biochem. 34:123–132. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
South PF, Fingerman IM, Mersman DP, Du HN
and Briggs SD: A conserved interaction between the SDI domain of
Bre2 and the Dpy-30 domain of Sdc1 is required for histone
methylation and gene expression. J Biol Chem. 285:595–607. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ali A and Tyagi S: Diverse roles of
WDR5-RbBP5-ASH2L-DPY30 (WRAD) complex in the functions of the SET1
histone methyltransferase family. J Biosci. 42:155–159. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang H, Shukla A, Wang X, Chen WY,
Bernstein BE and Roeder RG: Role for Dpy-30 in ES cell-fate
specification by regulation of H3K4 methylation within bivalent
domains. Cell. 144:513–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Simboeck E, Gutierrez A, Cozzuto L,
Beringer M, Caizzi L, Keyes WM and Di Croce L: DPY30 regulates
pathways in cellular senescence through ID protein expression. EMBO
J. 32:2217–2230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Shah K, Khodadadi-Jamayran A and
Jiang H: Dpy30 is critical for maintaining the identity and
function of adult hematopoietic stem cells. J Exp Med.
213:2349–2364. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Z, Augustin J, Chang C, Hu J, Shah K,
Chang CW, Townes T and Jiang H: The DPY30 subunit in SET1/MLL
complexes regulates the proliferation and differentiation of
hematopoietic progenitor cells. Blood. 124:2025–2033. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao T, Hong Y, Ming GL and Song H: Loss
of chromatin modulator Dpy30 compromises proliferation and
differentiation of postnatal neural stem cells. J Mol Cell Biol.
12:2–3. 2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee YJ, Han ME, Baek SJ, Kim SY and Oh SO:
Roles of DPY30 in the proliferation and motility of gastric cancer
cells. PLoS One. 10:e01318632015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Zhang S, Li A, Zhang A, Zhang S
and Chen L: DPY30 is required for the enhanced proliferation,
motility and epithelial-mesenchymal transition of epithelial
ovarian cancer cells. Int J Mol Med. 42:3065–3072. 2018.PubMed/NCBI
|
|
16
|
He FX, Zhang LL, Jin PF, Liu DD and Li AH:
DPY30 regulates cervical squamous cell carcinoma by mediating
epithelial-mesenchymal transition (EMT). Onco Targets Ther.
12:7139–7147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paschalis A, Sheehan B, Riisnaes R,
Rodrigues DN, Gurel B, Bertan C, Ferreira A, Lambros MBK, Seed G,
Yuan W, et al: Prostate-specific membrane antigen heterogeneity and
DNA repair defects in prostate cancer. Eur Urol. 76:469–478. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin P, Tian P, Pang J, Lai L, He G, Song Y
and Zheng Y: Clinical significance of COL1A1 and COL1A2 expression
levels in hypopharyngeal squamous cell carcinoma. Oncol Lett.
20:803–809. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth EC, Lagergren J, Fitzgerald RC,
Lordick F, Shah MA, Lagergren P and Cunningham D: Oesophageal
cancer. Nat Rev Dis Primers. 3:170482017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu B: DPY30 functions in glucose
homeostasis via integrating activated histone epigenetic
modifications. Biochem Biophys Res Commun. 507:286–290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong ZF, Zhang WQ, Wang SJ, Li SY, Shang
J, Liu F and Shen DY: Upregulation of DPY30 promotes cell
proliferation and predicts a poor prognosis in cholangiocarcinoma.
Biomed Pharmacother. 123:1097662020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Lou Z, Dong X, Yang W, Peng Y, Yin
B, Gong Y, Yuan J, Zhou W, Bartlam M, et al: Crystal structure of
the C-terminal domain of human DPY-30-like protein: A component of
the histone methyltransferase complex. J Mol Biol. 390:530–537.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee
JW, Verdine GL, Allis CD and Roeder RG: Regulation of MLL1 H3K4
methyltransferase activity by its core components. Nat Struct Mol
Biol. 13:713–719. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song Y, Wu F and Wu J: Targeting histone
methylation for cancer therapy: Enzymes, inhibitors, biological
activity and perspectives. J Hematol Oncol. 9:492016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Z, Shah K, Busby T, Giles K,
Khodadadi-Jamayran A, Li W and Jiang H: Hijacking a key chromatin
modulator creates epigenetic vulnerability for MYC-driven cancer. J
Clin Invest. 128:3605–3618. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shah KK, Whitaker RH, Busby T, Hu J, Shi
B, Wang Z, Zang C, Placzek WJ and Jiang H: Specific inhibition of
DPY30 activity by ASH2L-derived peptides suppresses blood cancer
cell growth. Exp Cell Res. 382:1114852019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chockley PJ and Keshamouni VG:
Immunological consequences of epithelial-mesenchymal transition in
tumor progression. J Immunol. 197:691–698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu K, Tao H, Si X and Chen Q: The histone
H3 lysine 4 presenter WDR5 as an oncogenic protein and novel
epigenetic target in cancer. Front Oncol. 8:5022018. View Article : Google Scholar : PubMed/NCBI
|