1. Introduction
Staphylococcus aureus (S. aureus) is a
Gram-positive coccus with low-GC content (1). Due to variations in certain genes in
the bacterial genome and the introduction of exogenous genes, this
strain exhibits different degrees of drug resistance against
β-lactams, quinolones and aminoglycosides (1). Since osteomyelitis is caused by
methicillin-resistant S. aureus (MRSA), investigating novel
targets and methods for treating MRSA has become an urgent clinical
need (2).
Bacteria need to regulate their metabolism in
response to changes due to environmental stress, such as
antibiotics, temperature and oligotrophy (3). Two-component signal transduction
systems (TCSTs) are primary signaling mechanisms utilized by
bacteria to regulate metabolism in response to environmental
changes (3). The mechanisms
underlying TCSTs regulatory systems are activated following
physical or chemical stimuli from the external environment; the
histidine protein kinase receptor proteins anchored to the cell
membrane sense the change in the external environment and undergo
phosphorylation. The phosphate group is transferred to the response
regulator which undergoes phosphorylation, and this is mediated by
phosphotransferase. Subsequently, the phosphorylated response
regulators bind to the promoter sequences of the downstream target
gene, thereby regulating its expression, upregulating expression of
proteins which allow the bacteria to adapt to external
environmental changes and enhance the adaptive viability of
bacteria (4,5).
The two-component signaling pathways are widely
found in prokaryotic bacteria, such as S. aureus (6). The entire genome of S. aureus
has been analyzed and it has been demonstrated that there are 17
two-component signaling pathways involved in the regulation of
bacterial physiological metabolism (7). S. aureus resistance is
inextricably associated with the regulation of the two-component
signaling pathway (8). The present
review assesses the current body of literature available regarding
the mechanisms of two-component signaling pathways, and the
emergency of developing suitable therapeutics to combat drug
resistant S. aureus infections.
2. WalKR/VicSR/YycGF two-component signaling
pathway
WalKR is a highly conserved and specific
two-component signaling pathway in Gram-positive bacteria with low
GC-content (9). S. aureus is
Gram-positive with low GC-content (10). The walKR gene is an essential
gene that is involved in the regulation of cell wall synthesis and
other types of physiological metabolism (10). Howden et al (11) analyzed 10 types of
vancomycin-intermediate resistance in S. aureus (VISA) and
three types of heterogeneous VISA (hVISA) strains via
high-throughput sequencing. The results demonstrated that eight
types of VISA and two types of hVISA strains harbored site
mutations in the walKR gene, and that this gene exhibited
the highest degree of mutational frequency (11), suggesting that the walKR gene
serves an important role in the generation of VISA and hVISA
strains. The walKR protein not only participates in the regulation
of cell wall synthesis, but also specifically binds to the promoter
regions of the lytM, atlA, ssaA and isaA genes to
positively regulate the expression of autolysin and peptidoglycan
hydrolase, so as to modify the surface structure of the cell wall,
including shortening the length of sugar chains in the cell wall
and decreasing the degree of cross-linking of peptidoglycan
(11). By changing the phenotype of
the cell wall to increase bacterial resistance, the effectiveness
of antibiotics which target the cell wall, such as vancomycin, is
suppressed (11,12).
Vancomycin is a glycopeptide antibiotic secreted by
Streptomyces orientalis, which inhibits cell wall synthesis
by binding to the D-alanyl-D-alanine residue on the cell wall
(13). In the absence of cell wall
protection, the soma is prone to rupture, thus acting as a
bactericide. A previous study showed that the insertion of the
strong promoter sequence IS256, in the promoter region of the
walKR gene of the S. aureus VISA strain upregulates
the expression of its downstream target protein and enhances
metabolic processes involving the strain's cell wall, resulting in
an increase in bacterial resistance (14). In addition, single nucleotide
polymorphisms are another important factor which underlie changes
in WalKR expression. Domains in the walKR gene of the VISA
strain carry a site mutation of a single nucleotide. The A96T
mutation in the walKR gene is the mutation of base G into
base A at site 24673 of S. aureus. The mutated base is
located in a conserved sequence of the walKR gene and is
associated with a conformational change of protein regulation by
phosphorylation (15). These site
mutations result in a decrease in the activity of the WalKR
protein, downregulating the expression of bacterial autolytic
enzymes, inhibiting autolytic processed in bacteria and decreasing
the sensitivity of the bacteria to the drug (11).
