Introduction
Urolithiasis is a complex and multifactorial
disorder (1) whose epidemiology
fluctuates due to geographical, cultural and ethnic groups
(2). Environmental, dietary and
genetic factors may affect disease progression (3). Although various molecules and
proteins, particularly ones involved in calcium metabolism are
suspected of involvement in the developmental stages of stone
formation, the genetic basis of stone formation remains to be
clarified (4,5). Supersaturation and crystallization of
urine ingredients affect the stone type, which may lead to
progression of stone formation. Results of recent studies have
shown that metabolic deficiency, which is effective in amino acid
metabolism may be crucial for the formation of several types of
stones including oxalate and uric acid types (6). In the same study, urine samples of
calcium oxalate patients exerted the decreased levels of ornithine
and tyrosine amino acids. L-ornithine is a dibasic key amino acid
involved in the urea cycle and polyamine biosynthetic pathway
(5,7). According to Kohri et
al(8), plasma values of several
amino acids including taurine, aspartic acid, hydroxyproline,
glutamic acid, glycine, alanine, cystathionine, ornithine and
lysine were found to be significantly higher in controls as
compared to patients with stone formation (8). It was suggested that the regulation of
amino acid metabolism in kidneys may promote stone formation by
altering microenviromental pH adjustment as well as the induction
of reactive oxygen species (9). In
addition, cystinuria, an autosomal recessive disease, which is
caused by a defective tubular reabsorption of cystine and the three
dibasic amino acids arginine, lysine and ornithine, result in a
lifelong risk of renal stone formation due to the low solubility of
cystine in urine (5).
Natural polyamines such as putrescine, spermidine
and spermine, are a family of aliphatic amines that are metabolized
in almost all living organisms (10). These polyamines have critical roles
in various activities such as cell proliferation, differentiation
and division, and stabilization of DNA, RNA and proteins (11). These polyamines also act as
anti-oxidant and anti-inflammatory agents that most likely act as
superoxide scavengers. Polyamines are also anti-glycating agents
that reduce the formation of end-products of glycation processes
(12). Clinical features of
polyamines were determined in carcinogenesis, atherosclerosis and
chronic diseases including chronic renal failure in patients who
showed a higher incidence of stone formation (13–15).
It is suggested that the alteration of polyamine metabolism
increases toxic products of the catabolic pathway and cause
significant uremic toxins such as acrolein to serve as a marker of
chronic renal failure (13). The
above findings demonstrated that the alteration of polyamine
homeostasis might serve for risk evaluation in the etiology of
acute or chronic degenerative diseases (16).
Putrescine synthesis from ornithine is regulated by
the rate-limiting enzyme ornithine decarboxylase (ODC) (10), which is expressed from the ODC gene
located at chromosome 2, band p25.1 (10). The expression of ODC may be altered
by the presence of several single-nucleotide polymorphisms (SNPs)
within the ODC gene intron 1 at position +316 (17). This SNP located between two c-myc
transcription factor binding sites and rare allele (allele A) leads
to a decrease in the ODC gene expression (18). However, Kohri et al(8) suggested that ODC polymorphism is
significantly associated with whole blood polyamines in females who
are A-allele homozygotes expressing more ODC than G-allele carrying
females (19).
Spermidine/spermine acetyltransferase (SSAT) is the
key polyamine catabolic enzyme that causes acetylation of
spermidine or spermine (10). Its
activity is required to provide polyamine homeostasis at tissue
level (20). The SSAT enzyme is
encoded by SAT-1 gene, which is located on the X chromosome, band
p22.1 (21). Various SNPs have been
determined within the SAT-1 gene near the polyamine-responsive
element (PRE) region (22). The SNP
within the SAT-1 gene is at position −1415 resulting in alteration
of the T nucleotide to C nucleotide (23). Findings of previous studies
(22–23) suggested that any chronic stress
factors might increase the polyamine synthesis and metabolization
of polyamines. It is hypothesized that due to proximity to the PRE
and the recognition sites of the transcription factors acting on
the promoter region of SAT-1 gene, the allelic variants of SAT-1
−1415T/C polymorphism may exert a significant impact on the
expression rate of SAT-1. Due to the presence of SAT-1 on the X
chromosome, the variance of allelic distribution of target SNP on
SAT-1 was found to be associated with anxiety disorders in a male
subpopulation (23). However, no
studies are available with regard to the presence of SAT-1 −1415T/C
polymorphism and the manner in which it affects other
pathophysiological conditions including urolithiasis.
