Introduction
Hodgkin's lymphoma (HL) is a tumor that has a
characteristic pathological behavior and requires specific careful
clinical management. Our understanding of the epidemiological
behavior of HL is constantly updated and data are available for
different regions; however, studies from the kingdom of Saudi
Arabia are scarce (1). At present,
genetic signatures identified through genetic screening are a
method of predicting and monitoring cancer prognosis by identifying
patterns in molecular genetics demonstrated to be associated with a
particular type of cancer (2,3). Previous findings have indicated that
patients with HL with unfavorable outcomes exhibited higher levels
of genetic instability compared with other types of cancer
(4). Indeed, HL incidence has
increased over the previous decade among the Cooperation Council
for the Arab States of the Gulf (5).
However, a systematic analysis for the global burden of disease
revealed that, despite the increase in the incidence of all types
of cancer, HL-associated mortalities had decreased between 2005 and
2015 (6). Based on data from Al-Diab
et al (7) on Saudi and Middle
Eastern populations, there is an elevated incidence of HL,
particularly in the younger age group of 12–40 years. This study
hypothesized that the rapid improvement in the living standards and
healthcare systems during the previous 2 decades has affected the
epidemiology of HL. In the Middle East and Asia, HL is more common
during childhood compared with Western countries, where its peak
incidence is in patients between 10 and 30 years old (8). A revised European-American Lymphoma
classification, which was adopted later by the World Health
Organization, uses morphologic, phenotypic, genotypic and clinical
data for the classification of HL (8). It encompasses two well defined entities:
Nodular lymphocyte predominant HL and the classical HL (CHL). CHL
comprises ~95% of HL cases worldwide (8).
Hodgkin Reed-Sternberg cells are surrounded by a
number of non-neoplastic immune cell subsets, which represent the
tumor microenvironment (TME) and typically dominates the cellular
milieu. However, the overall genetic expression profile in an
individual will contain a combination of the gene expression
patterns of the separate variable cellular components of the TME,
and that of the tumor cells (9).
Studies of candidate single nucleotide polymorphisms (SNPs) to
identify HL risk factors may be useful. Unless future studies
employ adequate sample sizes to reflect the etiological
heterogeneity within HL, identifying genotypes according to patient
age and tumor histology is essential (10). DNA mutation or damage of DNA repair
genes will subsequently decrease genomic integrity and stability.
Few studies have described an association between defective
proteins in DNA repair pathways and susceptibility to HL.
The DNA repair mechanism is genetically regulated,
and variation in the repair capacity was observed based on SNPs in
DNA repair genes (11). Several
epidemiological studies have identified hundreds of these SNPs, and
suggested a wide population variability in the DNA repair process,
with potential associations with higher risk of cancer and
hematological malignancies (4,11). A
polymorphism in the DNA repair gene may modulate the DNA repair
gene phenotype, particularly if these SNPs exist within the
regulatory region that alters the protein expression, and
consequently the functional properties of the repair enzymes
(11). At present, the effect of
these SNPs on altering the risk of HL has not been investigated
widely. Monroy et al (4)
evaluated the gene-gene interaction between candidate genes in
direct reversal, nucleotide excision repair (NER), base excision
repair (BER) and double strand break (DSB) pathways.
The X-ray repair cross-complementing (XRCC) gene is
involved in the repair of single strand DNA breaks created as a
result of exposure to ionizing radiation or alkylating agents. This
protein is active in the base excision repair (BER) pathway
(4). A micro satellite polymorphism
in this gene was identified to be associated with increased risk of
cancer in patients with radio sensitivity (4). Specifically, the XRCC1 gene is involved
in the BER pathway and is responsible for repairing oxidizing and
alkylating DNA lesions. The XRCC3 gene is important for maintaining
chromosome stability. In addition, this gene complements a repair
deficient mutant that is sensitive to numerous DNA damaging agents
and is unstable. The XRCC3 gene is involved in the DSB
recombination repair pathway and is able to restore DNA through a
recombination process between the damaged strand and the homologous
copy of the gene (4). XRCC3
(Thr241Met), XRCC1 (Arg194Trp) and XRCC1 (Arg399Gln) polymorphisms
serve a role in modulating the risk of HL (4). In addition, the cumulative genetic risk
score revealed a significant risk of HL with an increased number of
gene polymorphisms involved in BER and DSB (4). The XRCC gene SNPs result in an amino
acid substitution, which affects the structure and function of the
final intended protein (11), and
thereafter may affect the risk stratification or prognosis of the
HL.
