Introduction
Hepatitis A virus (HAV) infection, which is a major
cause of acute hepatitis, poses an important public health problem
worldwide (1-3). The
virus can spread through the faecal-oral route, e.g. ingestion of
contaminated food and water or direct contact with an infected
person (4,5). The manifestation can be asymptomatic or
symptomatic, ranging between mild and fulminant hepatitis, which is
rare (6,7). Lack of safe water, as well as poor
sanitation and hygiene are risk factors for HAV infection.
Epidemics can be prolonged and cause substantial economic loss
(7-10).
HAV belongs to the family Picornaviridae and the
genus Hepatovirus. HAV contains a 7.5 kb genome encoded by a
positive-sense, single-stranded RNA. HAV has six genotypes (I-VI);
genotypes I-III are infectious to humans (7,11). The
nucleotide variation between isolates of different genotypes is
~15%, and variation between subgenotypes ranges between 7 and 7.5%
in the VP1 capsid protein-P2A protease junction (7,12-15).
Previous studies have demonstrated that the HAV genotype present in
Indonesia is IA (16-18).
Outbreaks of hepatitis A in Indonesia have been
consecutively reported in the following cities: Bogor (West Java)
in 1998, Jember and Bondowoso (East Java) in 2006, Tangerang (West
Java) in 2007, Yogyakarta (Special Region of Yogyakarta) in 2008,
Ngawi (East Java) in 2009, Lamongan and Bangkalan (East Java) in
2018 and Pacitan (East Java) in 2019 according to the
Sub-directorate of Surveillance and Outbreak Response, Directorate
General of Disease Control and Environmental Health, Ministry of
Health (personal communication) (17).
The present study focused on the recent hepatitis A
outbreaks at a senior high school in Lamongan and at a boarding
school in Bangkalan in East Java in 2018. Habits, attitude and
knowledge level may serve a role in the incidence of hepatitis A in
affected regions. Indonesia is a country with high endemicity of
hepatitis A, but genetic information on HAV is still limited. The
aim of the present study was to obtain molecular epidemiological
data on HAV-caused outbreaks in the two affected areas. The
knowledge and incidence of hepatitis A infection were also
analysed.
Materials and methods
Study population
This study was an observational and cross-sectional
study. Serum samples were obtained from 88 individuals with
clinical manifestations of acute hepatitis in Lamongan (n=54) in
January 2018 and Bangkalan (n=34) in March 2018, with a mean age of
16 years (range, 15-55 years). The inclusion criterion of case
group was a clinical manifestation of hepatitis, such as fever,
sweating, headache, malaise, flatulence, nausea, vomiting, lack of
appetite, heartburn, jaundice and dark-coloured urine. The subjects
did not receive any drug treatments that may have affected the
results of the study. No antiviral treatments were administered.
The outbreak investigation was started one day after the outbreak
was reported by the Public Health Offices in Lamongan and
Bangkalan. A senior high school in Lamongan, termed ‘affected area
I’ in this study, is a half-day school, and the students return
home every day after school time. ‘Affected area II’ is a full-day
boarding school in Bangkalan, where students live in a dormitory in
the school area.
HAV serological test
The serum samples were screened for IgM anti-HAV
using a SD BIOLINE HAV IgG/IgM rapid test (Standard Diagnostics,
Inc.) according to the manufacturer's instructions.
RNA extraction and reverse
transcription (RT)-PCR amplification
Viral RNA was extracted from 140 µl serum using a
QIAamp viral RNA mini kit (Qiagen GmbH) according to the
manufacturer's instructions. The purified RNA samples were used to
generate cDNA using ReverTra Ace® (Toyobo Co., Ltd.)
according to the manufacturer's instructions. The VP1-P2A and
VP3-VP1 junctions were amplified using RT-PCR. The primers are
presented in Table I (17-19).
The primers for the VP1-P2A region were the basic region (BR)-5 and
BR-9 primers for first-round PCR and the RJ-3 and BR-6 primers for
second-round PCR. The primers for the VP3-VP1 region were the HAV1
and HAV2 primers for the first-round PCR and the HAV3 and HAV4
primers for the second-round PCR. The thermocycling conditions for
the HAV1 and HAV2 primers were as follows: 5 min at 94˚C; 40 cycles
of 30 sec at 94˚C, 30 sec at 57˚C and 45 sec at 72˚C; and 7 min at
72˚C. The thermocycling conditions for HAV3 and HAV4 were as
follows: 5 min at 94˚C; 40 cycles of 30 sec at 94˚C, 30 sec at 55˚C
and 45 sec at 72˚C; and 7 min at 72˚C. The thermocycling conditions
of the first and second rounds of PCR in the VP1-P2A region using
the BR-5 and BR-9 primers and the RJ-3 and BR-6 primers were the
same as those of the second-round PCR using the HAV3 and HAV4
primers. A total of 5 µl PCR product was analysed using 2% agarose
gel electrophoresis and stained with ethidium bromide to visualize
the bands.
 | Table IPrimers used for HAV RNA
amplification. |
Table I
Primers used for HAV RNA
amplification.
| Region | Primers | Sequences
(5'-3') | Nucleotide no. | Product size,
bp |
|---|
| VP1-P2A | BR-5 |
TTGTCTGTCACAGAACAATCAG | 2950-2972 | 361 |
| | BR-9 |
AGTCACACCTCTCCAGGAAAACTT | 3310-3286 | |
| | RJ-3 |
TCCCAGAGCTCCATTGAA | 2984-3002 | 234 |
| | BR-6 |
AGGAGGTGGAAGCACTTCATTTGA | 3217-3193 | |
| VP3-VP1 | HAV1 |
GCTCCTCTTTATCATGCTATGGAT | 2172-2196 | 244 |
| | HAV2 |
CAGGAAATGTCTCAGGTACTTTCT | 2415-2391 | |
| | HAV3 |
ATGTTAACTACACAAGTTGGAGAT | 2195-2218 | 186 |
| | HAV4 |
GATCCTCAATTGTTGTGATAGCT | 2380-2357 | |
Sequence and phylogenetic
analysis
The nucleotide sequences of sample HAVs were
determined using a BigDye Terminator v3.1 Cycle Sequencing kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with an
Applied Biosystems 3500xL Genetic Analyzer (Thermo Fisher
Scientific, Inc.). The results of the sequencing were compared with
data from the international DNA databank (GenBank; https://www.ncbi.nlm.nih.gov/genbank/).