The aim of the present study was to examine the
association of polyamine metabolism key enzymes ODC +316 G/A and
SAT-1 −1415 T/C gene polymorphisms and the risk of developing
urolithiasis.
Materials and methods
Study population
A total of 65 patients (42 male and 23 female) aged,
25–61 years (average age, 42.9±10.2 years) with recurrent
idiopathic calcium oxalate stone disease were enrolled in this
study. Blood and urine biochemistry tests were performed to
evaluate the hypercalcemia, hyperuricemia, hyperoxaluria or
hyperuricosuria cases for exclusion from the study. Patients who
showed symptoms of urinary tract infections, pregnancy, vascular
heart disease, acute or chronic infections, immunologic conditions
and history of malignancies, neoplastic, coagulation disorders or
chronic renal failure during the period of stone treatment were
also excluded. There were 74 males and 30 females in the control
group (age range, 20–58 years) with no family history for stone
formation. These subjects were evaluated by renal ultrasonography
and routine tests for urinary microscopic hematuria to exclude
individuals with renal calcifications. Inclusion criteria for the
two groups included use of non estrogen, progesterone preparation
or menopausal drug or vitamin-D, antiacid drugs, or heparin
preparation within the previous 2 months (Table I). Patients and control subjects
were informed about the study procedure. This study was approved by
the Ethics Committee of Yeditepe University in Turkey (March 22,
no. 2010/095). Human rights of the subjects were protected and any
necessary approval was obtained from the ethics committee.
Experiments performed on human subjects were conducted in
accordance with the Declaration of Helsinki. All the procedures
were carried out with the adequate understanding and written
consent of the subjects.
 | Table IStudy group characteristics. |
Table I
Study group characteristics.
| Control | Patients |
|---|
| Gender (n, %) |
| Female | 30 (29%) | 23 (35%) |
| Male | 74 (71%) | 42 (65%) |
| Age (mean ± SD) | 40.8±9.9 | 42.9±10.2 |
| BMI
(kg/m2) (mean) | 24.1 | 24.9 |
| Smoking (n, %) | 25 (24%) | 32 (49%) |
| Recurrence (n) | 0 | 44 |
| Familial history of
urolithiasis (n) | 0 | 65 |
| Blood chemistry
(mean) |
| Creatinine
(mg/dl) | 0.38 | 1.05 |
| PTH (pg/dl) | 38.5 | 66.94 |
| Ca (mg/dl) | 9.2 | 9.42 |
| Mg (mg/dl) | 1.8 | 2.22 |
| P (mg/dl) | 3.4 | 3.09 |
| Urine chemistry
(mean) |
| Uric acid
(mg/l) | 5.8 | 72.81 |
| pH | 5.52 | 5.76 |
| Creatinine (mg/24
h) | 0.28 (male)
0.69 (female) | 114.46 |
| Oxalate
(mg/day) | 38 | 42 |
| Citrate
(mg/day) | 118 | 230 |
Genotyping
A total of 169 venous blood samples were obtained
and placed in EDTA tubes and genomic DNA content was isolated from
blood mononuclear cells by using the salting-out method (24).