The xeroderma pigmentosum, complementation group C
(XPC) gene encodes for a protein that is a component of the NER
pathway. Concurrently, the XP complementation group G (XPG) gene
encodes for a specific single strand DNA endonuclease enzyme that
enables the 3′ incision step in the DNA excision repair pathway to
repair UV-induced damage. Mutations in the XP gene may result in
xeroderma pigmentosum, with a susceptibility to early development
of carcinomas (11). In HL, previous
study has investigated the correlation between HL risk and
polymorphisms in DNA repair protein XP group C, D and G (11). XPC, also known as transmembrane
Protein 43, XPD, also known as group 2 excision repair
cross-complementing, and XPG genes participate in the NER pathway,
which is responsible for repairing bulky DNA adducts and
helix-distorting lesions (11). A
previous study observed that allelic variants in the mutant DNA
repair protein XPC, in particular XPC Ala499Val, were directly
associated with HL risk (4).
Nevertheless, El-Zein et al (11) identified a direct association between
the XRCC1 gene polymorphism Arg399Gln and the risk of HL.
Conversely, analysis of the combined effect of XRCC1/XRCC3 and
XRCC1/XPC polymorphisms identified a significant association with
the risk of HL (11). In addition,
gene variants in XRCC1 Gln/Gln, XRCC3 Thr/Thr and XRCC1 Arg/Gln,
collectively with XPC Lys/Lys, led to a significant rise in the
risk of the HL (11).
In the present study, the aim was to define an SNP
molecular profile, based on DNA repair genes mutations, as a
predictive biomarker for the prognosis of CHL patients in Saudi
Arabia. This was achieved through analyzing gene expression by
polymerase chain reaction and determining associations with disease
relapse and overall survival of the CHL cohort.
Materials and methods
Sample collection
The present study was a retrospective case-control
study in which formalin-fixed paraffin embedded (FFPE) tissue
samples from patients with CHL were studied. A total of 100 FFPE
samples from patients diagnosed with CHL from January 1990 to
December 2015 were obtained. Samples from all the histological CHL
sub classes and stages were included, according to the World Health
Organization scheme, which sub classifies CHL into four subtypes:
Nodular sclerosis (NS), mixed cellularity (MC), lymphocyte depleted
(LD) and lymphocyte rich (LR) (12,13).
Diagnosing HL is recommended followed staging, and is made
according to the Ann Arbor staging scheme or its Cotswold
modification. In the present study the latter was used to classify
HL as low grade (stage I or II) and high grade (stage III or IV)
(14).
Patient tissue samples were recruited to examine 4
different DNA repair gene SNPs (2 XRCC1 SNPs, XRCC3, 2 XPC SNPs and
XPG) using a polymerase chain reaction (PCR) assay. DNA repair gene
information is presented in Table
I.
 | Table I.Information on DNA repair genes
SNPs. |
Table I.
Information on DNA repair genes
SNPs.
| Gene | Codon no. | rs no. | Chromosome no. | SNP type | Nucleotide
change | Codon change | Polymorphism
type | SNP sequence |
|---|
| XRCC1 | 399 | rs25487 | 19 | Missense | C/T | CCG→CTG | Transition,
substitution |
GGGTTGGCGTGTGAGGCCTTACCTC[C/T]
GGGAGGGCAGCCGCCGACGCATGCG |
| XRCC1 | 194 | rs1799782 | 19 | Missense | C/T | CGG→TGG | Transition,
substitution |
GAGGCCGGGGGCTCTCTTCTTCAGC[C/T]
GGATCAACAAGACATCCCCAGGTGA |
| XRCC3 | 241 | rs861539 | 14 | Missense | C/T | ACG→ATG | Transition,
substitution |
AGGCATCTGCAGTCCCTGGGGGCCA[C/T]
GCTGCGTGAGCTGAGCAGTGCCTTC |
| XPC | 499 | rs2228000 | 3 | Missense | C/T | GCG→GTG | Transition,
substitution |
CATCGTAAGGACCCAAGCTTGCCAG[C/T]
GGCATCCTCAAGCTCTTCAAGCAGT |
| XPC | 939 | rs2228001 | 3 | Missense | A/C | CAG→AAG | Transition,
substitution |
AGCTTCCCACCTGTTCCCATTTGAG[A/C]
AGCTGTGAGCTGAGCGCCCACTAGA |
| XPG | 1104 | rs17655 | 13 | Missense | C/G | CAT→GAT | Transition,
substitution |
TTCATTAAAGATGAACTTTCAGCAT[C/G]
TTCACTTGAAGATCCATCAGATGAT |
Concomitantly, peripheral blood (PB) samples were
collected from normal healthy blood donors, with no known personal
or family history of cancer, to be used as the ‘study control
group’. In addition to the study control group, 2 normal fresh
tonsil tissues were obtained to serve as ‘CHL negative controls’
(i.e., negative for HL). A total of 2 fresh lymph node tissues from
newly-diagnosed patients with CHL were also obtained to serve as
‘CHL positive controls’ (i.e., positive for the presence of
HL).
