Introduction
Oral squamous cell carcinoma (SCC) is the most
common malignant tumor in India, and each year affects ~300,000
people worldwide (1). Most of the oral
cancer cases in India are diagnosed with advanced clinical stage
with 5-year survival rate >50% (2).
Tobacco chewing/smoking, alcohol consumption and human
papillomavirus (HPV) infections, especially HPV 16 and 18 are the
major risk factor for oral cancer (3).
Oral carcinogenesis is a multistep process that involves the
acquisition of many genetic and epigenetic alterations and dynamic
changes in the gene expression of coding and non-coding RNAs.
Long non-coding RNAs are >200 nucleotides in
length with limited or no protein-coding capacity, and serve
primarily as the regulatory components in the cell. LncRNAs are
reported as implicated in a vast number of cellular processes, such
as cell proliferation, cell cycle progression, apoptosis, cell
survival and motility. Dysregulation of several lncRNAs is related
to metastasis, recurrence and prognosis in various cancer types
(4). Enhanced expression of the lncRNA
UCA1 has been suggested to promote cancer metastasis in head and
neck cancers (5). Colon Cancer
Associated Transcript 1 (CCAT1), an lncRNA, located in chromosome
8q24, close to c-Myc, a well-known oncogenic transcription factor
(6). Previous studies have indicated
that CCAT1 is differentially expressed in several types of cancers,
including colon cancer, gastric cancer, gall bladder cancer and
hepatocellular carcinoma (7–9). The CCAT1 was identified to be
significantly overexpressed and clinically correlated with colon
cancer patients' disease stage, lymph node metastasis and survival
after surgery (10). In addition,
CCAT1 was demonstrated to correlate with overall survival and
progression-free survival in breast cancer (11). Although the CCAT1 has been reported to
be involved in many human cancers, to date, its role in oral cancer
is completely unknown.
Recently, lncRNAs have been indicated to function as
a competing endogenous RNAs (ceRNA) by competitively binding to
microRNAs (miRNAs) through their miRNA response elements (MRE)
(12). These ceRNAs usually share MRE
with other coding transcripts targeted by miRNAs and thereby act as
sponges for that group of miRNAs, preventing the targeted mRNA
transcripts from degradation. The upregulation of CCAT1 has been
reported to downregulate the miRNAs miR155-5p, let7b-5p, miR490-3p
through a sponging mechanism and miR218-5p through epigenetic
silencing (9,13–15). In the
current report, the authors studied the role of CCAT1 in oral SCC
by analyzing their expression and correlating the expression level
with clinical characteristics. The expression of miR155-5p,
let7b-5p, miR490-3p and miR218-5p was further analyzed in order to
understand sponging activity of overexpressed CCAT1 and their role
in oral carcinogenesis.
Materials and methods
Tumor tissue samples
The present study was approved by the Institutional
Ethics Committee (IEC), Madras Medical College (Chennai, India; no.
04092010) and was conducted within the ethical framework of the Dr
ALM PG Institute of Basic Medical Sciences (Chennai, India). A
total of 60 oral cancer and 8 normal tissue samples were collected
from Royapettah Government Hospital (Chennai, India). Tissue
samples were maintained in RNAlater® solution (Ambion;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and transported
to the laboratory on ice. Patient's contextual and
clinicopathological characteristics were documented with standard
questionnaire as per the IEC guidelines, and written informed
consent was obtained from each patient.
Isolation of RNA from tumor
samples
Tissues stored in RNAlater® solution were
washed twice with freshly prepared ice-cold PBS buffer. Tissue
samples were homogenized in MicroSmash MS-100 automated homogenizer
(Tomy, Tokyo, Japan) with zirconium beads for 1 min and the
RNAeasy® mini kit (Qiagen GmbH, Hilden, Germany) was
used for RNA extraction following supplier instructions.
Expression profiling by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Expression profiling was performed for CCAT1 by
using a custom-designed universal probe assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.). c-Myc was amplified by TaqMan
assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
microRNAs by a custom-designed stem loop real-time assay (Applied
Biosystems; Thermo Fisher Scientific, Inc.) (16). Briefly, cDNA synthesis for CCAT1 and
GAPDH was conducted using custom-designed universal oligo reverse
primers (Table I) (17) and using random hexamer primer
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for c-Myc, and
specific stem-loop primers for each miRNA (Table I). Total RNA samples were pre-incubated
at 65°C for 20 min to denature the RNA secondary structures, and
were then placed on ice. All cDNA conversion reactions were
conducted using reagents from the Invitrogen Reverse Transcription
kit (Invitrogen; Thermo Fisher Scientific, Inc. USA) following the
manufacturer's protocol. Following reverse transcription, the cDNAs
were diluted 25 times and RT-qPCR was carried out in 384-well
optical plates (in triplicate) in 10 µl reactions with
TaqMan® 2X Universal Master mix (No AmpErase Uracil
N-Glycosylase; Thermo Fisher Scientific, Inc.), a specific forward
primer, universal reverse primer and universal FAM-labelled MGB
probe (Applied Biosystems; Thermo Fisher Scientific, Inc.) for
miRNA/lncRNA, and a custom designed TaqMan® assay was
used for c-Myc (Table II). Forward
primers specific to the gene/miRNAs and universal reverse primers
used for RT-qPCR are provided in Table
II. The experiments were carried out using a 7900HT Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). A
negative control without cDNA was also included in parallel for all
assays. GAPDH was used as an endogenous control for lncRNA, and
c-Myc and RNU44 as an endogenous control for miRNAs. Each
experiment was conducted in triplicate and relative quantification
of the expression was carried out using
2−ΔΔCt calculation (18).
 | Table I.List of reverse transcription primers
used for cDNA synthesis. |
Table I.