Genotypes of ODC +316 G/A polymorphism were
determined by the polymerase chain reaction (PCR)-restriction
fragment length polymorphism procedure. PCR reaction contained 1 μg
genomic DNA, 2 pmol each primer, 1X AmpliTaq 360 DNA Polymerase PCR
buffer (PN4398818; Applied Biosystems, Foster City, CA, USA), 200
μM dNTPs (R1121; Fermentas, Vilnius, Lithuania,) and 1 unit
AmpliTaq 360 DNA polymerase in a volume of 25 μl. The primers for
the ODC +316 G/A region were 5′-ATCGTGGCTGGTTTGAGCTG-3′ and
5′-GTCATCTGCTCTGTAGACACAGCG-3′. The protocol included an initial
denaturation step at 94°C for 3 min, followed by 30 cycles with 30
sec of denaturation at 94°C, 30 sec of annealing at 55°C and 45 sec
of elongation at 72°C, followed by a final elongation step 72°C for
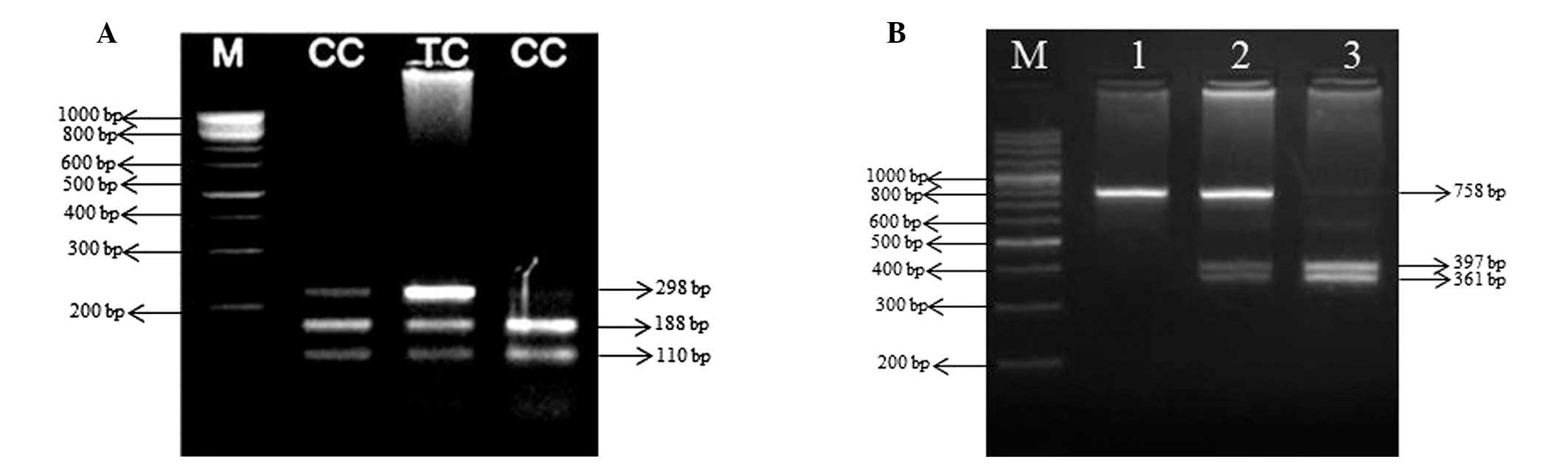
10 min. PCR product (10 μl of 757 bp long) of case and control
groups was digested with 5 units PstI (ER0611; Fermentas)
restriction enzymes overnight at 37°C. The digestion products were
subjected to 3.5% agarose gel electrophoresis. DNA from individuals
carrying A allele yielded two fragments of 397 and 361 bp, whereas
DNA from individuals carrying the G allele yielded only the uncut
758 bp. The presence of all three fragments was indicative of
heterozygotes (Fig. 1A).
Genotypes of SAT-1 −1415 T/C were determined using
PCR and restriction digestion with MspI restriction enzyme.
Primers for the amplification of SAT-1 −1415 T/C polymorphic site
were 5′-GAAGGCCTTTTCCTCCTCTG-3′ (forward) and
5′-GATAGGGCCTCACCATCTTG-3′ (reverse). The protocol included an
initial denaturation step at 94°C for 3 min, followed by 30 cycles
with 30 sec of denaturation at 94°C, 30 sec of annealing at 60°C
and 45 sec of elongation at 72°C, followed by a final elongation
step 72°C for 10 min. PCR amplification of a 298-bp fragment from
the promoter of SAT-1 gene was digested with 5 units MspI
(ER1271; Fermentas) restriction enzymes overnight at 37°C. The
digestion products were subjected to 2.5% agarose gel
electrophoresis. Restriction fragments of 188 and 110 bp revealed
the presence of C allele, whereas the T allele could not be
determined at the MspI recognition site (Fig. 1B).