The protein expressed by the yycHI gene as a
downstream target gene of WalKR, binds to the WalKR
histidine kinase receptor to form a complex to inhibit the
activation of WalKR, and thus negatively regulates the WalKR
pathway (16). The mutation rate of
the yycHI gene in the VISA strain isolated from a clinical
trial was significantly higher compared with that of the
vancomycin-sensitive strain (16).
This may due to the fact that the mutation of yycHI gene
resulted in weakening of the negative regulatory mechanism on the
WalKRK pathway, enhanced cell wall modification and cell wall
synthesis and improved bacterial resistance (11).
3. AirSR/YhcSR two-component signaling
pathway
The AirSR two-component signaling pathway is closely
associated with S. aureus resistance towards vancomycin. In
S. aureus (NCTC8325, a prototypical strain of S.
aureus stored at National Collection of Type Cultures) with an
airSR gene mutation, the expression levels of ~30 genes
associated with cell wall synthesis, such as cap, ddl and
pbp1, is significantly decreased and the minimum inhibitory
concentration of the airSR gene mutant against vancomycin is
significantly decreased. Further investigation using EMSA
technology showed that the AirR protein directly binds to the
promoter sequences of cell wall-forming genes such as cap,
ddl and pbp1. By positively regulating the expression of
these genes, the anabolic process of the cell wall was promoted,
which in turn increased the sensitivity of the bacteria to
vancomycin (17).
The YhcSR gene is an essential gene in S.
aureus that is primarily involved in the regulation of
important physiological processes, such as nitrate respiratory
metabolism (18). The nitrate
respiratory metabolic pathway is a key metabolic pathway required
for the survival of S. aureus in a hypoxic environment
(19). In addition, the YhcR protein
directly binds to the promoter regions of the opuC and
lac genes, and positively regulates the expression of these
two genes. The opuC gene primarily participates in the
synthesis of ABC transporters, whereas the lac gene is
primarily involved in the metabolic regulation of lactose and
galactose (20).
4. Vancomycin resistance associated
regulator/sensor (VraRS) two-component signaling pathway
VraRS two-component signaling pathway is another
important pathway associated with vancomycin resistance. The
thickness of the S. aureus cell wall is positively
correlated with the degree of vancomycin resistance (21). Proteins such as PBP2, SgtB and MurZ
are key proteins involved in the synthesis of the cell wall of
S. aureus and are positively regulated by the VraRS
two-component signaling pathway (22,23).
Kuroda et al (24) found that
the decrease in vancomycin sensitivity was associated with the
increased expression of the vraRS gene in the resistant
S. aureus strain, and the authors showed that the expression
levels of the vraRS gene was significantly increased by
placing S. aureus in a medium containing vancomycin. Further
investigations of several other antibiotics that act on S.
aureus and inhibit cell wall synthesis, such as teicoplanin,
ceftizoxime, imipenem, bacitracin and cycloserine, were performed,
and it was shown that the expression levels of the vraS gene
were increased (24). Based on these
results, it was hypothesized that when S. aureus is
stimulated by cell wall synthesis inhibitors, the cell wall
synthesis process is positively regulated through upregulating
expression of the vraRS gene, thereby reducing the
sensitivity to the antibiotics that inhibit cell wall
synthesis.