List of reverse transcription primers
used for cDNA synthesis.
| Gene | Primer sequence
(5′-3′) |
|---|
| Oligo primer for
mRNA/lncRNA |
CAGTGCAGGGTCCGAGGTACAGAGCCACCTGGGCAA |
|
| TTTTTTTTTTTVN |
| RNU44 |
GTCGTATCCAGTGCGTCGAGTGACACGAGAGCCACCTGGGCAA |
|
|
TTTGCACTGGATACGACAGTCAG |
| hsa-miR-let7b-5p |
GTCGTATCCAGTGCGTCGAGTGACACGAGAGCCACCTGGGCAA |
|
|
TTTGCACTGGATACGACAACCAC |
| hsa-miR-218-5p |
GTCGTATCCAGTGCGTCGAGTGACACGAGAGCCACCTGGGCAA |
|
|
TTTGCACTGGATACGACACATGG |
| hsa-miR-490-3p |
GTCGTATCCAGTGCGTCGAGTGACACGAGAGCCACCTGGGCAA |
|
|
TTTGCACTGGATACGACCAGCAT |
| hsa-miR-155-5p |
GTCGTATCCAGTGCGTCGAGTGACACGAGAGCCACCTGGGCAA |
|
|
TTTGCACTGGATACGACACCCCT |
 | Table II.List of gene specific primers and
sequences of universal primer and probe used in RT-qPCR. |
Table II.
List of gene specific primers and
sequences of universal primer and probe used in RT-qPCR.
| Gene | Forward primer
(5′-3′) |
|---|
| CCAT1 |
GTGTATCTTAGTTCAACCAAATTGTAATCATCTG |
| GAPDH |
GAAGAGGGGAGGGGCCTAGG |
| RNU44 |
GCAAATGCTGACTGAACATGA |
| let7b-5p |
GTGAGGTAGTAGGTTGTGTG |
| miR-218-5p |
CAGTTGTGCTTGATCTAACCA |
| miR-490-3p |
CAACCTGGAGGACTCCAT |
| miR-155-5p | CGCAGTTAATGCTA
ATCGTGATAG |
| Universal reverse
primer used for lncRNA/mRNA |
CAGTGCAGGGTCCGAGGT |
| Universal reverse
primer used for miRNA/snoRNA |
TCGTATCCAGTGCGTCGAGT |
| c-Myc | Forward:
ATTCTCTGCTCTCCTCGACG |
|
| Reverse:
CCTTACGCACAAGAGTTCCG |
| Universal probe for
RT-qPCR assay |
CAGAGCCACCTGGGCAATTTT |
Clinical evaluation
All 60 oral SCC patients were treated with 50–60 Gy
radiation with concurrent three courses of cisplatin (Naprod Life
Sciences, Mumbai, India) and 5-fluorouracil (Celon Laboratories
Ltd, Hyderabad, India)]. The tumor response to this
chemo/radio-therapy regimen was evaluated following 4 weeks of
treatment. Patients who were evaluated as a complete and partial
response to treatment were categorized as treatment responders, and
the remaining patients were designated as poor responders. The
patients were followed post-treatment for 13 months to study the
tumor recurrence and therapeutic outcome.
Statistical analysis
The correlation between lncRNA CCAT1 expression and
clinicopathological features of the patients were calculated by the
chi-squared test and the differences between means were analyzed by
Student's t-test (Mann-Whitney) using GraphPad Prism software
(version, 6.0; GraphPad Software, Inc., La Jolla, CA, USA).
Numerical data are presented as mean and ± standard error of the
mean in graphs. P<0.05 was considered to indicate a
statistically significant difference
Results
Clinical characteristics
A total of 60 oral squamous cell carcinoma tumors
and 8 normal tissue samples were used to study the expression of
CCAT1. Of 60 oral SCC cases, a 42% (n=25) of the patients were
>50 years of age and males were more predominant [(71.67%) n=43]
than females [(28.33%; n=17)]. Patients with any one or all habits
(tobacco chewing/smoking and alcohol) account for 81.6% (n=49) of
the cohort. 80% of the patients (n=48) presented tumors grades of
>T2 stage and 86.7% (n=52) were positive for lymph node
invasion. In the histological grading, 71.67% (n=43) of the cases
were undifferentiated (Table
III).
 | Table III.Relationship between CCAT1 expression
and clinicopathological characteristics in oral cancer
patients. |
Table III.