Statistical analysis
The Hardy-Weinberg analysis was used to ensure that
the allele frequencies of the controls and cases were in
equilibrium within the population. Allele distribution and genotype
frequencies among the cases and controls were compared with
predictions using the χ2 test. The odd ratios (OR) with
95% confidence intervals (CI) were calculated for the association
between ODC +316 G/A and SAT-1 −1415 T/C genotypes and the risk of
development of urolithiasis. P<0.05 was considered to indicate a
statistically significant difference. SPSS 13.0 software program
was used for the statistical analysis.
Results
In this study, the genotype distribution and allele
frequencies were evaluated according to the Hardy-Weinberg
equilibrium for polymorphic sites of polyamine metabolic key
enzymes ODC +316 G/A and SAT-1 −1415 T/C in the urolithiasis and
disease-free control group. As shown in Table II, the genotype distribution of ODC
+316 G/A SNP was not significantly different between urolithiasis
patients and the control group (χ2=0.676, df=2,
P=0.713). Similarly, the comparison of allele frequencies between
the urolithiasis and control groups did not exert any association
for ODC +316 G/A gene polymorphism alleles (χ2=0.024,
df=1, P=0.877) (Table III).
 | Table IIGenotype distribution of ornithine
decarboxylase (ODC) +316 G/A gene polymorphism between urolithiasis
cases and healthy controls. |
Table II
Genotype distribution of ornithine
decarboxylase (ODC) +316 G/A gene polymorphism between urolithiasis
cases and healthy controls.
| Cases (n=65) | Controls
(n=104) | | |
|---|
|
|
| | |
|---|
| Genotypes | No. | Percentage (%) | No. | Percentage (%) | Odds ratio (95%
CI) | P-value |
|---|
| GG | 36 | 55.4 | 56 | 53.8 | 1.00
(Reference) | 0.713 |
| GA | 22 | 33.8 | 40 | 38.5 | 1.662
(0.522–5.291) | |
| AA | 7 | 10.8 | 8 | 7.7 | 1.363
(0.453–4.096) | |
 | Table IIIAllele frequencies of ornithine
decarboxylase (ODC) +316 G/A gene polymorphism and
urolithiasis. |
Table III
Allele frequencies of ornithine
decarboxylase (ODC) +316 G/A gene polymorphism and
urolithiasis.
| Alleles | Cases (%) | Controls (%) | Odds ratio (95%
CI) | P-value |
|---|
| G | 0.723 | 0.731 | 1.011
(0.883–1.156) | 0.877 |
| A | 0.277 | 0.269 | 0.972
(0.680–1.389) | |
Genotype distributions and allele frequencies of
SAT-1 −1415 T/C in the urolithiasis and control groups are shown in
Table IV and V. The genotype distribution of SAT-1 −1415
T/C did not exert any significant difference between urolithiasis
patients and the control group (χ2=0.318, df=2,
P=0.853). When the allelic distribution of SAT −1415 T/C was
compared between the patients and control group, no significant
difference was observed (χ2=0.214, df=1, P=0.644)
(Table V).
 | Table IVGenotype distribution of SAT-1 −1415
T/C gene polymorphism among urolithiasis cases and healthy
controls. |
Table IV
Genotype distribution of SAT-1 −1415
T/C gene polymorphism among urolithiasis cases and healthy
controls.