The GenBank accession numbers used for comparison were as follows:
AB020566, KX151445, LC049340, AB020567, AB623053, AF485328,
AB839694, AB839693, AB839697, AB839696, AB918714, M14707, AF314208,
M20273, AY644676, AY644670, DQ991030, AB279732, FJ360732, AJ299464,
JQ655151, AB258387, AB279735, AB425339, M59286, D00924, AF485328,
AY294048, DQ114888, AY343856, DQ114866 and AJ519486.Phylogenetic
trees were constructed by the neighbour-joining method; to confirm
the reliability of phylogenetic tree analysis, bootstrap resampling
and reconstruction were performed 1,000 times using the Molecular
Evolutionary Genetics Analysis (MEGA) X software (https://www.megasoftware.net/).
Association between subject knowledge
of hepatitis A infection and incidence
The pre-designed questionnaire was pre-tested on a
group of students not included in the present study and for
validation. A self-administered structured questionnaire was used
to collect information about the sociodemographic characteristics
(age, sex, occupation) and knowledge of HAV including causes,
symptoms, transmission, target organ, treatment and prevention
(hygiene and sanitation) methods.
Statistical analysis
The samples were divided into two groups, i.e., the
control and case groups. The control group included healthy
students and staff members in affected areas I (n=51) and II
(n=33), whereas the case group included individuals with acute
hepatitis in affected areas I (n=54) and II (n=34). A total of 172
samples were included in the statistical analysis. Descriptive data
are presented as numbers and percentages. Chi square test was used
to analyse the association between knowledge of hepatitis A and the
incidence of infection in affected areas I and II. SPSS Statistics
for Windows version 22.0 (IBM Corp.) was used for statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Subject sex, age, occupation and
symptoms in the two affected areas
The characteristics of patients with hepatitis A
from the two affected areas are summarized in Table II. Among the 88 subjects enrolled
during the outbreaks, more female patients were present in affected
area I, whereas more male patients were present in affected area
II. Age ≤17 years was predominant in both affected areas. The most
prominent symptom was nausea (59%), accompanied by other symptoms.
All participants from the two affected areas were tested for
Anti-HAV IgM, HAV-RNA detection, and their serum samples were
subjected to sequencing. The results revealed that all 18 strains
belonged to the HAV IA subtype. The workflow and results obtained
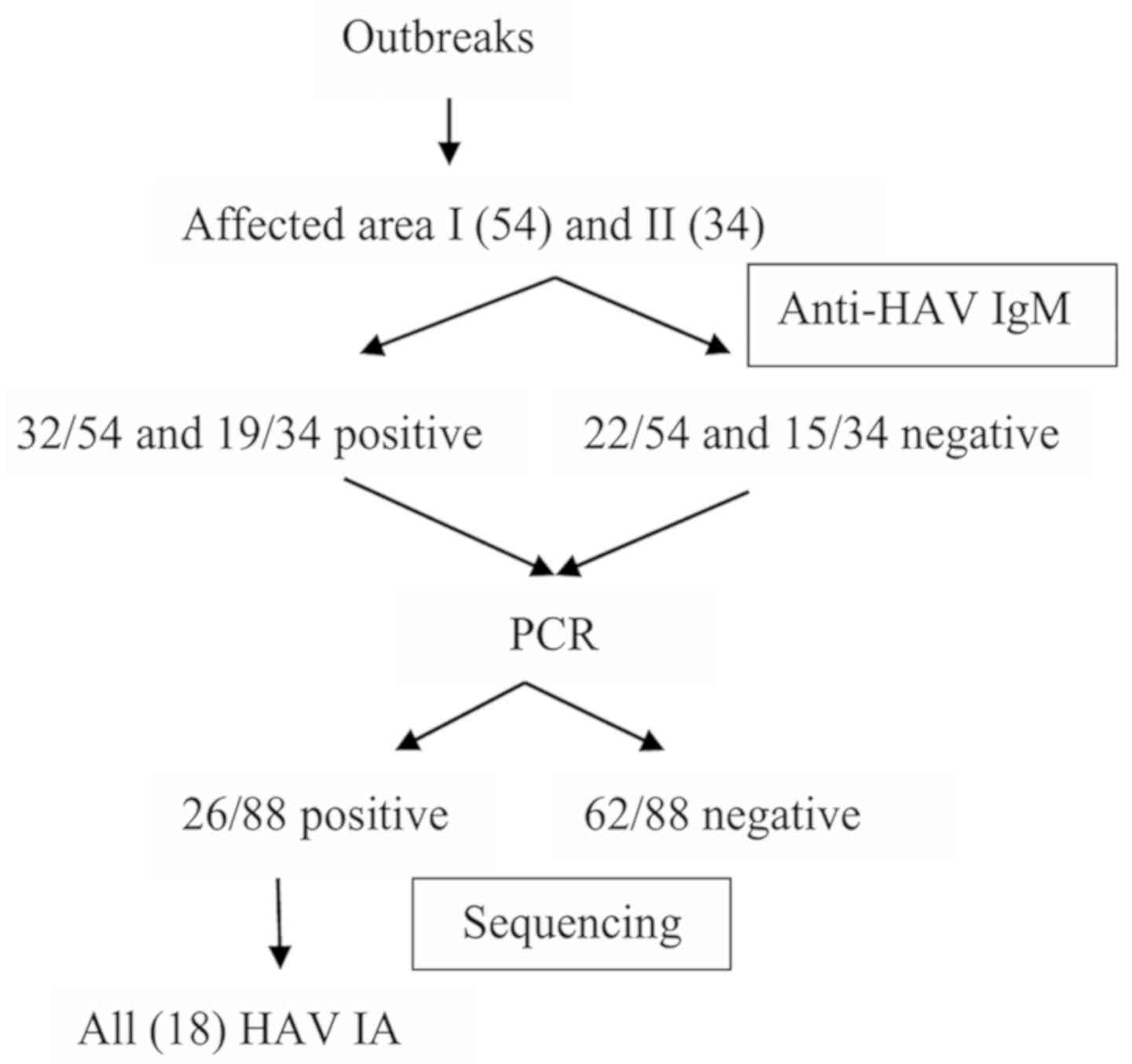
from the genetic analysis are presented in Fig. 1.