Ethical approval was obtained from the Institutional
Review Board at John Hopkins Aramco Healthcare Center (JHAH)
(Dhahran, Saudi Arabia) and from the Research and Ethics Committee
at Arabian Gulf University (Manama, Bahrain). All subjects
providing control tissues gave written informed consent.
Deparaffinization of FFPE tissue
samples and DNA extraction for molecular PCR
Total genomic DNA was extracted from the
paraffin-embedded patient tissues, control group tissues and the
study control group PB samples using QIAamp DNA FFPE Tissue (250)
kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol.
Determination of concentration and
purity of extracted DNA
DNA yields were determined by measuring the light
absorbance at 260 nm. The purity was determined by calculating the
ratio of absorbance at 260 nm to absorbance at 280 nm. Pure DNA has
an A260/A280 ratio of 1.7–1.9. At 260 nm, optimum absorbance
readings should measure between 0.1 and 1.0; dilutions of the
samples in the present study were adjusted accordingly.
PCR
PCR was then performed for all CHL tissue samples
and controls using the 96-well plate TaqMan® GTXpress™
Master Mix system (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's protocol.
The PCR and genotyping assays were performed using the Applied
Biosystems 7500 Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Designed primers were produced by
TaqMan® GTXpress™ (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and are listed in Table II. The reaction conditions were as
follows: A cycle at 95°C for 20 sec, followed by 40 cycles at 95°C
for 15 sec and 60°C for 60 sec.
 | Table II.DNA repair genes single nucleotide
polymorphism assay ID. |
Table II.
DNA repair genes single nucleotide
polymorphism assay ID.
| Gene | rs no. | Assay ID |
|---|
| XRCC1 | rs25487 | C____622564_10 |
| XRCC1 | rs1799782 | C__11463404_10 |
| XRCC3 | rs861539 | C___8901525_10 |
| XPC | rs2228000 | C__16018061_10 |
| XPC | rs2228001 | C____234284_1_ |
| XPG | rs17655 | C___1891743_10 |
Statistical analysis
SPSS software v.20 (IBM Corporation, Armonk, NY,
USA) was used to perform all statistical analysis. The CHL disease
relapse (DR) was calculated as the time from starting the treatment
until disease recurrence, or relapse. The overall survival (OS) was
calculated as the time from receiving the treatment until
mortality. Survival curves were estimated using the Kaplan-Meier
analysis method and assessed for statistical significance using the
log-rank test incorporating hazard function analysis and cumulative
hazard rate estimation compared with OS. Genotype and allele
frequencies analysis for all studied SNPs were performed using the
SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) (15) and frequencies between patients and
controls were assessed by Fisher's exact test and Pearson's
χ2 test. The χ2 test was also used to assess
association between all the studied SNPs with the disease relapse
and OS of the patients with CHL.
Results
Demographic characteristics of CHL
patients at JHAH
A review of the records at JHAH indicated that CHL
represented >85% of the HL cases diagnosed during the study
period. The study population comprised 56 males and 44 females,
with a male: female ratio of 1.3:1. The age of the patients ranged
from 3–80 years, with a median age of 42 years. In addition, 78% of
the patients with CHL were <45 years old (Table III). The data from the study
population indicated that 27/100 (27%) patients were ≥45 years
old.
 | Table III.Characteristics of the classical
Hodgkin's lymphoma cases at John Hopkins Aramco Healthcare Center
during 1990 to 2015. |
Table III.
Characteristics of the classical
Hodgkin's lymphoma cases at John Hopkins Aramco Healthcare Center
during 1990 to 2015.
|
| Total |
|---|
|
|
|
|---|
| Characteristics | n | % |
|---|
| Age, years | 100 | – |
| (<45) | 78 | 78.0 |
| (≥45) | 27 | 27.0 |
| Sex | 100 | – |
| Male | 56 | 56.0 |
| Female | 44 | 44.0 |
| Treatment received
(response) | 100 | – |
| Curative | 98 | 98.0 |
| Palliative | 2 | 2.0 |
| Relapse | 100 | – |
| No relapse | 84 | 84.0 |
| Relapse | 16 | 16.0 |
| Overall survival | 100 | – |
| Did not survive | 11 | 11.0 |
| Survived | 89 | 89.0 |
Furthermore, a total of 16 CHL patients (16%) had
relapsed within the median period of 3.2 years, whilst 11 patients
(11%) had succumbed from the disease within the same period. The
majority of the patients with CHL succumbed within the first 5
years, with an OS median of 3.5 years.