| Cases (n=65) | Controls
(n=104) | | |
|---|
|
|
| | |
|---|
| Genotypes | No. | Percentage (%) | No. | Percentage (%) | Odds ratio (95%
CI) | P-value |
|---|
| TT | 49 | 75.4 | 77 | 74 | 1.00
(Reference) | 0.853 |
| TC | 6 | 9.2 | 8 | 7.7 | 1.313
(0.416–4.141) | |
| CC | 10 | 15.4 | 19 | 18.3 | 1.581
(0.415–6.020) | |
 | Table VAllele frequencies of SAT-1 −1415 T/C
gene polymorphism and urolithiasis. |
Table V
Allele frequencies of SAT-1 −1415 T/C
gene polymorphism and urolithiasis.
| SAT-1 −1415
alleles | Cases (%) | Controls (%) | Odds ratio (95 %
CI) | P-value |
|---|
| T | 0.800 | 0.779 | 0.974
(0.870–1.089) | 0.644 |
| C | 0.200 | 0.221 | 1.106
(0.721–1.697) | |
The association of ODC +316 G/A and SAT-1 −1415 T/C
gene polymorphism and urolithiasis cases were compared in gender
subgroups. When the male population was compared according to ODC
+316 G/A and SAT-1 −1415 T/C gene polymorphisms and allele
frequencies between the urolithiasis cases and control group, the
genotype distribution of ODC +316 G/A (χ2=1.334, df=2,
P=0.520) and SAT-1 −1415 T/C (χ2=0.975, df=2, P=0.614)
did not reach a statistically significant difference between the
urolithiasis and control subjects. In accordance with genotype
distribution, allele frequencies of ODC +316 G/A and SAT-1 −1415
T/C SNP were not significant between the urolithiasis and control
groups (χ2=0.851, df=1, P=0.607, χ2=1.752,
df=1, P=0.255), respectively (Table
VI).
 | Table VIGenotype distribution and allele
frequencies of ornithine decarboxylase (ODC) +316 and SAT-1 −1415
T/C gene polymorphisms in gender subgroups. |
Table VI
Genotype distribution and allele
frequencies of ornithine decarboxylase (ODC) +316 and SAT-1 −1415
T/C gene polymorphisms in gender subgroups.
| ODC +316 |
|---|
|
|---|
| Male subgroup | Female
subgroup |
|---|
|
|
|
|---|
| Genotypes | Cases | Controls | Odds ratio (95%
CI) | P-value | Cases | Controls | Odds ratio (95%
CI) | P-value |
|---|
| GG | 25 (59.5%) | 44 (59.3%) | 1.00 | 0.520 | 11 (47.8%) | 12 (40%) | 1.00 | 0.825 |
| GA | 11 (26.2%) | 24 (32.4%) | 1.760
(0.513–6.042) | | 11 (47.8) | 16 (53.3%) | 0.219
(0.639-0.545) | |
| AA | 6 (14.3%) | 6 (8.1%) | 2.182
(0.573–8.314) | | 1 (4.3 %) | 2 (6.7%) | 0.061
(0.804-0.727) | |
| G allele | 0.726 | 0.757 | 1.042
(0.888–1.223) | 0.607 | 0.717 | 0.667 | 0.929
(0.720–1.199) | 0.674 |
| A allele | 0.274 | 0.243 | 0.888
(0.567–1.393) | | 0.283 | 0.333 | 1.179
(0.658–2.113) | |
|
| SAT-1 −1415 |
|
| Male subgroup | Female
subgroup |
|
|
|
| Genotypes | Cases | Controls | Odds ratio (95 %
CI) | P-value | Cases | Controls | Odds ratio (95%
CI) | P-value |
|
| TT | 35 (83.3%) | 56 (75.7%) | 1.00 | 0.614 | 14 (60.9%) | 21 (70%) | 1.00 | 0.402 |
| TC | 2 (4.8%) | 6 (9.1%) | 0.667
(0.216–2.055) | | 4 (17.4%) | 2 (6.7%) | 0.680
(0.919–1.071) | |
| CC | 5 (11.9%) | 12 (16.2 %) | 1.250
(0.185–8.444) | | 5 (21.7%) | 7 (23.3%) | 0.330
(0.566–1.400) | |
| T allele | 0.857 | 0.798 | 0.930
(0.826–1.048) | 0.255 | 0.696 | 0.733 | 1.057
(0.825–1.346) | 0.669 |
| C allele | 0.143 | 0.202 | 1.419
(0.768–2.621) | | 0.304 | 0.267 | 0.876
(0.478–1.606) | |
Discussion
Accumulating evidence suggests that in addition to
inorganic substances, organic substances may be important in the
development urolithiasis, following their identification from
calcium salts in the 1950s by Boyce and Sulkin (25). Those authors noted that in
quantities of <3% of total stone weight, this organic ‘matrix’
directed the biomineralization process in an orderly manner
(26). Urinary supersaturation with
respect to stone salts is regulated by the urinary concentration of
various participating ions such as calcium, oxalate and citrate,
which lead to hypercalciuria, hyperoxaluria or hypocitraturia
(4). Besides the urinary
microenvironment, several damaging agents in kidney cells or
dysfunction may affect the supersaturation and lead to stone
formation.