 | Table IICharacteristics of patients in the
two affected areas. |
Table II
Characteristics of patients in the
two affected areas.
| | Affected area I
(n=54) | Affected area II
(n=34) |
|---|
|
Characteristics | n | % | n | % |
|---|
| Sex |
|
Female | 35 | 65 | 6 | 18 |
|
Male | 19 | 35 | 28 | 82 |
| Age, years |
|
≤17 | 33 | 61 | 16 | 47 |
|
18-25 | 5 | 9 | 12 | 35 |
|
26-45 | 10 | 19 | 6 | 18 |
|
≥46 | 6 | 11 | 0 | 0 |
| Occupation |
|
Student | 34 | 63 | 27 | 79 |
|
Teacher | 8 | 15 | 3 | 9 |
|
Chef | 12 | 22 | 4 | 12 |
| Symptoms |
|
Fever | 23 | 43 | 18 | 53 |
|
Sweating | 7 | 13 | 14 | 41 |
|
Headache | 21 | 39 | 10 | 29 |
|
Malaise | 13 | 24 | 12 | 35 |
|
Flatulence | 14 | 26 | 13 | 38 |
|
Nausea | 32 | 59 | 16 | 47 |
|
Vomiting | 24 | 44 | 10 | 29 |
|
Lack of
appetite | 28 | 52 | 14 | 41 |
|
Heartburn | 13 | 24 | 19 | 56 |
|
Jaundice | 27 | 50 | 15 | 44 |
|
Dark-coloured
urine | 23 | 43 | 10 | 29 |
Prevalence of anti-HAV IgM
Serum samples with clinically suspected hepatitis A
were tested for the presence of anti-HAV IgM. Among the patients
from affected areas I and II, 32 of 54 (59.25%) and 19 of 34
(55.9%), respectively, were positive for anti-HAV IgM.
HAV RNA analysis
The VP3-VP1 and VP1-P2A junction regions were
amplified from all anti-HAV IgM-positive and negative serum
samples. The HAV RNA analysis of the VP3-VP1 junction was positive
in 8 of 54 and 6 of 34 patients from affected areas I and II,
respectively. The analysis of the VP1-P2A junction was positive in
16 of 54 patients from affected area I and in 2 of 34 patients from
affected area II. A total of 26 patients were HAV RNA-positive in
either the VP3-VP1 or VP1-P2A junction.
Sequencing and phylogenetic
analyses
VP1-P2A region
A total of seven nucleotide sequences of HAV strains
obtained in the present study, of which six were from affected area