XPG allele frequency analysis
The allele frequency analysis revealed that the XPG
repair gene SNP polymorphism rs17655 (C/G) demonstrated a
significant difference in allele frequency distribution between the
patients with CHL and the healthy controls (P=0.002). However, the
OR calculated for the wild allele (C) was low [OR=0.50; 95%
confidence interval (CI), 0.33–0.764; Table IV].
 | Table IV.Allele frequencies of 4 DNA repair
gene SNPs in 100 patients with classical HL and 100 controls. |
Table IV.
Allele frequencies of 4 DNA repair
gene SNPs in 100 patients with classical HL and 100 controls.
|
|
| Allele frequency |
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| DNA repair genes
SNPs | rs no. | HL patients | Control group | OR | CI (95%) | χ2 | df |
P-valuea |
P-valueb |
|---|
| XRCC1 | rs25487 | C | T | C | T | – | – | – | – | – | – |
|
|
| 0.76 | 0.25 | 0.78 | 0.22 | 0.87 | 0.546–1.383 | 0.35 | 1 | 0.554 | 0.554 |
| XRCC1 | rs1799782 | C | T | C | T | – | – | – | – | – | – |
|
|
| 0.03 | 0.97 | 0.07 | 0.93 | 0.44 | 0.16–1.94 | 2.094 | 1 | 0.1 | 0.1 |
| XRCC3 | rs861539 | C | T | C | T |
| – | – | – | – | – |
|
|
| 0.3 | 0.7 | 0.38 | 0.63 | 0.71 | 0.47–1.08 | 2.515 | 1 | 0.113 | 0.113 |
| XPC | rs2228001 | A | C | A | C | – | – | – | – | – | – |
|
|
| 0.57 | 0.43 | 0.58 | 0.43 | 0.98 | 0.65–1.46 | 0.01 | 1 | 0.919 | 0.919 |
| XPC | rs2228000 | C | T | C | T | – | – | – | – | – | – |
|
|
| 0.15 | 0.86 | 0.11 | 0.89 | 1.37 | 0.75–2.48 | 1.101 | 1 | 0.294 | 0.293 |
| XPG | rs17655 | C | G | C | G | – | – | – | – | – | – |
|
|
| 0.28 | 0.73 | 0.43 | 0.57 | 0.5 | 0.33–0.764 | 10.53 | 1 | 0.002 | 0.002 |
DNA repair genes association with DR
and OS
χ2 analysis of the 3 allele genotype
patterns of the DNA repair genes SNPs revealed that there is no
statistically significant association between any of the studied
SNPs with CHL DR or OS, with the exception of XPG SNP (rs17655),
which demonstrated statistical significance with the OS of the
patients with CHL (P=0.036) but not with DR. It was also identified
that this association was independent of patient age (P=0.415); by
contrast, XPG SNP was demonstrated to be significantly associated
with the male sex (P=0.034; Table
V).
 | Table V.Summary of significant associations
between DNA repair gene SNPs with DR, OS, age groups and sex of the
Saudi patients with CHL at John Hopkins Aramco Healthcare Center
during 1990 to 2015. |
Table V.
Summary of significant associations
between DNA repair gene SNPs with DR, OS, age groups and sex of the
Saudi patients with CHL at John Hopkins Aramco Healthcare Center
during 1990 to 2015.