Relationship between CCAT1 expression
and clinicopathological characteristics in oral cancer
patients.
|
|
| CCAT1 expression
level |
|
|---|
|
|
|
|
|
|---|
| Clinical
characters | No. | Low (n=44) | High (n=16) |
P-valuea |
|---|
| Age |
|
|
| 0.5558 |
|
<50 | 25 | 17 | 8 |
|
|
≥51 | 35 | 27 | 8 |
|
| Gender |
|
|
| 0.1199 |
|
Male | 43 | 29 | 14 |
|
|
Female | 17 | 15 | 2 |
|
| Risk habits |
|
|
| 1.0000 |
|
Positive | 49 | 36 | 13 |
|
|
Negative | 11 | 8 | 3 |
|
| Tumor stage |
|
|
| 1.0000 |
|
≤T2 | 12 | 9 | 3 |
|
|
>T2 | 48 | 35 | 13 |
|
| Nodal invasion |
|
|
| 0.4293 |
|
Positive | 52 | 39 | 13 |
|
|
Negative | 8 | 5 | 3 |
|
|
Differentiation |
|
|
| 0.5181 |
|
Differentiated | 17 | 14 | 3 |
|
|
Undifferentiated | 43 | 30 | 13 |
|
LncRNA CCAT1 is overexpressed in oral
SSCs
Expression level of CCAT1 was analyzed in 60 oral
tumors and 8 normal tissue samples by RT-qPCR. CCAT1 overexpression
was observed in 27% (16/60) of oral cancer samples (Fig. 1) and the expression was identified to
be exponentially increased across the tumor samples. Therefore, the
CCAT1 overexpressed samples were characterized into three
categories, based on the expression levels. In group 1, the mean
CCAT1 expression level was equal to the normal samples [73%
(44/60)], in group 2 the expression was >15 fold higher than
normal tissues [13.5% (8/60)] and in group 3 the expression was
>30-fold higher than normal tissues [13.5% (8/60)]. Group 2 and
group 3 tumors presented statistically significant overexpression
of CCAT1 (P=0.0002 and P=0.001, respectively), when compared to
control tissue samples.
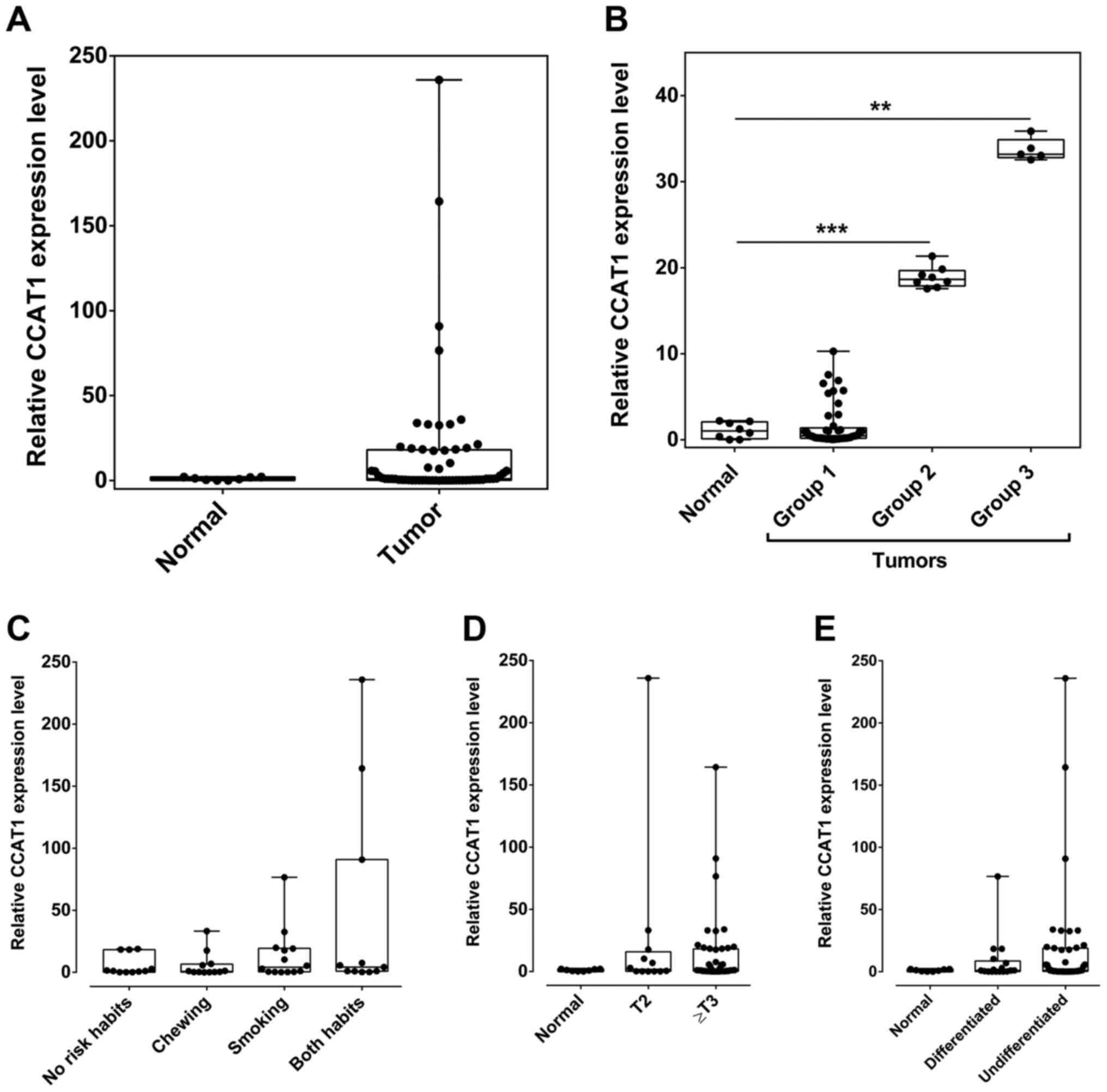 | Figure 1.Expression of CCAT1 in oral squamous
cell carcinomas. (A) Expression of CCAT1 in oral tumors compared
with normal tissues. (B) Expression of CCAT1 was identified to be
exponentially higher in oral tumors. The samples are categorized
into groups depending on the expression clusters. (C) Oral cancer
patients with a smoking habit presented a higher, but not
significant, expression of CCAT1 compared to those who chew
tobacco. (D) CCAT1 was overexpressed in tumors with tumor grade
>T2, compared with normal tissues. (E) Undifferentiated oral
tumors expressed higher levels of CCAT1, when compared with tumors
of differentiated cellular pathology. The groups used in the study
are as follows: Normal, unaffected tissue samples (n=8); group 1,
mean CCAT1 expression level=mean CCAT1 expression level in the
normal samples (n=44); group 2, the mean CCAT1 expression was
>15-fold higher than normal tissues (n=8); group 3, mean CCAT1
expression was >30 fold higher than normal tissues (n=8). Data
are presented as the mean ± standard error of the mean.