I and one was from affected area II, were 99-100% identical to each
other in the VP1-P2A region and were closely related to the HAV
subgenotype IA. Samples from affected area I exhibited 100%
homology with a strain with the accession number AB918714
(Surabaya-Indonesia) (Fig. 2).
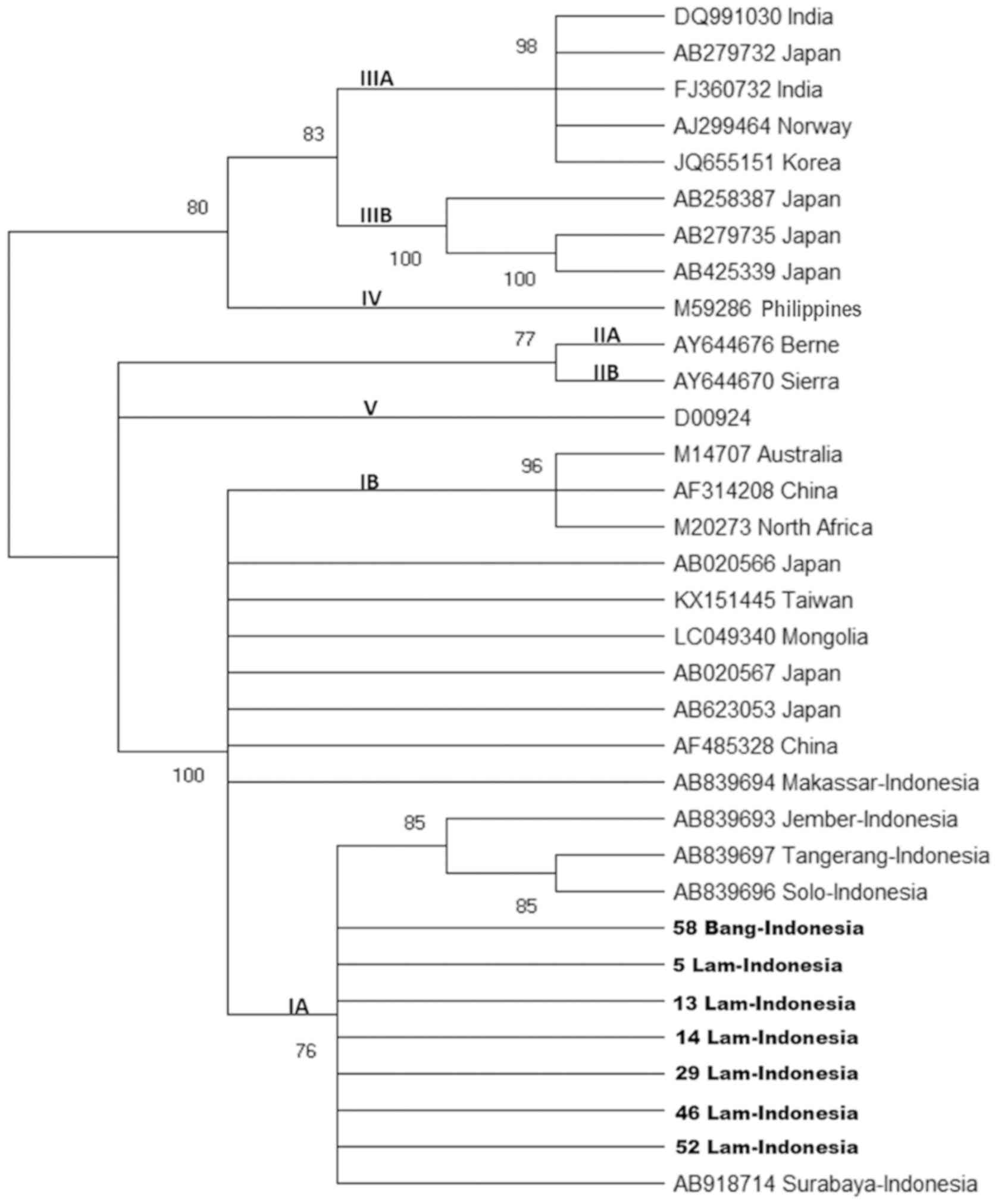 | Figure 2.Phylogenetic tree generated using the
nucleotide sequences obtained from the VP1-P2A junction region. HAV
strains isolated from Lam (affected area I) and Bang (affected area
II) (number of samples presented) and 26 reported HAV isolates of
genotypes/subgenotypes IA, IB, IIA, IIB, IIIA, IIIB, IV and V with
complete or nearly complete sequences are included for comparison.
Numbers in the tree indicate bootstrap reliability. The length of
each horizontal bar indicates the number of nucleotide
substitutions per site. Isolates from the Genbank database are
indicated by the accession number, and relevant (town and) country
names have been listed for each HAV strain. HAV, hepatitis A virus;
Lam, Lamongan; Bang, Bangkalan. |
VP3-VP1 region
A total of 11 nucleotide sequences of HAV strains
obtained in the present study, including seven samples from
affected area I and four samples from affected area II, were 99%
identical to each other in the VP3-VP1 junction region sequence and
were closely related to HAV strains of subgenotype IA (Fig. 3).
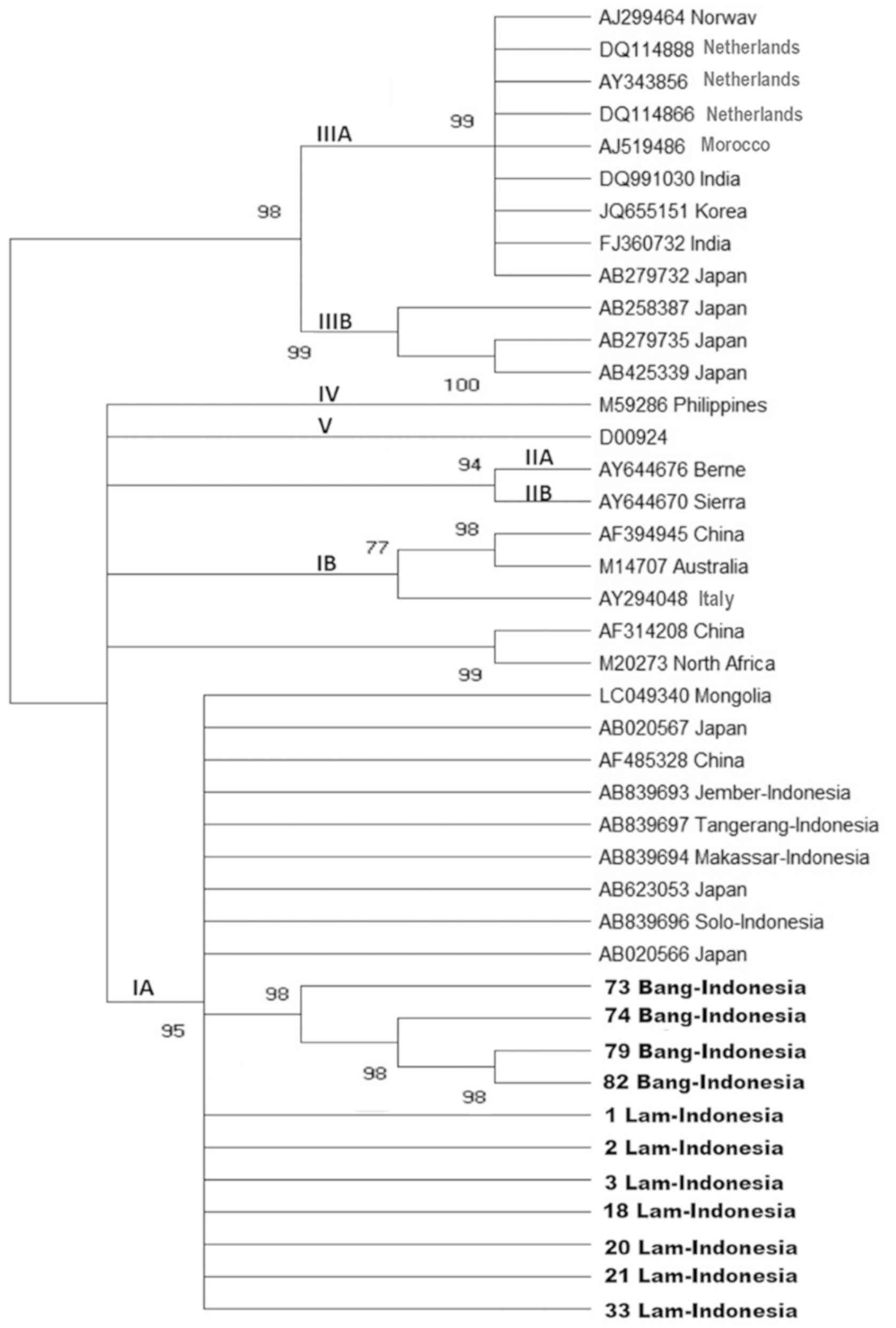 | Figure 3.Phylogenetic tree generated using the
nucleotide sequences obtained from the VP3-VP1 junction region. HAV
strains isolated from Lam (affected area I) and Bang (affected area
II) (number of samples presented) and 30 reported HAV isolates of
genotypes/subgenotypes IA, IB, IIA, IIB, IIIA, IIIB, IV and V with
complete or nearly complete sequences are included for comparison.
