Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer (1) and 880,792
CRC-associated deaths were reported in 2018(2). The prevalence of CRC is 9.2% in women
and 10% in men, and it is the second most common type of cancer in
women and third most common type of cancer in men (3). In ~90% of cases, CRC is sporadic, and
patients do not have any family history of the disease (4). Risk factors for CRC include age (higher
in individuals >50 years), and a family history of inflammatory
bowel disease (3.7% risk for CRC) and Crohn's disease (2.5% risk
for CRC) (5-7).
Diet and lifestyle may increase the risk of CRC (8), such as smoking (9,10),
consumption of red meat (11,12) and
low levels of physical activity (13).
Mutations in various genes, alterations in DNA
methylation and chromosomal instability have been identified as
genetic causes of CRC. Analysis of gene expression profiles of
cancer cells serves an important role in the diagnosis and
treatment of patients with cancer and may result in improved
insight into the dysregulated mechanisms associated with specific
types of cancer. HOX genes are involved in the regulation of
different cellular processes, including differentiation,
angiogenesis, signaling, apoptosis, mobility and metastasis
(14,15). HOXC genes (HOXC4,
HOXC5, HOXC6, HOXC8, HOXC9,
HOXC10, HOXC11, HOXC12 and HOXC13) are
members of the HOX family of genes and are located on
chromosome 12q13.3(16), and
HOXC13 is involved in the growth and formation of hair and
nails (17,18).
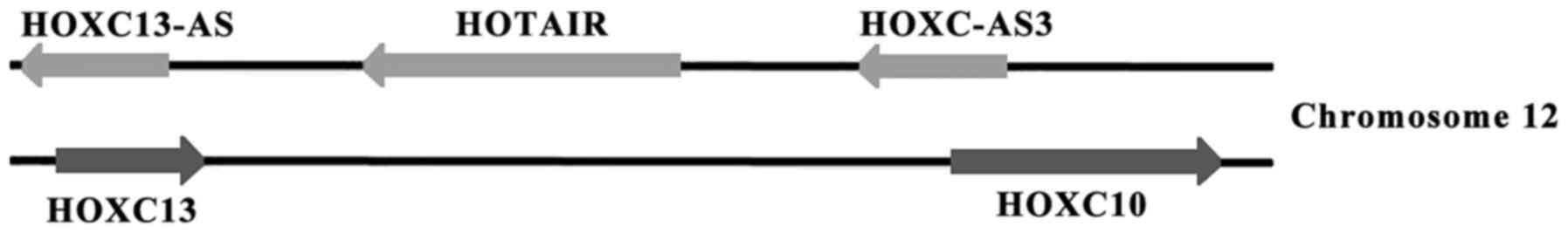
Three long non-coding RNAs (lncRNAs), HOTAIR,
HOXC13-AS and HOXC-AS3 are located on the antisense
strand of the HOXC gene cluster (Fig. 1). HOTAIR is a polyadenylated
RNA that binds to certain protein complexes, such as PRC2, and
regulates the conformation of chromatin (19-21).
HOXC10, HOXC-AS3, HOXC13 and HOXC13-AS
are located in close proximity to the oncogenic lncRNA,
HOTAIR. Thus, it was hypothesized that there may be an
association between these genes with HOTAIR and development
of cancer. In the present study, the expression levels of these
genes were assessed using reverse transcription-quantitative (RT-q)
PCR in tumor tissues and matching healthy adjacent tissues in
patients with CRC.
Materials and methods
Patients and tissue samples
A total of 39 pairs of tumor tissue and healthy
adjacent tissue was obtained from patients. The median age of
patients was 54 years old (range, 30-79 years) and included 20
males and 19 females. Tissues were obtained from patients with CRC
following surgery and immediately stored in liquid nitrogen. The
present study was approved by the Ethics Committee of Shahid
Chamran University of Ahvaz (Ahvaz, Iran) and written informed
consent was obtained from all patients.