5. LytRS two-component signaling
pathway
Cationic antimicrobial peptides (CAPs) are a class
of active peptides produced by host cells, which are involved in
the body's innate immune system (25). CAPs bind to the bacterial cell
membrane through electrostatic attraction. Due to the oil-water
amphipathy, the small molecule active peptide is inserted into the
cell membrane of bacteria, and destroy the integrity and increase
the permeability of the cell membrane, leading to the lysis and
death of cells (25). When the
surface potential of the bacterial cell membrane is decreased by
CAPs, the LytS receptor in the LytRS two-component signal undergoes
autophosphorylation and activates the corresponding response
regulatory protein, LytR. The LytR protein binds to the
corresponding promoter regions of the downstream target genes, such
as the irgAB gene (26). The
irgAB gene is a downstream gene adjacent to the lytRS
gene and is crucially involved in the synthesis of anti-perforin,
which inhibits programmed cell death and lysis (27). Thus, the LytRS two-component signal
regulates the upregulation of the irgAB gene, inhibits
programmed cell death and autolytic enzyme activity of bacteria,
and therefore enhances the drug resistance of bacteria.
Furthermore, the lytRS gene serves a role in
regulating bacterial extracellular DNA, which is an important
component of biofilms (28). When
the surface potential of the bacterial cell membrane is decreased,
the activation of this pathway subsequently decreases the secretion
of extracellular DNA and inhibits the formation of biofilms
(26). As the formation of the
bacterial membrane improves bacterial resistance (29), the lytRS gene may decrease
bacterial resistance by negatively regulating the secretion of
extracellular DNA. Thus, the LytRS two-component signaling pathway
may exhibit dual-regulation in the resistance of S. aureus.
The yhcSR and vraRS genes in S. aureus are
homologous to the yhcYZ and yvqEC genes in
Bacteroides subtilis (B. subtilis). The yhcYZ
and yvqEC genes in B. subtilis interact and regulate
cell wall synthesis (30). Thus, it
is hypothesized that yhcSR and vraRS genes may also
employ similar mechanisms, namely, regulating cell wall
synthesis.
6. GraRS/ApsRS two-component signaling
pathway
Bacteriocin is a peptide substance that is
self-synthesized by bacteria to inhibit the proliferation of other
types of bacteria. These peptides are released into the cytoplasm
with the assistance of the ATP-binding cassette (ABC) transport
system (31). In addition to
releasing its own bacteriocin, the ATP transport system also pumps
exogenous bacteriocins out of the cytoplasm as a defense mechanism
(32). Multiple ATP gene loci
adjacent to the bicomponent signal were identified following the
analysis of the gene locus of S. aureus (32). However, S. aureus has not been
confirmed to possess bacteriocin synthesis-associated genes, and
therefore these structural sites are hypothesized to be primarily
associated with immune defense (33). High expression of genes associated
with the two-component signaling pathway in GraRS is associated
with VISA production (34). The
GraRS two-component signaling pathway upregulates the expression of
VraFG ATP transporter-associated genes, enhances efflux transport
mechanisms in bacteria, increases transport of intracellular
antibiotics, such as vancomycin and mitomycin B, out of the
cytoplasm and prevents antibiotics from exerting antibacterial
effects (35).
Conversely, the dlt and mprF genes
decrease the negative charge on the surface of the cell membrane
through modifying components, such as teichoic acid and
phosphatidylglycerol on the surface of the cell membrane (36,37). The
GraRS gene regulates the expression of dlt and
mprF genes, and alters the negative potential on the cell
wall surface, thereby decreasing the binding capacity of positively
charged antibiotics (such as vancomycin and mitomycin B), and
weakens their antibacterial effect (35). However, currently, the mechanism
underlying the activation of the GraRS receptor protein is unknown,
and the mechanism underlying the effect of the GraRS pathway on
drug resistance requires further investigation.