Numbers in the tree indicate bootstrap reliability. The length of
each horizontal bar indicates the number of nucleotide
substitutions per site. Isolates from the Genbank database are
indicated by their accession number, and relevant (town and)
country names have been listed for each HAV strain. HAV, hepatitis
A virus; Lam, Lamongan; Bang, Bangkalan. |
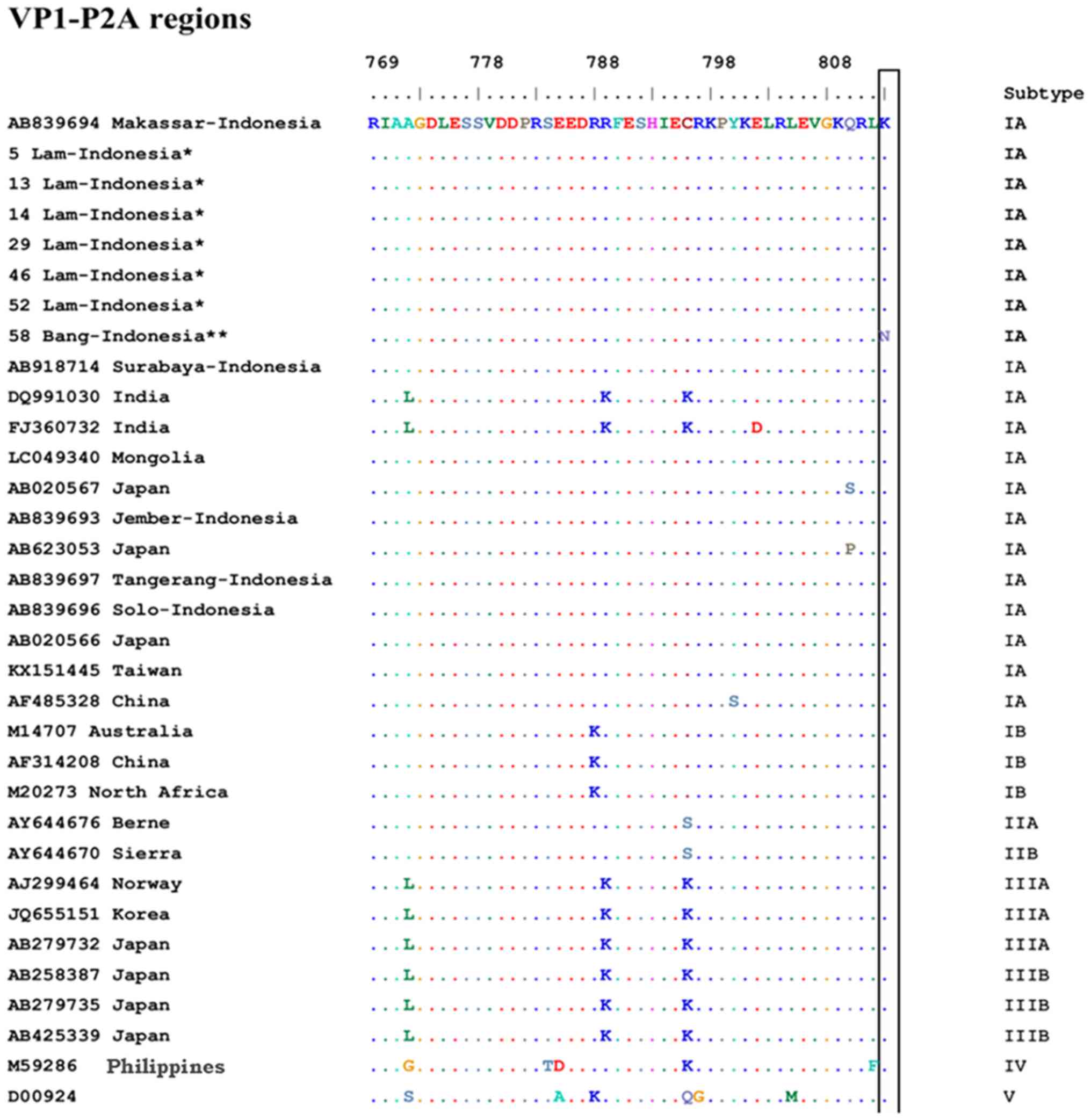
Alignment of amino acid sequences
The predicted amino acid sequences of the VP1-P2A
and VP3-VP1 regions from the samples collected in the present study
were compared with those from previously reported strains (Figs. 4 and 5).
Representative HAV isolates of all genotypes and subgenotypes are
described in Figs. 4 and 5. Although all affected area I and II
strains belonged to subgenotype IA, the samples from affected area
II had amino acids that were not present in any other strain from
area I, i.e., K813N in the VP1-P2A junction and M519T in the
VP3-VP1 junction.