RT-qPCR
A total of 50 mg frozen tissue from each patient was
crushed and homogenized. Total RNA was extracted using
TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and RNA was dissolved in diethyl pyrocarbonate. The RNA
concentration of each sample was measured using a NanoDrop™ 2000/c
spectrophotometer (Thermo Fisher Scientific, Inc.) and stored at
-80˚C. RNA integrity was assessed using electrophoresis on a 1%
agarose gel containing SafeStain (CinnaGen), and the 28S, 18S and
5S bands were observed. Extracted RNAs were treated using DNaseI
(Takara Bio, Inc.). Primescript™ RT reagent kit (Takara Bio, Inc.)
was used for reverse transcription of RNA to cDNA. A total of 1 µl
random hexamers (100 µM) and 1 µl oligo(dT) primer (50 µM) were
added to 1 µg DNase treated RNA and RNase free water was added to a
final volume of 5 µl and incubated at 65˚C for 5 min. Subsequently,
1 µl reverse transcriptase enzyme (1 U/µl) and 4 µl 5X buffer was
added, and RNase free H2O was added to a final volume of
20 µl and incubated at 37˚C for 30 min for cDNA synthesis, followed
by incubation at 80˚C for 5 sec to inactivate the enzyme and cDNA
was stored at -20˚C. For qPCR, the following reagents were mixed as
follows: 10 µl SYBR Premix Ex Taq II (2X) (Takara Bio, Inc.), 0.5
µl each forward and reverse primers (10 pmol each; Macrogen, Inc.),
2 µl cDNA and 7 µl DNase free water. The primer sequences used are
presented in Table I. β-actin
was used as the loading control. The thermocycling conditions were:
95˚C for 30 sec; 40 cycles of 95˚C for 5 sec and 60˚C for 32 sec;
and a dissociation stage of 95˚C for 15 sec, 60˚C for 60 sec and
95˚C for 1 sec. Relative expression of HOXC10,
HOXC-AS3, HOTAIR, HOXC13 and HOXC13-AS
was measured using the 2-∆∆Cq method (22) for the 39 pairs of the tumor tissues
and marginal normal tissues in patients with CRC. β-actin
was used as the reference gene.
 | Table ISequence of primers used in this
study. |
Table I
Sequence of primers used in this
study.
| Genes | Primer sequences
(5'-3') |
|---|
| HOXC10 | F:
CCAGACACCTCGGATAACG |
| HOXC10 | R:
GGCACCTCTTCTTCCTTCC |
| HOXC13 | F:
TCTCCCTTCCCAGACGTGGT |
| HOXC13 | R:
CGCTCAGAGAGGTTCGTGGT |
| HOTAIR | F:
GAAAGGTCCTGCTCCGCTTC |
| HOTAIR | R:
TCCTCTCGCCGCCGTCTG |
| HOXC-AS3 | F:
CGATAGGCGGCTTTGG |
| HOXC-AS3 | R:
CGTCTTGTGTGCTGGTTTCC |
| HOXC13-AS | F:
CGGACATCGGAGCACTATG |
| HOXC13-AS | R:
CGGCTGGTCTTCTTGAGG |
| β-actin | F:
ATTGGCAATGAGCGGTTC |
| β-actin | R:
TGAAGGTAGTTTCGTGGATG |
Data analysis
All analysis was performed using GraphPad Prism
version 5 (GraphPad Software, Inc.). Differences between two groups
were compared using a Wilcoxon test. A Spearman's rank correlation
coefficient was used for correlation analysis of relative gene
expression with clinical parameters.