**P<0.01, ***P<0.001 vs. normal tissues. CCAT1, Colon Cancer
Associated Transcript 1. |
Association of CCAT1 with
clinicopathological characteristics
In order to understand the significance of lncRNA
CCAT1 overexpression in oral cancer, CCAT1 overexpression was
correlated with clinicopathological features of the tumors. In
univariate analysis, except for age (P=0.0072), none of the
clinicopathological features tested were statistically significant
(Table IV), and this may be due to
the wide variation in the expression level of CCAT1 in tumor
samples of the clinical sub-groups. Patients with higher tumor
grade and undifferentiated pathology presented high level
expression of CCAT1. The majority of the CCAT1 overexpressing
samples (62.5%) were tumors from buccal mucosa and the tumor
samples with tobacco smoking and practising both the habit (tobacco
chewing and smoking) expressed a high level of CCAT1. The authors
then analyzed the head and neck cancer datasets from TCGA database
using the TANRIC interactive platform (16) and a similar CCAT1 overexpression was
observed in cases with the history of tobacco abuse (P<0.0001),
suggesting the role of tobacco smoking/chewing in oral
tumorigenesis.
 | Table IV.Relationship between CCAT1 expression
categories in groups and their clinicopathological characteristics
in oral cancer patients. |
Table IV.
Relationship between CCAT1 expression
categories in groups and their clinicopathological characteristics
in oral cancer patients.
|
| CCAT1 expression
categories |
|
|---|
|
|
|
|
|---|
| Clinical
characteristics | Group 1 (n=44)
(%) | Group 2 (n=8)
(%) | Group 3 (n=8)
(%) |
P-valuea |
|---|
| Age |
|
|
| 0.0072b |
|
<50 | 17 (38.6) | 7 (87.5) | 1 (12.5) |
|
|
≥51 | 27 (61.4) | 1 (12.5) | 7 (87.5) |
|
| Gender |
|
|
| 0.2601 |
|
Male | 29 (66) | 7 (87.5) | 7 (87.5) |
|
|
Female | 15 (44) | 1 (12.5) | 1 (12.5) |
|
| Risk habits |
|
|
| 0.1526 |
|
Positive | 36 (81.9) | 5 (62.5) | 8 (100) |
|
|
Negative | 8 (18.1) | 3 (37.5) | 0 (−) |
|
| Tumor stage |
|
|
| 0.7804 |
|
≤T2 | 9 (20.5) | 1 (12.5) | 1 (12.5) |
|
|
>T2 | 35 (79.5) | 7 (87.5) | 7 (87.5) |
|
| Nodal invasion |
|
|
| 0.5784 |
|
Positive | 39 (88.6) | 7 (87.5) | 6 (75) |
|
|
Negative | 5 (11.4) | 1 (12.5) | 2 (25) |
|
|
Differentiation |
|
|
| 0.1723 |
|
Differentiated | 14 (31.8) | 2 (25) | 0 (−) |
|
|