Association between knowledge level of
subjects and incidence of hepatitis A infection
The knowledge level was divided into three
categories; the category ‘high’ included responses with >75%
correct answers, ‘moderate’ included responses with 50 to 74%
correct answers and ‘low’ included responses with <50% correct
answers. The distribution of knowledge of the subjects was mainly
high (84%) in the control group and moderate (41%) in the case
group in affected area I. In affected area II, the knowledge was
high in 55% of the control group and 56% in the case group. The
distribution of knowledge of the subjects in the present study is
presented in Table III. The
association between knowledge level and the incidence of hepatitis
A infection is presented in Table
IV. Significant differences were observed in the level of
knowledge and the incidence of Hepatitis A in affected area I
(P=0.001), whereas the knowledge of students in affected area II
was not associated with incidence.
 | Table IIIKnowledge of hepatitis A in infected
(case) and control groups. |
Table III
Knowledge of hepatitis A in infected
(case) and control groups.
| | Affected area
I | Affected area
II |
|---|
| | Control (n=51) | Case (n=54) | Control (n=33) | Case (n=34) |
|---|
| Knowledge
level | n | % | n | % | n | % | n | % |
|---|
| High | 43 | 84 | 21 | 39 | 18 | 55 | 19 | 56 |
| Moderate | 7 | 14 | 22 | 41 | 14 | 42 | 12 | 35 |
| Low | 1 | 2 | 11 | 20 | 1 | 3 | 3 | 9 |
 | Table IVAssociation between knowledge level
and incidence of hepatitis |
Table IV
Association between knowledge level
and incidence of hepatitis
| | | OR | | |
|---|
| Knowledge
level | P-value | Low-high | Low-moderate | Association between
variables (incidence of hepatitis A infection) | α (CI=95%) |
|---|
| Affected area
I | 0.001a | 22.523 | 3.5 | 42.8% | 0.05 |
| Affected area
II | 0.558 | - | - | 13.1% | |
Discussion
The present study determined and analysed the
genomic sequence of HAV isolates from the Lamongan and Bangkalan
hepatitis A outbreak areas in 2018. The major advantage of this
study was the acquired genetic information of HAV from the latest
outbreaks in two affected areas. The subtype of HAV in the two
studied affected areas was IA, similar to those previously
identified (16-18),
although the viruses did not originate from the same strain.
Lamongan and Bangkalan are cities ~109 km apart on two different
islands. Of the 88 patients suspected to have hepatitis A from the
two affected areas, 51 were positive for anti-HAV IgM and 26
exhibited positive PCR results in the VP3-VP1 and/or VP1-P2A
region. Anti-HAV IgM is a routine laboratory diagnostic test for
hepatitis A (20). Anti-HAV IgM is
detectable at or prior to the onset of clinical symptoms, and the
levels decline in 3 to 6 months (11). Of note, acute hepatitis A may also
occur without the production of detectable IgM antibodies (21). In the present study, cases of acute
HAV infection were be diagnosed by detection of HAV RNA; this
molecular marker is detectable ~14 days prior to the appearance of
the acute-phase serological markers and remains persistently
detectable for an average of 79 days following symptom onset and
peak hepatic enzyme levels (7,20-22).
The establishment of early laboratory diagnosis of HAV infection is
important for the guidance and implementation of measures for the
prevention and control of outbreaks. Rapid tests have been widely
used as screening tools in developing countries (5,23).
Viral genotypic profiles are required to identify
foodborne outbreaks, implement preventative measures and recognize
transmission routes (3,24-26).
The VP1 region of HAV is an area that contains variable amino
acids, which is why this region was used as one of the areas for
molecular detection in the present study (19,27).
Amplification and sequencing of variable regions within the capsid
proteins, including the VP3/VP1 and VP1/P2A junctions of wild-type
HAV isolates from different regions of the world, revealed
significant nucleotide sequence heterogeneity, but limited amino
acid heterogeneity (28). These
junctions have been used in the analyses of a number of sequences,
especially for comparing sequences of isolates obtained from
several countries (17,18,28-31).
The results of the present study demonstrated that the HAV genotype
of all strains in affected areas I and II belonged to subgenotype
IA, although the causative strain of HAV in affected area I was
different from that identified in affected area II. This result was
similar to that of our previous study and other studies from other
regions in Indonesia, which identified clustering with genotype IA
strains (16-18).
Worldwide, genotype I is the most prevalent, with
subtype IA more common compared with IB. As subgenotype IA is
prevalent, genotyping/subgenotyping alone can rarely be utilized to
identify the source of an HAV outbreak or the chain of transmission
(1,11). The HAV isolates that have been
identified thus far display a high degree of genetic conservation,
and modest genetic heterogeneity exists in several genomic regions,
with the exception of the 5' untranslated region, which has
demonstrated high levels of conservation, supporting the use of
RT-PCR for the sensitive detection of HAV RNA (1,3,11,32,33).
Amino acid sequences of VP3-VP1 were compared with
diverse subgenotype strains reported in various countries, and all
samples from affected area II were identified to contain a unique
amino acid, M519T, compared with those from affected area I and
other reported IA strains from the DNA Data Bank. The samples from
affected area II also had a unique amino acid, K813N, in VP1-P2A,
whereas samples from affected area I did not exhibit K813
mutations, similar to previous reports from Indonesia and other
countries (27,34,35). Thus,
although these epidemics in the two areas occurred at a similar
time and the causative epidemic agents were HAV-IA, they were
different strains of HAV.
The results of the questionnaire on hepatitis A
infection in relation to hygiene in affected area I revealed that
the control group possessed a high level of knowledge of hepatitis
A, whereas the case group exhibited a moderate level of knowledge.