Results
Comparison of expression of HOX genes
between tumor and normal tissues
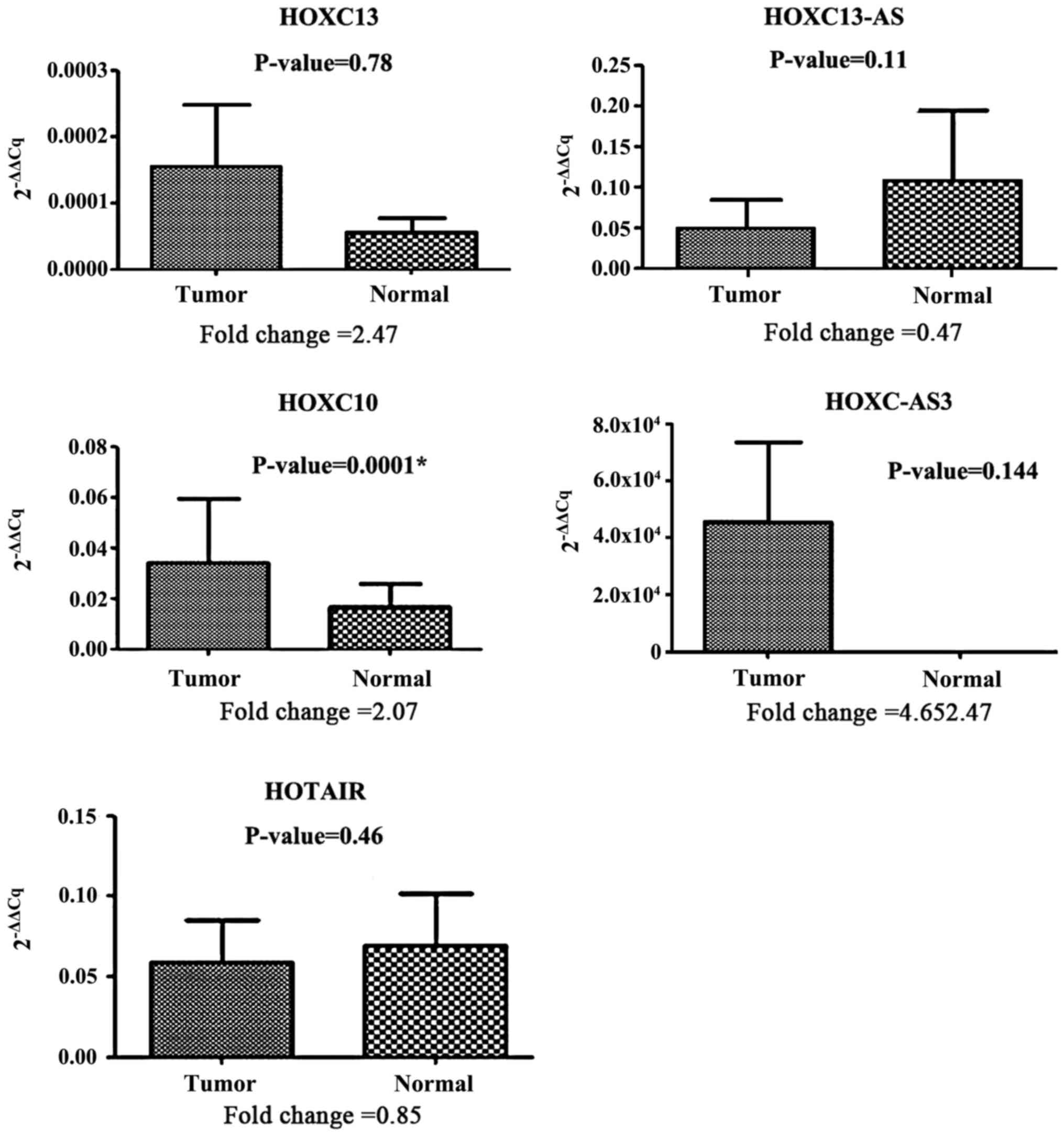
Expression of HOX expression of HOXC-AS3 and
HOXC13 in the tumor tissues was upregulated compared with
the marginal normal tissues with a fold change of 4.65 and 2.47,
respectively. Although HOXC-AS3 and HOXC13 were
upregulated in the tumor tissues, statistical analysis showed the
difference was not significant (P=0.144 and P=0.78, respectively;
Fig. 2). However, expression of
HOXC10 was significantly upregulated in the tumor tissues
compared with the marginal normal tissues with a fold change of
2.07 (P=0.0001; Fig. 2). Comparison
of the expression of HOXC10, HOXC-AS3, HOTAIR,
HOXC13 and HOXC13-AS in the tumor and normal tissues
is shown in Fig. 2.