Undifferentiated | 30 (68.2) | 6 (75) | 8 (100) |
|
CCAT1 overexpression is associated
with treatment response
The CCAT1 overexpression has been indicated to be
associated with the clinical features and therapeutic response in
various human cancers (10,11). Therefore, the authors correlated the
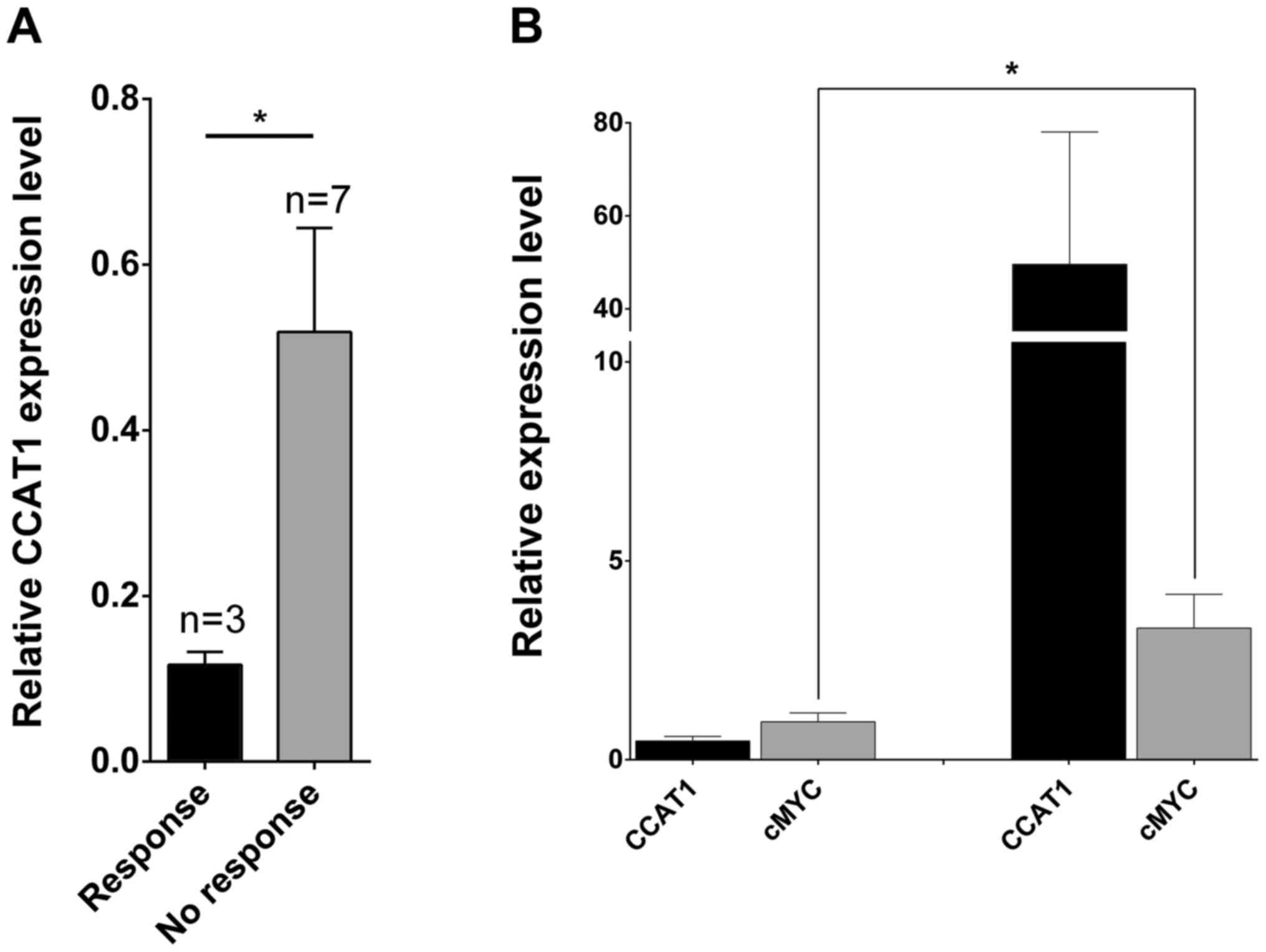
CCAT1 expression level with the patient's treatment responses. Most
of the patients participated in the presented study are from rural
areas and are uneducated, and 50 of 60 patients failed to appear
for the monthly check up. Only 10 patients were followed-up
post-treatment for 13 months to analyze the treatment response. As
illustrated in Fig. 2A, patients with
significant overexpression of CCAT1 presented a poor therapeutic
response (P=0.01). Furthermore, when overall survival of the
high/low level of CCAT1 expressing group of head and neck cases
from the TCGA database was analyzed, a high level expression of
CCAT1 presented strong association with poor survival (P=0.008;
data not shown).
The c-Myc oncogene is overexpressed in
oral squamous cell carcinomas
To understand the functional significance of c-Myc,
an oncogenic transcription factor reported to be associated with
various cancers including oral cancer and located in the same
chromosome loci with CCAT1 (7,19,20), c-Myc
expression was analyzed in all samples. Interestingly, c-Myc was
demonstrated as overexpressed in the tumor samples that expressed a
high level of CCAT1 (P=0.0473; Fig.
2B).
Expression of miR155-5p, let7b-5p,
miR218-5p and miR490-3p in oral SSCs
CCAT1 was reported to function as ceRNA in various
cancers (13–15). Therefore, to understand whether a
CCAT1-mediated ceRNA network is functioning in oral SCCs, the
expression of miR155-5p, let7b-5p, miR218-5p and miR490-3p was
profiled and a significant downregulation of miR155-5p (P=0.037)
and let7b-5p (P<0.0001) was observed in oral SCCs compared to
normal tissues (Fig. 3A). The miRNAs,
miR218-5p and miR490-3p were upregulated in oral SCC tumors
compared with normal samples but it was identified as being
downregulated significantly in CCAT1 overexpressing tumor group 2
and 3 (Fig. 3B). let7b-5p was
significantly downregulated across all the tumor groups (group 1,
P=0.0001; group 2, P=0.004; and group 3, P=0.0002) compared with
the normal tissues (Fig. 3C).