|
| Patients with
CHL | Control group |
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| DNA repair gene
SNPs | n | % | n | % | DR | OS | Males | Females | Age (<45
years) | Age (≥45
years) |
|---|
| XRCC1
(rs25487) | 100 | – | 100 | – | 0.696 | 0.275 | – | – | – | – |
| Hom for
Wt allele | 55 | 55 | 62 | 62 | – | – | – | – | – | – |
|
Het | 41 | 41 | 32 | 32 | – | – | – | – | – | – |
| Hom for
Mut allele | 4 | 4 | 6 | 6 | – | – | – | – | – | – |
| XRCC1
(rs1799782) | 100 |
| 100 |
| 0.32 | 0.504 | – | – | – | – |
| Hom for
Wt allele | 1 | 1 | 0 | 0 | – | – | – | – | – | – |
|
Het | 4 | 4 | 13 | 13 | – | – | – | – | – | – |
| Hom for
Mut allele | 95 | 95 | 87 | 87 | – | – | – | – | – | – |
| XRCC3
(rs861539) | 100 |
| 100 |
| 0.207 | 0.519 | – | – | – | – |
| Hom for
Wt allele | 12 | 12 | 11 | 11 | – | – | – | – | – | – |
|
Het | 36 | 36 | 53 | 53 | – | – | – | – | – | – |
| Hom for
Mut allele | 52 | 52 | 36 | 36 | – | – | – | – | – | – |
| XPC
(rs2228001) | 100 | – | – | – | 0.572 | 0.078 | – | – | – | – |
| Hom for
Wt allele | 29 | 29 | 30 | 30 | – | – | – | – | – | – |
|
Het | 56 | 56 | 55 | 55 | – | – | – | – | – | – |
| Hom for
Mut allele | 15 | 15 | 15 | 15 | – | – | – | – | – | – |
| XPC
(rs2228000) | 100 | – | 100 | – | 0.885 | 0.449 | – | – | – | – |
| Hom for
Wt allele | 1 | 1 | 2 | 2 | – | – | – | – | – | – |
|
Het | 27 | 27 | 18 | 18 | – | – | – | – | – | – |
| Hom for