Correlation between the expression of
genes
To determine the correlation coefficients, the fold
change ratio of each gene was used. There was a significant
positive correlation between expression of HOTAIR and
HOXC13 (r=0.75, strong correlation; P=0.00000004); between
HOTAIR and HOXC13-AS (r=0.57, moderate correlation;
P=0.001); and between HOXC13 and HOXC13-AS (r=0.43,
moderate; P=0.006). There was positive correlation between
HOTAIR and HOXC10 (r=0.306, moderate; P=0.058), and
between HOTAIR and HOXC-AS3 (r=0.384, moderate;
P=0.016). There was a significantly negative correlation between
HOXC10 and HOXC-AS3 (r=-0.331, moderate;
P=0.039).
Association between gene expression
and clinicopathological features
The association between the expression of the five
genes with clinicopathological characteristics of the patients was
calculated. Statistical analysis showed that there was no
significant association between sex, histological grade, tumor size
(cm), Tumor-Node-Metastasis (TNM) stage (23), lymphatic invasion, vascular invasion
and the expression of HOXC10, HOXC-AS3,
HOTAIR, HOXC13 or HOXC13-AS in the patients
with CRC (all P>0.05; Table
II).
 | Table IIAssociation between expression of the
selected genes and clinicopathological characteristics. |
Table II
Association between expression of the
selected genes and clinicopathological characteristics.
|
Characteristics | n | HOXC13 |
HOXC13-AS | HOTAIR | HOXC10 |
HOXC-AS3 |
|---|
| Sex | | 0.66 | 0.37 | 0.56 | 0.42 | 0.83 |
|
Male | 20 | | | | | |
|
Female | 19 | | | | | |
| Histological
grade | | 0.3 | 0.57 | 0.4 | 0.07 | 0.57 |
|
≤2 | 33 | | | | | |
|
>2 | 6 | | | | | |
| Tumor size, cm | | 0.078 | 0.81 | 0.74 | 0.32 | 0.66 |
|
≤5 | 20 | | | | | |
|
>5 | 19 | | | | | |
| TNM stage | | 0.16 | 0.28 | 0.22 | 0.85 | 0.06 |
|
≤3 | 20 | | | | | |
|
>3 | 19 | | | | | |
| Lymphatic
invasion | | 0.41 | 0.66 | 0.6 | 0.81 | 0.36 |
|
Yes | 20 | | | | | |
|
No | 19 | | | | | |
| Vascular
invasion | | 0.53 | 0.42 | 0.83 | 0.6 | 0.15 |
|
Yes | 19 | | | | | |
|
No | 20 | | | | | |
Discussion
In 2012, ~1.4 million new cases of CRC were
diagnosed. By 2035, it is estimated that there will be >2.4
million new cases of colorectal cancer (24). In the present study, the expression
levels of HOXC10, HOXC-AS3, HOTAIR,
HOXC13 and HOXC13-AS in matching normal and tumor
tissues from 39 patients with CRC were assessed using RT-qPCR.
HOXC10, HOXC-AS3, HOXC13, and HOXC13-AS
are located in proximity to the oncogenic lncRNA HOTAIR.
Thus, it was hypothesized that there may be an association between
these genes and HOTAIR in carcinogenesis. To evaluate this
hypothesis, the correlation between the expression of these five
genes with HOTAIR, and the association between these genes
and certain clinicopathological characteristics were calculated.
The results showed that the expression of HOTAIR was not
significantly altered in tumor tissues compared with marginal
normal tissues, contrasting with previous studies which reported a
significant increase in the expression of HOTAIR in CRC
(25-29).
Kogo et al (30) studied 100
pairs of tumor and normal CRC samples and reported that there was
no significant increase in expression of HOTAIR. Svoboda
et al (31) also reported
that the relative expression of HOTAIR between the tumor and
adjacent normal tissue was not significantly different in patients
with CRC. These differences may be related to the ethnicity of the
individuals studied, their lifestyle, diet or other lifestyle
factors. Upregulation of HOXC13 has been reported in
odontogenic tumors, liposarcoma, metastatic melanoma, esophageal
squamous cell carcinoma, lung adenocarcinoma and ameloblastoma
(32-37).