Consistent with the previous reports, a downregulation of let7b-5p,
and miR155-5p was observed in tumor samples that overexpressed
CCAT1, compared with normal tissues. These results suggested that
CCAT1 acts as sponge for miR155-5p, let7b-5p, miR218-5p and
miR490-3p, resulting in the degradation of these microRNAs.
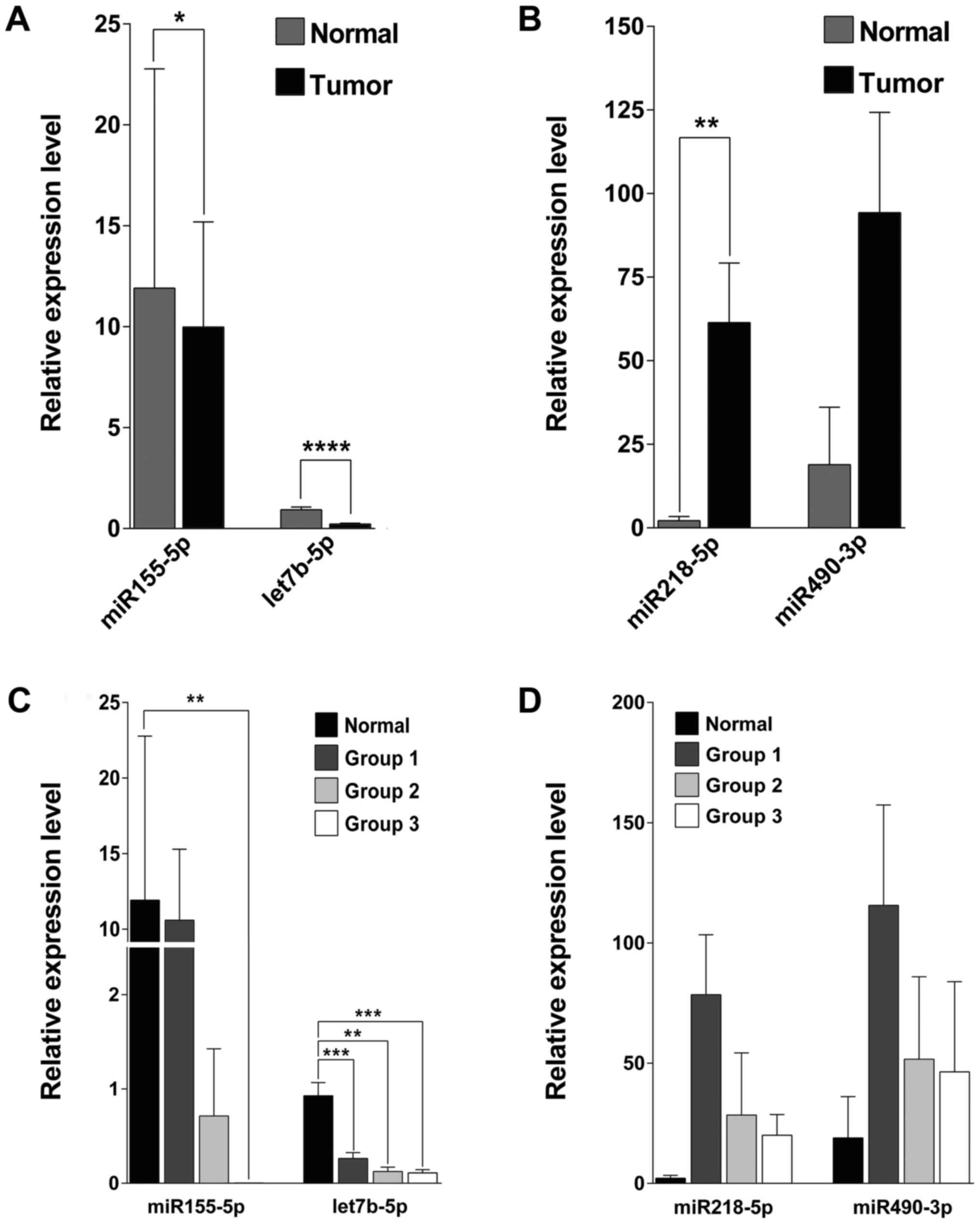 | Figure 3.Expression of miR155-5p, let7b-5p,
miR218-5p and miR490-3p. (A) miR155-5p and let7b-5p were
significantly downregulated in oral tumors, when compared with
normal tissues. (B) miR218-5p and miR490-3p were upregulated in
oral tumors, when compared with normal tissues. (C) miR155 and
let7b were downregulated in oral tumor groups and miR-let7b was
significantly downregulated in all three groups of the CCAT1
overexpressed samples, when compared with the normal tissues. (D)
miR218 and miR490 were downregulated in tumor groups overexpressing
CCAT1, but were overexpressed when compared with normal tissue
levels. Notably, group 1 samples expressed CCAT1 at levels equal to
normal, group 2 samples expressed CCAT1 at 15-fold higher and group
3 samples expressed CCAT1 at >30-fold. The groups used in the
study are as follows: Normal, unaffected tissue samples (n=8);