7. BceRS/BraRS/NsaRS two-component signaling
pathway
Similar to the GraRS/ApsRS two-component signaling
pathway, the BceRS two-component signaling pathway gene is adjacent
to the bceAB and vraDE genes of the ABC transport
system (38). bceAB is
located upstream of the bceRS gene, whereas vraDE is located
~80 bases downstream of the bceRS gene (38). The BceS protein activates the BceR
protein following stimulation by external bacteriocin, and this
results in the upregulation of the expression of bceAB and
vraDE genes, thereby increasing transport of exogenous
bacteriocins out of the cytoplasm by BceAB and VraDE proteins,
preventing bacteriocin from acting as an antibacterial agent, and
thus resulting in an increase in bacterial resistance (39). The minimum inhibitory concentration
of S. aureus towards bacteriocin is decreased by 2-4-fold by
reducing bceAB and vraDE gene expression (which
encode the APC transporter gene) in S. aureus (38).
Nisin A. and Nukacin ISK-1 are type I bacteriocins
secreted by Staphylococcus warner and Lactococcus
lactis, respectively (40,41).
Nisin A. exerts its bacteriostatic effects by acting on the cell
membrane of bacteria to form pore complexes, which cause leakage
and dissolution of cell fluid. Additionally, Nisin A also serves a
role in inhibiting cell wall synthesis (40). Nukacin ISK-1 serves an antimicrobial
role primarily via inhibiting the synthesis of the cell wall
(41). When the two bacteriocins
were co-cultured with the BraRS gene mutant S.
aureus, the growth of the mutant strain was significantly
inhibited (50-33%) (42). Thus, the
BraRS gene exhibits significant regulatory effects on the symbiosis
of S. aureus and the type I bacteriocin strain.
8. Hexose phosphate transporter
regulator/sensor (HptRS) two-component signaling pathway
The HptRS two-component signaling pathway is
primarily composed of hptA, hptRS and uhpT.
HptA initiates autophosphorylation of the HptS protein by sensing
changes in the concentration of surrounding phosphates, such as
3-phosphoric acid glucose, glucose-6-phosphate and Fosfomycin.
uhpT is a downstream regulatory gene of the HptRS
two-component signaling pathway. UhpT protein transports the
aforementioned phosphates into the cytoplasm of bacteria to provide
a source of carbon for physiological metabolism in bacteria
(43). In addition to extracellular
growth, S. aureus may invade host epithelial cells and
acquire phosphate hexose from the cytoplasm of the host cell to
maintain physiological metabolism in the bacteria (44) via the phosphoenolpyruvate
phosphotransferase pathway (45).
The molecular structure of Fosfomycin, a broad-spectrum antibiotic
in clinical trials, is similar to phosphoenolpyruvate. Instead of
being metabolized by the body, the antibiotic is excreted in its
original form, thus it is widely used for the treatment of
osteomyelitis due to its lower toxicity and fewer side effects
(46). This type of antibiotic may
be transported into the cytoplasm via the bacterial phosphohexose
transporter to exert its antibiotic effects through competitively
binding to UDP-N-acetylglucosamine-3-O-enolpyruvate transferase
(encoded by the murA gene), inhibiting the synthesis of the
peptidoglycan precursor, as well as interfering with the synthesis
of the cell wall (47). However,
when mutations occur in the HptPS two-component signaling pathway,
the expression of UhpT protein is decreased, accompanied by a
decrease in the uptake of Fosfomycin, and thus a decrease in the
effectiveness of the drug, resulting in enhanced bacterial
resistance (43).