However, the expression levels of HOXC13 in CRC was not
significantly altered in the present study. Tatangelo et al
(38) reported a significant
upregulation in the expression of HOXC13 and HOTAIR
in right (proximal) side of CRCs tissues.
Overexpression of HOXC13-AS has been reported
in nasopharyngeal carcinoma (39).
To the best of our knowledge, the present study is the first to
evaluate the expression of HOXC13-AS in CRC, and the results
showed that there was no significant difference in the expression
of HOXC13-AS between normal and cancer tissues.
Studies have shown that the expression of
HOXC10 is significantly increased in thyroid cancer, breast
carcinoma and lung cancer (40-43).
Kim et al (44) reported that
expression of HOXC10 was upregulated in gastric cancer. They
showed that upregulation of HOXC10 increases proliferation
and migration of gastric cancer cells (44). In the present study the expression
levels of HOXC10 in CRC were assessed and the results showed
that the relative expression in the tumor tissues was significantly
higher compared with marginal normal tissues in CRC (P=0.0001).
Zhang et al (45) showed that
expression of HOXC-AS3 was upregulated in gastric cancer
tissues compared with normal tissues, but in the present study,
expression of HOXC-AS3 was not changed significantly altered
in tumor tissues of patients with CRC compared with normal tissues.
To the best of our knowledge, the present study is the first to
assess the expression of HOXC-AS3 in patients with CRC.
The association between the expression
HOXC10, HOXC-AS3, HOTAIR, HOXC13 and
HOXC13-AS with clinicopathological characteristics,
including sex, histological grade, tumor size (cm), TNM stage,
lymphatic invasion and vascular invasion were calculated. Although
there was no significant relationship between any of the genes
evaluated and any of the clinicopathological characteristics, the
correlation coefficients were positive for all the clinical
parameters and expression data, suggesting that, whilst an
association may exist it is not strong and not statistically
significant. Therefore, upregulation of the studied genes may have
weak or moderate effects on these characteristics. For protein
coding genes, it may be possible that the expression levels of the
protein products may be directly correlated with these
characteristics. Additionally, upregulated genes may affect the
initiation and progression of tumorigenesis, including increasing
proliferation and reducing apoptosis (46).
To determine the correlation coefficient, the fold
change ratio of each gene was used. There was a strong and
significant correlation between the expression of HOTAIR and
HOXC13; and a moderate but significant correlation between
the expression of HOTAIR and HOXC13-AS, and between
HOXC13 and HOXC13-AS.
The correlation between the expression of
HOTAIR and HOXC10 and between the expression of
HOTAIR and HOXC-AS3 were moderate but not
significant. There was a moderate negative correlation between the
expression of HOXC-AS3 and HOXC10 and this difference
was significant.
HOTAIR and HOXC13 appeared to exhibit
similar expression pattern as there was a strong correlation
between them. The results of the present study suggest that these
genes may affect expression of each other expression in cis;
however, additional functional studies are required to determine
whether this is the case. Due to the significant upregulation of
HOCX10 in the tumor tissues, it may be used as a biomarker
for the diagnosis and treatment of CRC.
Future studies should use larger cohorts and
evaluate the expression of the studied genes in the serum and blood
of patients. Additionally, the expression of these genes in other
types of cancer and the expression of other HOX family genes in CRC
should be assessed. To determine whether HOTAIR influences
expression of any of the HOX family genes, HOTAIR expression should
be knocked down and the expression of surrounding genes
assessed.
In conclusion the present study showed that
HOXC10 expression was significantly higher in CRC samples
compared with the normal adjacent tissues, but expression of
HOXC-AS3, HOTAIR, HOXC13 and HOXC13-AS
did not differ significantly. Based on these results, HOXC10 may be
considered as a biomarker for diagnosis of CRC. Additionally
the expression of HOTAIR and HOXC13 were strongly
correlated and thus may share a regulatory mechanism of expression
or one of these genes may regulate the expression of the other.