group 1, mean CCAT1 expression level=mean CCAT1 expression level in
the normal samples (n=44); group 2, the mean CCAT1 expression was
>15-fold higher than normal tissues (n=8); group 3, mean CCAT1
expression was >30-fold higher than normal tissues (n=8). Data
are presented as the mean ± standard error of the mean. *P<0.05,
**P<0.01, ***P<0.001, ****P<0.0001 vs. normal tissues.
CCAT1, Colon Cancer Associated Transcript 1; miR, microRNA. |
Discussion
lncRNAs are >200 nt in length and serves critical
regulatory roles in diverse cellular processes. Differential
expression of lncRNAs is increasingly documented as a hallmark
feature in cancer (7–9). In the present study, the authors analyzed
the expression level of lncRNA CCAT1 and explored its clinical
significance in oral cancer. It was identified that CCAT1
expression was several folds higher in oral cancer samples, when
compared with the normal tissue samples. Interestingly, as the
expression of CCAT1 in tumor samples is demonstrated to be
exponentially increased, the tumors were characterized into three
groups in the increasing order of expression and correlated the
CCAT1 expression levels with patients' clinicopathological
features. The CCAT1 amplified samples congregated into the tumors
with aggressive phenotypes and the CCAT1 overexpression was
observed in the patients with cigarette smoking (P=0.68).
Consistent with these results, head and neck cancer datasets from
TCGA database also presented overexpression of CCAT1 in tumors with
tobacco smoking history. Furthermore, patients with overexpression
of CCAT1 demonstrated a poor response to treatment (P=0.01;
Figs. 1 and 2A) and similar results were also presented in
the head and neck cancer TCGA datasets. Copy number alterations of
CCAT1 have been reported to be frequent in breast cancer (21) and, in ER+/HER2−
cases, CCAT1 amplification was reported to be strongly associated
with early relapse (22).
Myc has been demonstrated to serve a key role in the
progression of cell proliferation and tumorigenesis in oral cancer
(23). As the c-Myc gene is located
515 kb upstream of CCAT1 in the same chromosome loci, the authors
tested the c-Myc expression and observed significant high level of
c-Myc in CCAT1 overexpressing tumors. To the best of the authors'
knowledge, the present study was the first to observe the
concomitant overexpression of both c-Myc and CCAT1 in oral cancer,
although the c-Myc protein has already been reported as
overexpressed in 80% of the oral SCCs and correlated with poor
prognosis (24). Furthermore, c-Myc
was reported to contribute microenvironment-mediated drug
resistance in AML and oral cell lines (25,26). The
CCAT1 locus has been presented to harbor a super-enhancer that
could control the c-Myc expression. The CCAT1 was demonstrated to
interact with a transcriptional enhancer MYC-335 via chromatin
looping and consequently, could interact with the MYC-promoter
resulting regulation of c-Myc expression in cis (10,27). In
addition, knockdown of CCAT1 led to a decrease in c-Myc levels in
colon cancer-derived cell lines (28).
In addition, aberrant expression of CCAT1 controlled by c-Myc
predicted the poor prognosis in hepatocellular carcinoma and
gastric cancer (29,30).
Functioning as ceRNA, CCAT1 has been reported to
downregulate miR155-5p, let7b-5p and miR490-3p through
sequestration and miR218-5p by epigenetic regulation (9,13–15). Therefore, the expression of miR155-5p,
let7b-5p, miR218-5p and miR490-3p in oral tumors and normal tissues
was analyzed. Interestingly, miR155-5p and let7b-5p were
significantly downregulated in all four tumor groups compared with
normal tissues, but miR218-5p and miR490-3p only in tumor groups 2
and 3 (Fig. 3). In lung cancer, CCAT1
was reportedly overexpressed in cigarette smokers, and indicated to
alter the cell cycle transition through epigenetic silencing of
miR281 and activation of EMT-associated gene BMI1 (13). CCAT1 was reported to promote
hepatocellular carcinoma progression by sponging the microRNA let7
family, tumor suppressor miRNAs well associated with oral cancer
(9). The CCAT1 has been reported to
regulate miR490 in gastric cancer by similar mechanism mentioned
above and favors tumor cell migration and accounts for poor
therapeutic outcome (14). The
presented results suggested that CCAT1 overexpression was likely to
affect the downstream target miRNA level in oral tumors via
sponging mechanism as shown in other cancers (9,14). In
addition, CCAT1 overexpressed samples expressed a low level of
miR155 and let7b and the same has been frequently reported to be
deregulated in several cancer cell lines and cancers (31). All these observations suggest that
CCAT1 overexpression may serve an important role in oral
carcinogenesis and treatment response. Further research is
warranted to confirm CCAT1 as a potential target for oral cancer
diagnosis and treatment.
Acknowledgements
The authors would like to thank the patients for
their consent to provide clinical samples. G.A. gratefully
acknowledges the University Grant Commission (UGC), Government of
India for providing research fellowship. The authors also
gratefully acknowledge the Department of Science and Technology
Fund for Improvement of S&T Infrastructure, UGC-Special
Assistance Programme and the Department of Health Multidisciplinary
Research Unit infrastructure facility. The study was supported by
grants received by the corresponding author from the Department of
Atomic Energy, Board of Research in Nuclear Sciences (grant no.