In affected area II, the control and case groups possessed a high
level of knowledge. No statistically significant differences were
observed in the level of knowledge and the incidence of hepatitis
A. Investigations on the two affected area found that poor hygiene
and sanitation in canteens (no available washbasins) and the close
proximity of septic tanks to wells may have contributed to the
spread of HAV. It may be assumed that in affected area II, these
facilities were used by all occupants; thus, when infection was
present, it could spread quickly. By contrast, in affected area I,
the students had a choice of facilities and did not live together
in a dormitory, which may have reduced the risk of transmission of
hepatitis A infection. Therefore, the level of knowledge in
affected area II did not affect behaviours to avoid hepatitis A
infection. This result is similar to that of another study
(36), which indicated that although
public awareness was high, practical knowledge regarding
differences in the mode of transmission, consequences and
prevention was low in highly endemic countries, especially among
those with a lower level of education. Additionally, no differences
were observed in the prevention of hepatitis in an intervention
group (37). Age, sex and geographic
location are not associated with the level of knowledge and
practice to avoid hepatitis A infection (37,38).
The limitation of the present study was that
clinical symptoms rather than laboratory tests were used as
inclusion criteria for the control groups; further studies with
larger samples are needed to acquire more information about HAV in
the affected areas.
Acknowledgements
Not applicable.
Funding
This study was funded by the Progam Pendidikan
Magister Menuju Doktor Untuk Sarjana Unggul (PMDSU) Batch III of
the Ministry of Research, Technology and Higher Education of the
Republic of Indonesia (grant no. 134/UN3.14/LT/2018), in part by
The Japan Initiative for Global Research Network on Infectious
Diseases (J-GRID) programme from the Ministry of Education,
Culture, Sports, Science, and Technology (MEXT), Japan, and in part
by a Grant in Aid from Dato' Sri Professor Dr Tahir through the
Tahir Professorship Program, Indonesia.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
This study was conducted and designed by MIL, JJ,
TU, SS, and DS. TM, YAM, DP and DS performed sample collection. DS,
YAM, DP and MA performed the laboratory experiments. DS, JJ, MIL,
TU, SS analysed data and wrote the manuscripts. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the Faculty of Medicine, Airlangga University
(Surabaya, Indonesia). The ethical approval number is
158/EC/KEPK/FKUA/2018. All patients provided written informed
consent. Informed consent for participations <18 years in this
study was obtained from the parents of each individual.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vaughan G, Goncalves Rossi LM, Forbi JC,
de Paula VS, Purdy MA, Xia G and Khudyakov YE: Hepatitis A virus:
Host interactions, molecular epidemiology and evolution. Infect
Genet Evol. 21:227–243. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shin E, Kim JS, Oh KH, Oh SS, Kwon M, Kim
S, Park J, Kwak HS, Chung GT, Kim CJ, et al: A waterborne outbreak
involving hepatitis A virus genotype IA at a residential facility
in the Republic of Korea in 2015. J Clin Virol. 94:63–66.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Quiñones B, Lee BG, Martinsky TJ, Yambao
JC, Haje PK and Schena M: Sensitive genotyping of
foodborne-associated human noroviruses and hepatitis A virus using
an array-based platform. Sensors (Basel). 17:1–16. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lin KY, Chen GJ, Lee YL, Huang YC, Cheng
A, Sun HY, Chang SY, Liu CE and Hung CC: Hepatitis A virus
infection and hepatitis A vaccination in human immunodeficiency
virus-positive patients: A review. World J Gastroenterol.
23:3589–3606. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
de Almeida Ribeiro CR, Amado LA, Tourinho
RS, Pinto Lima LR, Melgaço JG, de Almeida AJ, Bastos LS,
Lewis-Ximenez LL and de Paula VS: Accuracy of rapid test for
diagnosis of hepatitis A with different infection rate settings and
with predictive modeling. Future Microbiol. 14:247–258.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Walker BW: Hepatitis A infection: On alert
for outbreaks. Nursing. 48:66–69. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lemon SM, Ott JJ, Van Damme P and Shouval
D: Type A viral hepatitis: A summary and update on the molecular
virology, epidemiology, pathogenesis and prevention. J Hepatol.
68:167–184. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
World Health Organization (WHO): Hepatitis
A. https://www.who.int/news-room/fact-sheets/detail/hepatitis-a,
2019.
|
|
9
|
Ouardani I, Turki S, Aouni M and Romalde
JL: Detection and molecular characterization of hepatitis A virus
from Tunisian wastewater treatment plants with different secondary
treatments. Appl Environ Microbiol. 82:3834–3845. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Walczak-Galezewska MK, Skrypnik D,
Szulinska M, Skrypnik K and Bogdanski P: Conservative management of
acute calculous cholecystitis complicated by pancreatitis in an
elderly woman: A case report. Medicine (Baltimore).
97(e11200)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nainan OV, Xia G, Vaughan G and Margolis
HS: Diagnosis of hepatitis A virus infection: A molecular approach.
Clin Microbiol Rev. 19:63–79. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ishii K, Kiyohara T, Yoshizaki S, Kawabata
K, Kanayama A, Yahata Y, Takahashi T, Kinoshita H, Saitou T,
Sunagawa T, et al: Epidemiological and genetic analysis of a 2014
outbreak of hepatitis A in Japan. Vaccine. 33:6029–6036.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kanda T, Nakamoto S, Wu S, Nakamura M,
Jiang X, Haga Y, Sasaki R and Yokosuka O: Direct-acting Antivirals
and Host-targeting agents against the hepatitis A virus. J Clin
Transl Hepatol. 3:205–210. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
McKnight KL and Lemon SM: Hepatitis A
virus genome organization and replication strategy. Cold Spring
Harb Perspect Med. 8:1–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Blanco Fernández MD, Torres C,
Riviello-López G, Poma HR, Rajal VB, Nates S, Cisterna DM, Campos
RH and Mbayed VA: Analysis of the circulation of hepatitis A virus
in Argentina since vaccine introduction. Clin Microbiol Infect.