9. Conclusion
As the incidence of MRSA infections is increasing
each year, the therapeutic effects of antibiotics are becoming
notably decreased. Although the emergence of novel treatment
methods face enormous challenges, research based on new treatment
concepts is required to combat emergent resistant MRSA strains. The
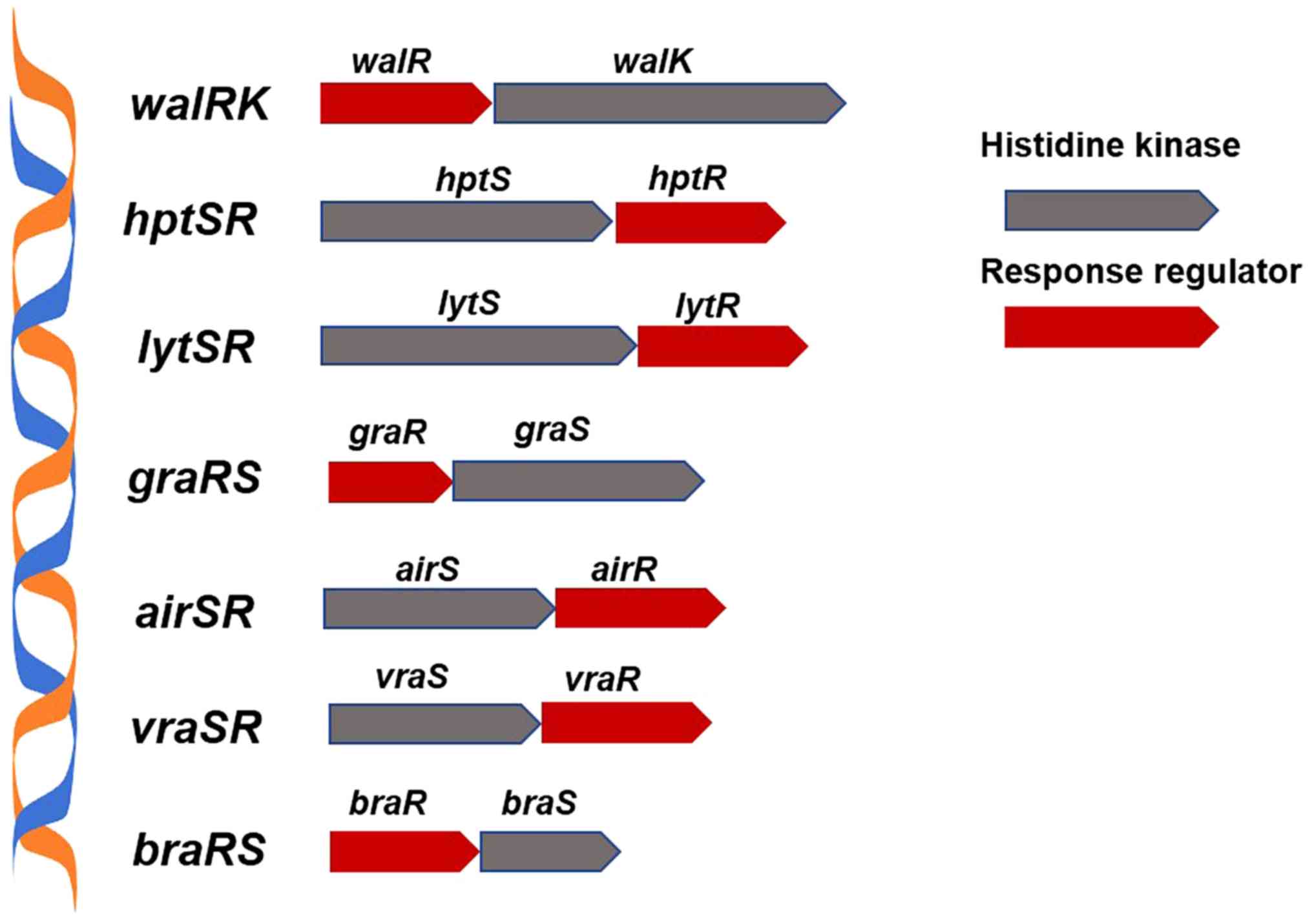
two-component signaling pathway of S. aureus (Fig. 1) regulates the sensitivity of strains
to antimicrobial agents through efflux mechanisms, regulation of
cell wall anabolic processes, and inhibition of drug uptake.
Furthermore, the two-component signaling pathways may regulate
bacterial physiological metabolism and virulence factors, and
improve the adaptability of bacteria to the external environment.
Hence, an in-depth study of the mechanisms involved in the
two-component signaling pathway of S. aureus may highlight
novel potential targets for the treatment of osteomyelitis.
Acknowledgements
Not applicable.
Funding
This study was supported by the Sichuan Provincial
Natural Science Foundation of China (grant no. 2018SZ0125), Sichuan
Provincial Natural Science Foundation of China (grant no.