35/14/10/2014-BRNS/0210), and the Department of Biotechnology,
Government of India (grant no. BT/PR4820/MED/12/622/2013).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bourhis J, Overgaard J, Audry H, Ang KK,
Saunders M, Bernier J, Horiot JC, Le M, aître A, Pajak TF, Poulsen
MG, et al: Hyperfractionated or accelerated radiotherapy in head
and neck cancer: A meta-analysis. Lancet. 368:843–854. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Manikandan M, Deva Magendhra, Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang H: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alaiyan B, Ilyayev N, Stojadinovic A,
Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink
B, et al: Differential expression of colon cancer associated
transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence.
BMC Cancer. 13:1962013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He X, Tan X, Wang X, Jin H, Liu L, Ma L,
Yu H and Fan Z: C-Myc-activated long noncoding RNA CCAT1 promotes
colon cancer cell proliferation and invasion. Tumour Boil.
35:12181–12188. 2014. View Article : Google Scholar
|
|
11
|
Zhang XF, Liu T, Li Y and Li S:
Overexpression of long non-coding RNA CCAT1 is a novel biomarker of
poor prognosis in patients with breast cancer. Int J Clin Exp
Pathol. 8:9440–9445. 2015.PubMed/NCBI
|
|
12
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu L, Xu H, Luo F, Liu X, Lu X, Yang Q,
Xue J, Chen C, Shi L and Liu Q: Epigenetic silencing of miR-218 by
the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle
transition in the malignant transformation of HBE cells induced by
cigarette smoke extract. Toxicol Appl Pharmacol. 304:30–41. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou B, Wang Y, Jiang J, Jiang H, Song J,
Han T, Shi J and Qiao H: The long noncoding RNA colon
cancer-associated transcript-1/miR-490 axis regulates gastric
cancer cell migration by targeting hnRNPA1. IUBMB Life. 68:201–210.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Wang W, Cao L, Li Z and Wang X:
Long Non-Coding RNA CCAT1 acts as a competing endogenous RNA to
regulate cell growth and differentiation in acute myeloid leukemia.
Mol Cells. 39:330–336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang K, Zhang X, Liu H, Wang Z, Zhong J,
Huang Z, Peng X, Zeng Y, Wang Y, Yang Y, et al: A novel real-time
PCR assay of microRNAs using S-Poly(T), a specific oligo(dT)
reverse transcription primer with excellent sensitivity and
specificity. PLoS One. 7:e485362012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Han L, Roebuck P, Diao L, Liu L,
Yuan Y, Weinstein JN and Liang H: TANRIC: An interactive open
platform to explore the function of lncRNAs in cancer. Cancer Res.
75:3728–3737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grisanzio C and Freedman ML: Chromosome
8q24-Associated cancers and MYC. Genes Cancer. 1:555–559. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Naylor TL, Greshock J, Wang Y, Colligon T,
Yu QC, Clemmer V, Zaks TZ and Weber BL: High resolution genomic
analysis of sporadic breast cancer using array-based comparative
genomic hybridization. Breast Cancer Res. 7:R1186–R1198. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bilal E, Vassallo K, Toppmeyer D, Barnard
N, Rye IH, Almendro V, Russnes H, Børresen-Dale AL, Levine AJ,
Bhanot G and Ganesan S: Amplified loci on chromosomes 8 and 17
predict early relapse in ER-positive breast cancers. PLoS One.
7:e385752012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mishra R and Das BR: Early overexpression
of Cdk4 and possible role of KRF and c-myc in chewing tobacco
mediated oral cancer development. Mol Biol Rep. 30:207–213. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pai R, Pai S, Lalitha R, Kumaraswamy S,
Lalitha N, Johnston R and Bhargava M: Over-expression of c-Myc
oncoprotein in oral squamous cell carcinoma in the South Indian
population. Ecancermedicalscience. 3:1282009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia B, Tian C, Guo S, Zhang L, Zhao D, Qu
F, Zhao W, Wang Y, Wu X, Da W, et al: c-Myc plays part in drug
resistance mediated by bone marrow stromal cells in acute myeloid
leukemia. Leuk Res. 39:92–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghosh RD, Ghuwalewala S, Das P, Mandloi S,
Alam SK, Chakraborty J, Sarkar S, Chakrabarti S, Panda CK and
Roychoudhury S: MicroRNA profiling of cisplatin-resistant oral
squamous cell carcinoma cell lines enriched with
cancer-stem-cell-like and epithelial-mesenchymal transition-type
features. Sci Rep. 6:239322016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pomerantz MM, Ahmadiyeh N, Jia L, Herman
P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M,
et al: The 8q24 cancer risk variant rs6983267 shows long-range
interaction with MYC in colorectal cancer. Nat Genet. 41:882–884.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Ma M, Liu W, Ding W and Yu H:
Enhanced expression of long noncoding RNA CARLo-5 is associated
with the development of gastric cancer. Int J Clin Exp Pathol.
7:8471–8479. 2014.PubMed/NCBI
|
|
30
|
Zhu HQ, Zhou X, Chang H, Li HG, Liu FF, Ma
CQ and Lu J: Aberrant expression of CCAT1 regulated by c-Myc
predicts the prognosis of hepatocellular carcinoma. Asian Pac J
Cancer Prev. 16:5181–5185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY,
Yan M, Qu QL, Zhu ZG and Liu BY: MicroRNA-155 is downregulated in
gastric cancer cells and involved in cell metastasis. Oncol Rep.
27:1960–1966. 2012.PubMed/NCBI
|