Mut allele | 72 | 72 | 80 | 80 | – | – | – | – | – | – |
| XPG (rs17655) | 100 | – | 100 | – | 0.078 | 0.036 | 0.034 | 0.159 | 0.415 | 0.41 |
| Hom for
Wt allele | 2 | 2 | 18 | 18 | – | – | – | – | – | – |
|
Het | 51 | 51 | 50 | 50 | – | – | – | – | – | – |
| Hom for
Mut allele | 47 | 47 | 32 | 32 | – | – | – | – | – | – |
In addition, analysis of the effect of different
SNPs combinations was considered in the present study. No
significant association with DR or OS in the patients with CHL was
observed during the combined analysis of XPG with all other studied
SNP polymorphisms, with the exception of the combination of XPG
(rs17655)/XRCC1 (rs1799782), for which a significant association
with the OS of CHL was identified (P=0.026).
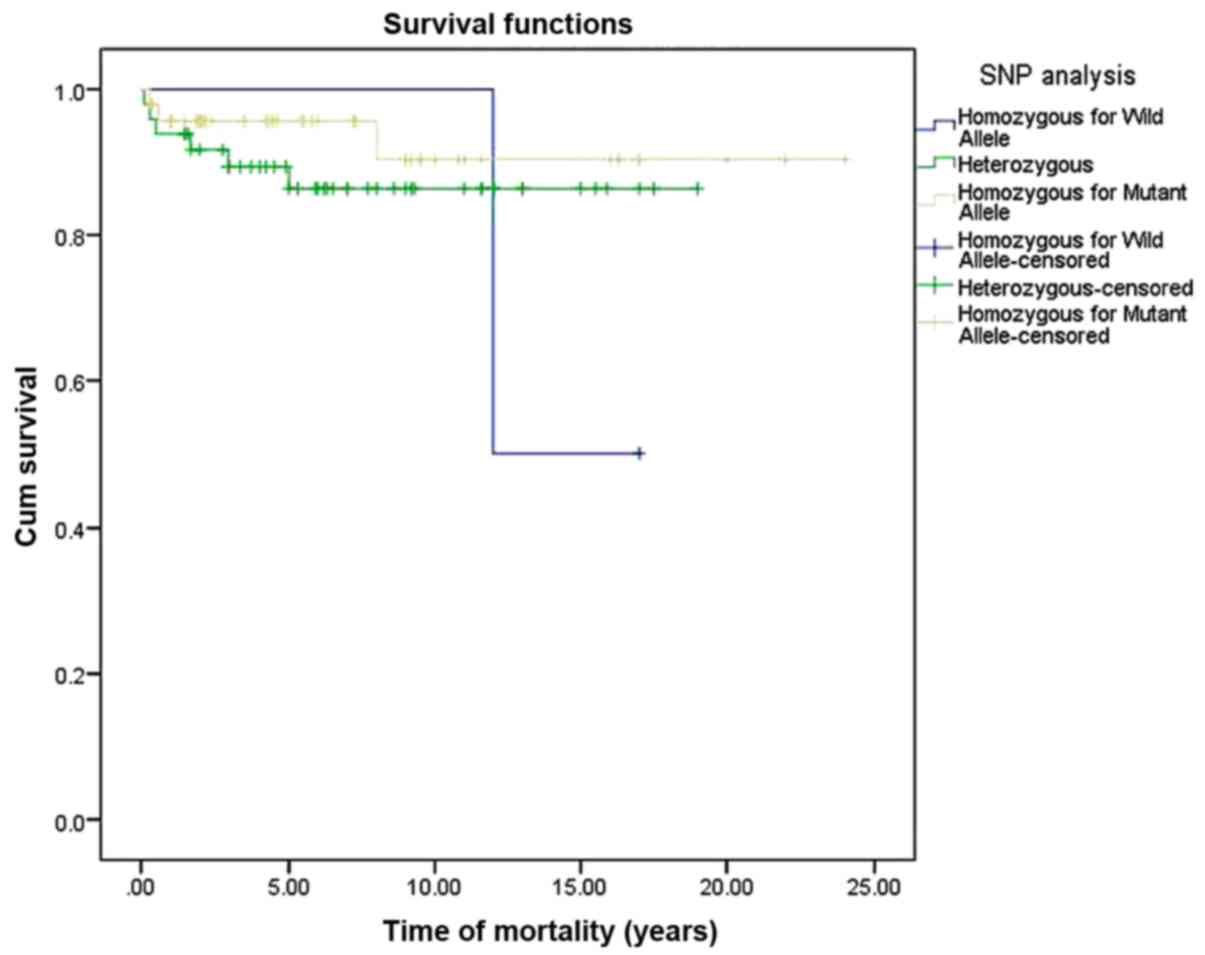
Survival curve estimation based on XPG
SNP analysis
The survival curve was estimated for the XPG SNP
using the Kaplan-Meier analysis method. Accordingly, an observable
decrease in the OS function of the patients with CHL, with
statistical significant log-rank test (P=0.0361), considerably with
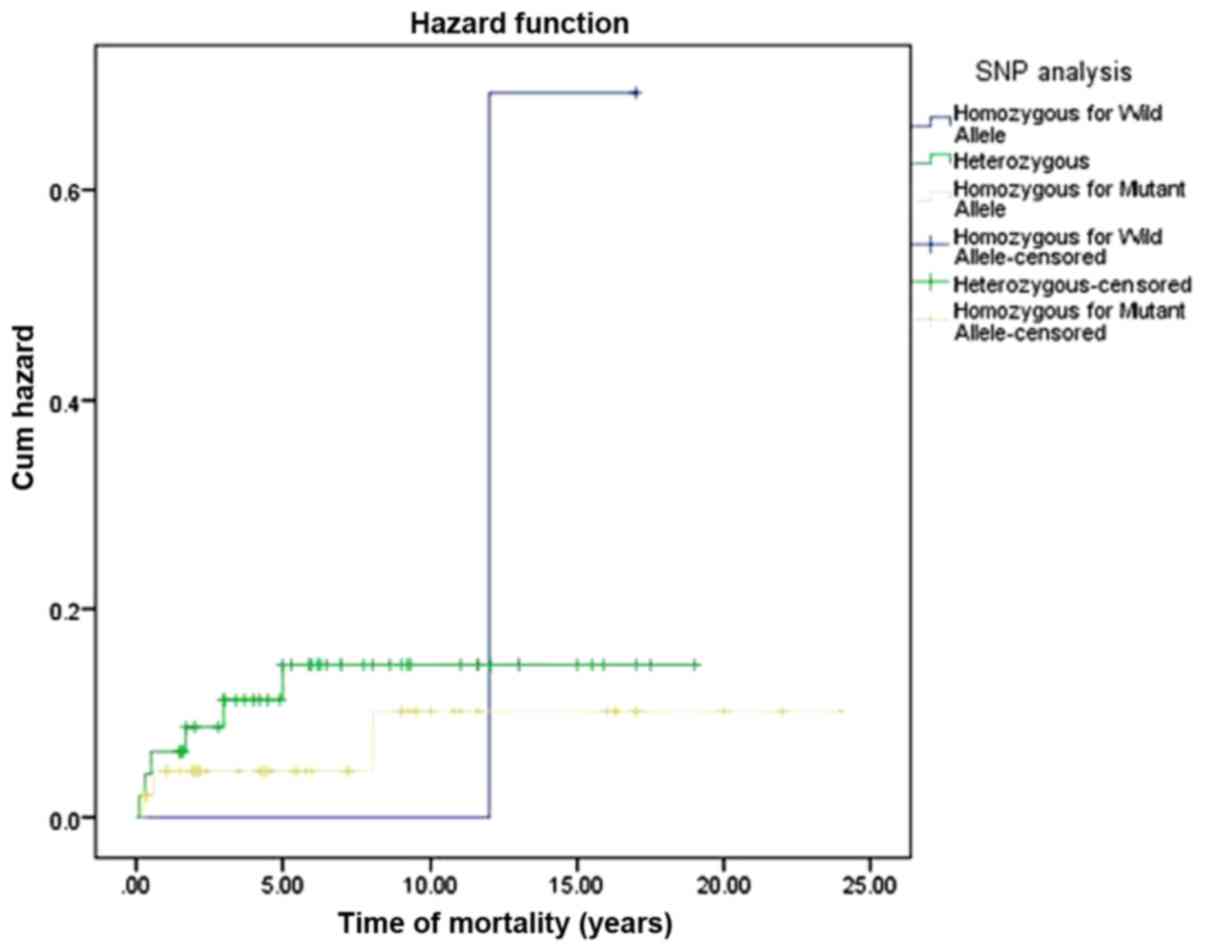
presence of homozygosity for wild allele (Fig. 1). Consequently, the hazard function
analysis indicated an increased cumulative hazard rate with the
different genotyping pattern of XPG SNP in association with CHL OS
(Fig. 2). However, the estimated mean
OS in patients with CHL with XPG SNPs was ~21 years (95% CI,
19–23).