There is no single theory that provides
understanding of the molecular basis and pathogenesis of stone
formation in kidneys. One of the identified mechanisms is free
solution crystallization, which has been utilized in cystinuria
patients who showed a higher recurrence rate for urinary stone
formation (27). Cystinuria is an
autosomal recessive disorder in renal tubular and intestinal
transport of dibasic amino acids, which results in increased
urinary excretion of cystine, ornithine, lysine and arginine
(28). Ornithine and arginine are
precursor amino acids of polyamine biosynthesis and ODC is the
metabolic enzyme that produces putrescine from ornithine. Polyamine
metabolism is regulated by a number of key enzymes, with ODC and
SSAT and expression of these genes regulating the homeostatic
tissue environment. According to previous studies (17,21)
polyamine derivatives and metabolic conjugates are critical to
provide balanced tissue environment. Circulating polyamines in
blood and polyamine content of urine also serve as candidate
biomarkers in the evaluation of disease progression for several
chronic diseases and malignancies (20). In this study, we investigated the
possible association of the altered metabolism of diamine
precursors of polyamine biosynthesis with urinary stone formation.
The findings from this study reveal that the ODC gene +316 G/A and
SAT-1 gene 1415 T/C polymorphisms cannot serve as candidate genetic
markers for screening the causes of stone disease as the frequency
of distribution in SNPs was similar in urolithiasis when compared
with the disease-free control group. A recent study has shown that
ODC >316 G/A polymorphism (rs2302615) was associated with the
progression of sporadic breast cancer and that the presence of one
copy of A allele has a protective role in the prevention of the
disease (29). In addition,
previous studies suggested that the G315A SNP in the ODC gene may
be a genetic marker for risk of colorectal neoplasia and may also
modify the association of aspirin use with risk (30,31).
Individuals homozygous for the minor A allele of ODC were low risk
for colorectal cancer recurrence compared to those with the major G
allele. For the specific inhibitor of ODC, eflornithine or sulindac
therapy received by colorectal cancer patients showed higher
ototoxicity due to the presence of AA allele. However, A allele
showed a protective effect against the recurrence of disease in
aspirin users, and two copies of G allele reduced the risk of
recurrence after treatment with eflornithine and sulindac (32). Therefore the distribution of ODC-1
G/A alleles in association with disease progression should be
evaluated using other genes that might be more effective.
Results of previous studies have shown that total
polyamine content in blood and fibroblast cultures of patients who
were suffering from schizophrenia were significantly higher
compared to the control group (33–35).
Although several reports indicated a strong association with
reduced SAT-1 expression and SAT-1 342 C allele among male suicide
completers (23), the allelic
distribution of SAT-1 342 C did not exert any relationship with
SAT-1 expression level in suicidal behavior as shown in a patient
group from another study (36).
Bermudo-Soriano et al(34)found that SAT-1 −1415 T/Chas a strong
association with anxiety but not schizophrenia. Therefore SAT-1
polymorphic alterations are unclear as to the possible association
of SNPs on SAT-1 gene expression profile. For this reason, cell and
animal models should be utilized to evaluate the functional role of
dbSNPs at the promoter region of SAT-1 (SNPs=6526342 and
92893).