2019YFS0270) and the National Undergraduate Training Programs for
Innovation and Entrepreneurship (grant no. C2019105797).
Availability of data and materials
Not applicable.
Authors' contributions
SW, KL, YL, HZ and LL conceived and designed the
present review. SW, KL, YL, HZ and LL drafted and critically
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ono D, Yamaguchi T, Hamada M, Sonoda S,
Sato A, Aoki K, Kajiwara C, Kimura S, Fujisaki M, Tojo H, et al:
Analysis of synergy between beta-lactams and
anti-methicillin-resistant Staphylococcus aureus agents from
the standpoint of strain characteristics and binding action. J
Infect Chemother. 25:273–280. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chuang YY and Huang YC: Molecular
epidemiology of community-associated meticillin-resistant
Staphylococcus aureus in Asia. Lancet Infect Dis.
13:698–708. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mattos-Graner RO and Duncan MJ:
Two-component signal transduction systems in oral bacteria. J Oral
Microbiol. 9(1400858)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Goulian M: Two-component signaling circuit
structure and properties. Curr Opin Microbiol. 13:184–189.
2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tierney AR and Rather PN: Roles of
two-component regulatory systems in antibiotic resistance. Future
Microbiol. 14:533–552. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cheng RR, Morcos F, Levine H and Onuchic
JN: Toward rationally redesigning bacterial two-component signaling
systems using coevolutionary information. Proc Natl Acad Sci USA.
111:E563–E571. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kuroda M, Ohta T, Uchiyama I, Baba T,
Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al:
Whole genome sequencing of meticillin-resistant Staphylococcus
aureus. Lancet. 357:1225–1240. 2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Matsuo M, Kato F, Oogai Y, Kawai T, Sugai
M and Komatsuzawa H: Distinct two-component systems in
methicillin-resistant Staphylococcus aureus can change the
susceptibility to antimicrobial agents. J Antimicrob Chemother.
65:1536–1537. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xu T, Wu Y, Lin Z, Bertram R, Götz F,
Zhang Y and Qu D: Identification of Genes controlled by the
essential YycFG Two-component system reveals a role for biofilm
modulation in Staphylococcus epidermidis. Front Microbiol.
8(724)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Villanueva M, García B, Valle J, Rapún B,
Ruiz de Los Mozos I, Solano C, Martí M, Penadés JR, Toledo-Arana A
and Lasa I: Sensory deprivation in Staphylococcus aureus.
Nat Commun. 9(523)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Howden BP, McEvoy CR, Allen DL, Chua K,
Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, et
al: Evolution of multidrug resistance during Staphylococcus
aureus infection involves mutation of the essential two
component regulator WalKR. PLoS Pathog. 7(e1002359)2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim T, Choi J, Lee S, Yeo KJ, Cheong HK
and Kim KK: Structural studies on the extracellular domain of
sensor Histidine Kinase YycG from Staphylococcus aureus and
its functional implications. J Mol Biol. 428:3074–3089.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Peng H, Rao Y, Yuan W, Zheng Y, Shang W,
Hu Z, Yang Y, Tan L, Xiong K, Li S, et al: Reconstruction of the
vancomycin-susceptible Staphylococcus aureus phenotype from
a vancomycin-intermediate S. aureus XN108. Front Microbiol.
9(2955)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jansen A, Türck M, Szekat C, Nagel M,
Clever I and Bierbaum G: Role of insertion elements and yycFG in
the development of decreased susceptibility to vancomycin in
Staphylococcus aureus. Int J Med Microbiol. 297:205–215.
2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bourret RB: Receiver domain structure and
function in response regulator proteins. Curr Opin Microbiol.