Discussion
The results from the present study suggested that
common DNA polymorphisms of DNA repair genes and their association
with HL disease progression and patient survival may be used as
predictive biomarkers. Furthermore, they may assist in the
identification of a curative therapy for CHL, or to improve the
current protocols. The present study highlighted the requirement
for establishing a molecular profile for CHL in a mixed population,
for example that of Saudi Arabia.
The primary aim of the present study was to identify
a SNP-based molecular profile, using DNA repair genes mutations,
which may assist in evaluating prognostic statuses and be used as
prognostic biomarker in CHL. The presence of selected DNA repair
gene polymorphisms in CHL was demonstrated to have an association
with the risk of the disease. In addition, the interaction between
DNA repair pathways was hypothesized to contribute to the HL
clinical outcome. Although the prognostic significance of the DNA
repair genes remains unclear, a number of retrospective reviews
suggested that genetic polymorphisms of certain DNA repair genes
may have a role in modifying the risk of HL (4,11). In the
present study 5 DNA repair genes were selected, which were
confirmed to be associated with HL risk, and the association
between these genes SNPs and CHL prognosis was examined in the
study population.
Analysis of the allele frequency distribution of the
repair gene SNPs in CHL tissues and normal controls indicated that
there was a significant difference in allele frequencies of the XPG
repair gene polymorphism (rs17655) between patients with CHL and
the healthy controls (P=0.002), besides its low OR (OR=0.50). This
OR estimates that the relative risk of developing HL in the patient
group with XPG nucleotide C SNP is 50%, compared with in those
patients with the G nucleotide. In addition, in comparison with
other DNA repair genes SNPs, XPG may be a useful predictive
indicator for the risk of HL.
Using the χ2 test, no statistical
significance between any of the DNA repair gene genotyping
expression levels and the DR was observed: XRCC1 rs25487 (P=0.696);
XRCC1 rs1799782 (P=0.320); XRCC3 (P=0.207); XPC rs2228001
(P=0.572); XPC rs2228000 (P=0.885); and XPG (P=0.078). Analysis of
the association between the expression levels of these DNA repair
genes with the OS rate demonstrated that only the XPG SNP (rs17655)
was significantly associated with OS (P=0.036). Regarding the
results of the allele frequency analysis of the XPG gene, a
significant difference in expression levels of the XPG gene among
the patients with CHL compared with the healthy controls was
observed. In addition, the association between the XPG SNP and CHL
survival was demonstrated to be independent of age, yet it was
identified to be modified by the sex of the patients with CHL. A
combined analysis of XPG/XRCC1 SNPs exhibited a significant
association with CHL OS in the Saudi patient population. In light
of the statistically significant association between XPG SNPs and
CHL patient survival, the data from the present study suggest a
potential use of XPG expression analysis as a single predictive
molecular biomarker for CHL clinical prognosis. To the best of our
knowledge, the present study is the first to describe an
association between molecular genetic patterns and HL in patients
from Saudi Arabia.
In conclusion, it was demonstrated that the XPG
repair gene (rs17655) SNP had a detrimental effect on CHL OS.
Therefore, the XPG repair gene may be a useful predictive genetic
biomarker for CHL clinical outcome in Saudi patients. These data
may provide a novel avenue for the improvement of the clinical
management of patients with CHL, through molecular analyses for DNA
repair gene expression, which may lead to improving therapeutic
protocols. However, a well-designed multi-center prospective study
with a larger number of subjects and controls is recommended.