In conclusion, the results obtained from this study
to determine ODC +316 G/A polymorphism and susceptibility to
urinary stone disease provide no evidence that particular ODC +316
G/A genotypes are associated with disease progression. In addition,
the distribution of alleles of SAT-1 −1415 T/C gene polymorphism
did not exert any significant association with urolithiasis.
Although polyamines have critical roles in cellular homeostasis and
are referred to as malignancy markers, other genotypic variations
are required to understand the genetic basis of urolithiasis. To
the best of our knowledge this is the first study to analyse the
effect of the polymorphisms of polyamine metabolic enzymes genes in
urolithiasis.
Acknowledgements
The authors thank to Tuğba Kızılboğa and Deniz
Coşkun for their technical assistance.
References
|
1
|
Muslumanoglu AY, Binbay M, Yuruk E, et al:
Updated epidemiologic study of urolithiasis in Turkey. I: changing
characteristics of urolithiasis. Urol Res. 39:309–314. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ertan P, Tekin G, Oger N, Alkan S and
Horasan GD: Metabolic and demographic characteristics of children
with urolithiasis in Western Turkey. Urol Res. 39:105–110. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koyuncu HH, Yencilek F, Eryildirim B and
Sarica K: Family history in stone disease: how important is it for
the onset of the disease and the incidence of recurrence? Urol Res.
38:105–109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khan SR and Canales BK: Genetic basis of
renal cellular dysfunction and the formation of kidney stones. Urol
Res. 37:169–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fjellstedt E, Harnevik L, Jeppsson JO,
Tiselius HG, Soderkvist P and Denneberg T: Urinary excretion of
total cystine and the dibasic amino acids arginine, lysine and
ornithine in relation to genetic findings in patients with
cystinuria treated with sulfhydryl compounds. Urol Res. 31:417–425.
2003. View Article : Google Scholar
|
|
6
|
Atanassova SS, Panchev P and Ivanova M:
Plasma levels and urinary excretion of amino acids by subjects with
renal calculi. Amino Acids. 38:1277–1282. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kepka-Lenhart D, Ash DE and Morris SM:
Determination of mammalian arginase activity. Methods Enzymol.
440:221–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kohri K, Takada M, Katoh Y, Kataoka K,
Iguchi M and Kurita T: Amino acids in urine and plasma of
urolithiasis patients. Int Urol Nephrol. 21:9–16. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Azoury R, Garti N and Sarig S: The amino
acid factor in stone formers’ and normal urines. Urol Res.
14:295–298. 1986.
|
|
10
|
Casero RA and Pegg AE: Polyamine
catabolism and disease. Biochem J. 421:323–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matthews HR: Polyamines, chromatin
structure and transcription. Bioessays. 15:561–566. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aziz SM, Toborek M, Hennig B, Endean E and
Lipke DW: Polyamine regulatory processes and oxidative stress in
monocrotaline-treated pulmonary artery endothelial cells. Cell Biol
Int. 21:801–812. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Igarashi K, Ueda S, Yoshida K and
Kashiwagi K: Polyamines in renal failure. Amino Acids. 31:477–483.
2006. View Article : Google Scholar
|
|
14
|
Sakata K, Kashiwagi K, Sharmin S, et al:
Increase in putrescine, amine oxidase, and acrolein in plasma of
renal failure patients. Biochem Biophys Res Commun. 305:143–149.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakata K, Kashiwagi K, Sharmin S, Ueda S
and Igarashi K: Acrolein produced from polyamines as one of the
uraemic toxins. Biochem Soc Trans. 31:371–374. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Igarashi K and Kashiwagi K: Use of
polyamine metabolites as markers for stroke and renal failure.