18:E548–E551. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mulyanto Wibawa ID, Suparyatmo JB,
Amirudin R, Ohnishi H, Takahashi M, Nishizawa T and Okamoto H: The
complete genomes of subgenotype IA hepatitis A virus strains from
four different islands in Indonesia form a phylogenetic cluster.
Arch Virol. 159:935–945. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Juniastuti Dedy WN, Mochamad A, Yamani LN,
Utsumi T, Sustini F, et al: Analysis of genetic and serology of
hepatitis A virus infection during and after outbreak in two junior
high schools in Surabaya, Indonesia. J Med Virol. 91:1048–1055.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Utsumi T, Yano Y, Amin M, Lusida MI,
Soetjipto Hotta H and Hayashi Y: Acute hepatitis due to hepatitis A
virus subgenotype IA as an imported infectious disease from
Indonesia. Kobe J Med Sci. 60:E43–E47. 2014.PubMed/NCBI
|
|
19
|
Yun H, Kim S, Lee H, Byun KS, Kwon SY, Yim
HJ, Lim YS, Jeong SH and Jee Y: Genetic analysis of HAV strains
isolated from patients with acute hepatitis in Korea, 2005-2006. J
Med Virol. 80:777–784. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schwarz NG, Revillion M, Dussaix E, Giraud
M, Liberpre C, Couturier E, et al: Surveillance and outbreak
reports food-borne outbreak of hepatitis A virus (HAV). Up
Normandy. 13:1–5. 2008.
|
|
21
|
Pondé RAA: The serological markers of
acute infection with hepatitis A, B, C, D, E and G viruses
revisited. Arch Virol. 162:3587–3602. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Aggarwal R and Goel A: Hepatitis A:
Epidemiology in resource-poor countries. Curr Opin Infect Dis.
28:488–496. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cha YJ, Park Q, Kang ES, Yoo BC, Park KU,
Kim JW, Hwang YS and Kim MH: Performance evaluation of the OraQuick
hepatitis C virus rapid antibody test. Ann Lab Med. 33:184–189.
2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vinjé J: Advances in laboratory methods
for detection and typing of norovirus. J Clin Microbiol.
53:373–381. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Verhoef L, Vennema H, van Pelt W, Lees D,
Boshuizen H, Henshilwood K and Koopmans M: Food-Borne Viruses in
Europe Network: Use of norovirus genotype profiles to differentiate
origins of foodborne outbreaks. Emerg Infect Dis. 16:617–624.
2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Verhoef L, Kouyos RD, Vennema H, Kroneman
A, Siebenga J, van Pelt W and Koopmans M: Foodborne Viruses in
Europe Network: An integrated approach to identifying international
foodborne norovirus outbreaks. Emerg Infect Dis. 17:412–418.
2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bruni R, Taffon S, Equestre M, Chionne P,
Madonna E, Rizzo C, Tosti ME, Alfonsi V, Ricotta L, De Medici D, et
al: Italian National Task Force on Hepatitis A: Key role of
sequencing to trace hepatitis a viruses circulating in Italy during
a large multi-country European foodborne outbreak in 2013. PLoS
One. 11(e0149642)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lee H, Jeong H, Yun H, Kim K, Kim JH, Yang
JM and Cheon DS: Genetic analysis of hepatitis A virus strains that
induced epidemics in Korea during 2007-2009. J Clin Microbiol.
50:1252–1257. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cristina J and Costa-Mattioli M: Genetic
variability and molecular evolution of hepatitis A virus. Virus
Res. 127:151–157. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
de Paula VS, Lu L, Niel C, Gaspar AMC and
Robertson BH: Genetic analysis of hepatitis A virus isolates from
Brazil. J Med Virol. 73:378–383. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chironna M, Grottola A, Lanave C, Villa E,
Barbuti S and Quarto M: Genetic analysis of HAV strains recovered
from patients with acute hepatitis from Southern Italy. J Med
Virol. 70:343–349. 2003.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bruni R, Taffon S, Equestre M, Cella E, Lo
Presti A, Costantino A, Chionne P, Madonna E, Golkocheva-Markova E,
Bankova D, et al: Hepatitis a virus genotypes and strains from an
endemic area of Europe, Bulgaria 2012-2014. BMC Infect Dis.
17(497)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu GD, Hu NZ and Hu YZ: Full-length
genome of wild-type hepatitis A virus (DL3) isolated in China.
World J Gastroenterol. 9:499–504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hamza H, Abd-Elshafy DN, Fayed SA, Bahgat
MM, El-Esnawy NA and Abdel-Mobdy E: Detection and characterization
of hepatitis A virus circulating in Egypt. Arch Virol.
162:1921–1931. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lee AR, Lee SG, Kang LH, Jheong WH and
Paik SY: Full-length genomic sequence of subgenotype IIIA hepatitis
A virus isolate in Republic of Korea. BioMed Res Int.
2013(426034)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Crutzen R and Göritz AS: Public awareness
and practical knowledge regarding Hepatitis A, B, and C: A
two-country survey. J Infect Public Health. 5:195–198.
2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tenner CT, Herzog K, Chaudhari S, Bini EJ
and Weinshel EH: Knowledge, attitudes and barriers regarding
vaccination against hepatitis A and B in patients with chronic
hepatitis C virus infection: A survey of family medicine and
internal medicine physicians in the United States. Int J Clin
Pract. 66:1009–1013. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Larios SE, Masson CL, Shopshire MS,
Hettema J, Jordan AE, McKnight C, Young C, Khalili M, Seewald RM,
Min A, et al: Education and counseling in the methadone treatment
setting improves knowledge of viral hepatitis. J Subst Abuse Treat.
46:528–531. 2014.PubMed/NCBI View Article : Google Scholar
|