Acknowledgements
The authors would like to thank colleagues at the
Arabian Gulf University and at Johns Hopkins Aramco Healthcare who
provided technical assistance and efficient support. In addition,
special thanks are given to Dr Ahmed Alsagheir and Dr Adel Alkhatti
from the Oncology Center at Johns Hopkins Aramco Healthcare.
Funding
The present study was supported by Arabian Gulf
University, College of Medicine and Medical Sciences, Kingdom of
Bahrain (grant no. 36-PI-01/15).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors contributions
HASA and WFR designed the experiments. HASA wrote
the manuscript, performed the experiments, validation and analysis.
WFR, AHSD and MDF reviewed and revised the manuscript and AHSD
acquired funding. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The appropriate research ethical approval for the
research and informed consent was obtained all patients providing
tissues and blood samples.
Patient consent for publication
Written informed consent was obtained from the
patients, guardians or next of kin for deceased patients, as
appropriate, for the publication of any associated data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rauf MS, Akhtar S and Maghfoor I: Changing
trends of adult lymphoma in the Kingdom of Saudi Arabia -
comparison of data sources. Asian Pac J Cancer Prev. 16:2069–2072.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leukemia and Lymphoma Society: American
Cancer Society Cancer Facts & Figures 2016; GLOBOCAN 2012.
simpleClinicalTrials.govCRI grantee progress
reports and other CRI grantee documents. 2016
|
|
3
|
Kron KJ, Bailey SD and Lupien M: Enhancer
alterations in cancer: A source for a cell identity crisis. Genome
Med. 6:772014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Monroy CM, Cortes AC, Lopez M, Rourke E,
Etzel CJ, Younes A, Strom SS and El-Zein R: Hodgkin lymphoma risk:
Role of genetic polymorphisms and gene-gene interactions in DNA
repair pathways. Mol Carcinog. 50:825–834. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Al-Madouj AN, Eldali A and Al-Zahrani AS:
Ten Year Cancer Incidence among nationals of the GCC states
1998-2007. Gulf Center for Cancer Control and Prevention, King
Faisal Specialist Hospital and Research Center. Riyadh. 2011.
|
|
6
|
Fitzmaurice C, Allen C, Barber RM,
Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O,
Dandona R, Dandona L, et al: Global, regional, and national cancer
incidence, mortality, years of life lost, years lived with
disability, and disability-adjusted life-years for 32 cancer
groups, 1990 to 2015: A Systematic Analysis for the Global Burden
of Disease Study. JAMA Oncol. 3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Diab AI, Siddiqui N, Sogiawalla FF and
Fawzy EM: The changing trends of adult Hodgkin's disease in Saudi
Arabia. Saudi Med J. 24:617–622. 2003.PubMed/NCBI
|
|
8
|
Maggioncalda A, Malik N, Shenoy P, Smith
M, Sinha R and Flowers CR: Clinical, molecular, and environmental
risk factors for hodgkin lymphoma. Adv Hematol. 2011:7362612011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scott DW and Steidl C: The classical
Hodgkin lymphoma tumor microenvironment: Macrophages and gene
expression-based modeling. Hematology Am Soc Hematol Educ Program.
2014:144–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sud A, Hemminki K and Houlston RS:
Candidate gene association studies and risk of Hodgkin lymphoma: A
systematic review and meta-analysis. Hematol Oncol. 35:34–50. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El-Zein R, Monroy CM, Etzel CJ, Cortes AC,
Xing Y, Collier AL and Strom SS: Genetic polymorphisms in DNA
repair genes as modulators of Hodgkin disease risk. Cancer.
115:1651–1659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piccaluga PP, Agostinelli C, Gazzola A,
Tripodo C, Bacci F, Sabattini E, Sista MT, Mannu C, Sapienza MR,
Rossi M, et al: Pathobiology of Hodgkin lymphoma. Adv Hematol.
2011:9208982011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smith LB: Nodular lymphocyte predominant
Hodgkin lymphoma: Diagnostic pearls and pitfalls. Arch Pathol Lab
Med. 134:1434–1439. 2010.PubMed/NCBI
|
|
14
|
Carbone PP, Kaplan HS, Musshoff K,
Smithers DW and Tubiana M: Report of the Committee on Hodgkin's
disease staging classification. Cancer Res. 31:1860–1861.
1971.PubMed/NCBI
|
|
15
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction, and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005. View Article : Google Scholar : PubMed/NCBI
|