13:142–149. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cameron DR, Jiang JH, Kostoulias X,
Foxwell DJ and Peleg AY: Vancomycin susceptibility in
methicillin-resistant Staphylococcus aureus is mediated by
YycHI activation of the WalRK essential two-component regulatory
system. Sci Rep. 6(30823)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sun H, Yang Y, Xue T and Sun B: Modulation
of cell wall synthesis and susceptibility to vancomycin by the
two-component system AirSR in Staphylococcus aureus
NCTC8325. BMC Microbiol. 13(286)2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yan M, Hall JW, Yang J and Ji Y: The
essential yhcSR two-component signal transduction system directly
regulates the lac and opuCABCD operons of Staphylococcus
aureus. PLoS One. 7(e50608)2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yan M, Yu C, Yang J and Ji Y: The
essential two-component system YhcSR is involved in regulation of
the nitrate respiratory pathway of Staphylococcus aureus. J
Bacteriol. 193:1799–1805. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yan M, Hall JW, Yang J and Ji Y: The
essential yhcSR Two-component signal transduction system directly
regulates the lac and opuCABCD operons of Staphylococcus
aureus. PLoS One. 7(e50608)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tajbakhsh G and Golemi-Kotra D: The
dimerization interface in VraR is essential for induction of the
cell wall stress response in Staphylococcus aureus: A
potential druggable target. BMC Microbiol. 19(153)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hanaki H, Kuwahara-Arai K, Boyle-Vavra S,
Daum RS, Labischinski H and Hiramatsu K: Activated cell-wall
synthesis is associated with vancomycin resistance in
methicillin-resistant Staphylococcus aureus clinical strains
Mu3 and Mu50. J Antimicrob Chemother. 42:199–209. 1998.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen H, Xiong Z, Liu K, Li S, Wang R, Wang
X, Zhang Y and Wang H: Transcriptional profiling of the
two-component regulatory system VraSR in Staphylococcus
aureus with low-level vancomycin resistance. Int J Antimicrob
Agents. 47:362–367. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kuroda M, Kuwaharaarai K and Hiramatsu K:
Identification of the up- and down-regulated genes in
vancomycin-resistant Staphylococcus aureus strains Mu3 and
Mu50 by cDNA differential hybridization method. Biochem Biophys Res
Commun. 269:485–490. 2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Omardien S, Brul S and Zaat SA:
Antimicrobial activity of cationic antimicrobial peptides against
gram-positives: Current progress made in understanding the mode of
action and the response of bacteria. Front Cell Dev Biol.
4(111)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Patel K and Golemi-Kotra D: Signaling
mechanism by the Staphylococcus aureus two-component system
LytSR: Role of acetyl phosphate in bypassing the cell membrane
electrical potential sensor LytS. Version 2. F1000Res.
4(79)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sadykov MR and Bayles KW: The control of
death and lysis in staphylococcal biofilms: A coordination
of physiological signals. Curr Opin Microbiol. 15:211–215.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mann EE, Rice KC, Boles BR, Endres JL,
Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR and
Bayles KW: Modulation of eDNA release and degradation affects
Staphylococcus aureus biofilm maturation. PLoS One.
4(e5822)2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
McCarthy H, Rudkin JK, Black NS, Gallagher
L, O'Neill E and O'Gara JP: Methicillin resistance and the biofilm
phenotype in Staphylococcus aureus. Front Cell Infect
Microbiol. 5(1)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kobayashi K, Ogura M, Yamaguchi H, Yoshida
K, Ogasawara N, Tanaka T and Fujita Y: Comprehensive DNA microarray
analysis of Bacillus subtilis two-component regulatory
systems. J Bacteriol. 183:7365–7370. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Crowe-McAuliffe C, Graf M, Huter P, Takada
H, Abdelshahid M, Nováček J, Murina V, Atkinson GC, Hauryliuk V and
Wilson DN: Structural basis for antibiotic resistance mediated by
the Bacillus subtilis ABCF ATPase VmlR. Proc Natl Acad Sci
USA. 115:8978–8983. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liao F, Mo Z, Gu W, Xu W, Fu X and Zhang
Y: A comparative genomic analysis between methicillin-resistant
Staphylococcus aureus strains of hospital acquired and
community infections in Yunnan province of China. BMC Infect Dis.