Methods Mol Biol. 720:395–408. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bello-Fernandez C, Packham G and Cleveland
JL: The ornithine decarboxylase gene is a transcriptional target of
c-Myc. Proc Natl Acad Sci USA. 90:7804–7808. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo Y, Harris RB, Rosson D, Boorman D and
O’Brien TG: Functional analysis of human ornithine decarboxylase
alleles. Cancer Res. 60:6314–6317. 2000.PubMed/NCBI
|
|
19
|
Kondo T, Hamajima N, Nishio K, et al:
Association of a polymorphism in the ornithine decarboxylase gene
with whole blood polyamine concentrations in a non-smoking healthy
population. J Health Sci. 53:406–412. 2007. View Article : Google Scholar
|
|
20
|
Seiler N, Atanassov CL and Raul F:
Polyamine metabolism as target for cancer chemoprevention (Review).
Int J Oncol. 13:993–1006. 1998.PubMed/NCBI
|
|
21
|
Xiao L, Celano P, Mank AR, et al:
Structure of the human spermidine/spermine N1-acetyltransferase
gene (exon/intron gene organization and localization to Xp22.1).
Biochem Biophys Res Commun. 187:1493–1502. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomitori H, Nenoi M, Mita K, Daino K,
Igarashi K and Ichimura S: Functional characterization of the human
spermidine/spermine N(1)-acetyltransferase gene promoter. Biochim
Biophys Acta. 1579:180–184. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sequeira A, Gwadry FG, Ffrench-Mullen JM,
et al: Implication of SSAT by gene expression and genetic variation
in suicide and major depression. Arch Gen Psychiatry. 63:35–48.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boyce WH and Sulkin NM: Biocolloids of
urine in health and in calculous disease. III. The mucoprotein
matrix of urinary calculi. J Clin Invest. 35:1067–1079. 1956.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Canales BK, Anderson L, Higgins L, et al:
Proteomic analysis of a matrix stone: a case report. Urol Res.
37:323–329. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Coe FL, Evan AP, Worcester EM and Lingeman
JE: Three pathways for human kidney stone formation. Urol Res.
38:147–160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ahmed K, Dasgupta P and Khan MS: Cystine
calculi: challenging group of stones. Postgrad Med J. 82:799–801.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brown I, Halliday S, Greig H, Heys SD,
Wallace HM and Schofield AC: Genetic polymorphism in ornithine
decarboxylase and risk of breast cancer. Fam Cancer. 8:307–311.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zell JA, Ziogas A, Ignatenko N, et al:
Associations of a polymorphism in the ornithine decarboxylase gene
with colorectal cancer survival. Clin Cancer Res. 15:6208–6216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barry EL, Baron JA, Bhat S, et al:
Ornithine decarboxylase polymorphism modification of response to
aspirin treatment for colorectal adenoma prevention. J Natl Cancer
Inst. 98:1494–1500. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zell JA, McLaren CE, Chen WP, Thompson PA,
Gerner EW and Meyskens FL: Ornithine decarboxylase-1 polymorphism,
chemoprevention with eflornithine and sulindac, and outcomes among
colorectal adenoma patients. J Natl Cancer Inst. 102:1513–1516.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Das D, De Fonseka N and Das I:
Interactions of nitric oxide, free radicals and polyamines in the
membrane pathology of schizophrenia. Biochem Soc Trans.
26:S1431998.PubMed/NCBI
|
|
34
|
Bermudo-Soriano CR, Vaquero-Lorenzo C,
Diaz-Hernandez M, et al: SAT-1 −1415T/C polymorphism and
susceptibility to schizophrenia. Prog Neuropsychopharmacol Biol
Psychiatry. 33:345–348. 2009.
|
|
35
|
Vaquero-Lorenzo C, Riaza Bermudo-Soriano
C, Perez-Rodriguez MM, et al: Positive association between SAT-1
−1415T/C polymorphism and anxiety. Am J Med Genet B Neuropsychiatr
Genet. 150B:515–519. 2009.PubMed/NCBI
|
|
36
|
Guipponi M, Deutsch S, Kohler K, et al:
Genetic and epigenetic analysis of SSAT gene dysregulation in
suicidal behavior. Am J Med Genet B Neuropsychiatr Genet.
150B:799–807. 2009. View Article : Google Scholar : PubMed/NCBI
|