20(137)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gardete S and Tomasz A: Mechanisms of
vancomycin resistance in Staphylococcus aureus. J Clin
Invest. 124:2836–2840. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zeng D, Debabov D, Hartsell TL, Cano RJ,
Adams S, Schuyler JA, McMillan R and Pace JL: Approved Glycopeptide
antibacterial drugs: Mechanism of action and resistance. Cold
Spring Harb Perspect Med. 6(a026989)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chaili S, Cheung AL, Bayer AS, Xiong YQ,
Waring AJ, Memmi G, Donegan N, Yang SJ and Yeaman MR: The GraS
Sensor in Staphylococcus aureus mediates resistance to host
defense peptides differing in mechanisms of action. Infect Immun.
84:459–466. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mechler L, Bonetti EJ, Reichert S,
Flötenmeyer M, Schrenzel J, Bertram R, François P and Götz F:
Daptomycin tolerance in the Staphylococcus aureus pitA6
mutant is due to Upregulation of the dlt operon. Antimicrob Agents
Chemother. 60:2684–2691. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kanesaka I, Fujisaki S, Aiba Y, Watanabe
S, Mikawa T, Katsuse AK, Takahashi H, Cui L and Kobayashi I:
Characterization of compensatory mutations associated with
restoration of daptomycin-susceptibility in daptomycin
non-susceptible methicillin-resistant Staphylococcus aureus
and the role mprF mutations. J Infect Chemother. 25:1–5.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kawada-Matsuo M, Yoshida Y, Nakamura N and
Komatsuzawa H: Role of two-component systems in the resistance of
Staphylococcus aureus to antibacterial agents. Virulence.
2:427–430. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang Y, Luo M, Zhou H, Li C, Luk A, Zhao
G, Fung K and Ip M: Role of Two-component system response regulator
bceR in the antimicrobial resistance, virulence, biofilm formation,
and stress response of group B streptococcus. Front Microbiol.
10(10)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hyde AJ, Parisot J, McNichol A and Bonev
BB: Nisin-induced changes in Bacillus morphology suggest a paradigm
of antibiotic action. Proc Natl Acad Sci USA. 103:19896–19901.
2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Islam MR, Nishie M, Nagao J, Zendo T,
Keller S, Nakayama J, Kohda D, Sahl HG and Sonomoto K: Ring A of
nukacin ISK-1: A lipid II-binding motif for type-A(II) lantibiotic.
J Am Chem Soc. 134:3687–3690. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kawada-Matsuo M, Yoshida Y, Zendo T, Nagao
J, Oogai Y, Nakamura Y, Sonomoto K, Nakamura N and Komatsuzawa H:
Three distinct two-component systems are involved in resistance to
the class I bacteriocins, Nukacin ISK-1 and nisin A, in
Staphylococcus aureus. PLoS One. 8(e69455)2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Park JY, Kim JW, Moon BY, Lee J, Fortin
YJ, Austin FW, Yang SJ and Seo KS: Characterization of a novel
two-component regulatory system, HptRS, the regulator for the
hexose phosphate transport system in Staphylococcus aureus.
Infect Immun. 83(1620)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Postma PW, Lengeler JW and Jacobson GR:
Phosphoenolpyruvate: Carbohydrate phosphotransferase systems of
bacteria. Microbiol Rev. 57:232–269. 1993.PubMed/NCBI
|
|
45
|
Fraunholz M and Sinha B: Intracellular
Staphylococcus aureus: Live-in and let die. Front Cell
Infect Microbiol. 2(43)2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Trinh TD, Smith JR and Rybak MJ:
Parenteral Fosfomycin for the treatment of multidrug resistant
bacterial infections: The rise of the epoxide. Pharmacotherapy.
39:1077–1094. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hrast M, Rožman K, Jukič M, Patin D, Gobec
S and Sova M: Synthesis and structure-activity relationship study
of novel quinazolinone-based inhibitors of MurA. Bioorg Med Chem
Lett. 27:3529–3533. 2017.PubMed/NCBI View Article : Google Scholar
|