Further functional studies are required to elucidate the mechanism
underlying this correlation.
Acknowledgements
Not applicable.
Funding
Funding
The present study was funded by the Shahid Chamran
University of Ahvaz (Ahvaz, Iran).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ and HG conceived and designed the study; MG and
ME acquired, analyzed and interpreted the data; ME, MG, MZ and HG
participated in drafting the manuscript. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Shahid Chamran University of Ahvaz (Ahvaz, Iran) and
written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108.
2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
International Agency for Research on
Cancer WHO. Cancer today. IARC Web site. https://gco.iarc.fr/today/. Accessed Jan 21, 2019.
|
|
3
|
Stewart BW and Wild CP (eds): World Cancer
Report 2014. IARC Nonserial Publication, Lyon, pp630, 2014.
|
|
4
|
Bogaert J and Prenen H: Molecular genetics
of colorectal cancer. Ann Gastroenterol. 27:9–14. 2014.PubMed/NCBI
|
|
5
|
Levin B, Lieberman DA, McFarland B,
Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S,
Johnson D, et al: Screening and surveillance for the early
detection of colorectal cancer and adenomatous polyps, 2008: A
joint guideline from the American cancer society, the US
multi-society task force on colorectal cancer, and the American
college of radiology. CA Cancer J Clin. 58:130–16. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Eaden JA, Abrams KR and Mayberry JF: The
risk of colorectal cancer in ulcerative colitis: A meta-analysis.
Gut. 48:526–535. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Canavan C, Abrams KR and Mayberry J:
Meta-analysis: Colorectal and small bowel cancer risk in patients
with crohn's disease. Aliment Pharmacol Ther. 23:1097–1104.
2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Robertson DJ: ABC of colorectal cancer.
Gastroenterology. 143:868–886. 2012.
|
|
9
|
Botteri E, Iodice S, Bagnardi V, Raimondi
S, Lowenfels AB and Maisonneuve P: Smoking and colorectal cancer: A
meta-analysis. JAMA. 300:2765–2778. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liang PS, Chen TY and Giovannucci E:
Cigarette smoking and colorectal cancer incidence and mortality:
Systematic review and meta-analysis. Int J Cancer. 124:2406–2415.
2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bastide NM, Pierre FH and Corpet DE: Heme
iron from meat and risk of colorectal cancer: A meta-analysis and a
review of the mechanisms involved. Cancer Prev Res (Phila).
4:177–184. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Santarelli RL, Pierre F and Corpet DE:
Processed meat and colorectal cancer: A review of epidemiologic and
experimental evidence. Nutr Cancer. 60:131–144. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Martinez-Useros J and Garcia-Foncillas J:
Obesity and colorectal cancer: Molecular features of adipose
tissue. J Transl Med. 14(21)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Grier DG, Thompson A, Kwasniewska A,
McGonigle GJ, Halliday HL and Lappin TR: The pathophysiology of HOX
genes and their role in cancer. J Pathol. 205:154–171.
2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371.
2010.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Apiou F, Flagiello D, Cillo C, Malfoy B,
Poupon MF and Dutrillaux B: Fine mapping of human HOX gene
clusters. Cytogenet Cell Genet. 73:114–115. 1996.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Godwin AR and Capecchi MR: Hoxc13 mutant
mice lack external hair. Genes Dev. 12:11–20. 1998.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kulessa H, Turk G and Hogan BL:
Inhibitionof Bmp signaling affects growth and differentiation in
the anagen hair follicle. EMBO J. 19:6664–6674. 2000.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Woo CJ and Kingston RE: HOTAIR lifts
noncoding RNAs to new levels. Cell. 129:1257–1259. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chu C, Qu K, Zhong FL, Artandi SE and
Chang HY: Genomic maps oflong noncoding rna occupancy reveal
principles of rna-chromatin interactions. Mol Cell. 44:667–668.
2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Galli F, Ruspi L, Marzorati A, Lavazza M,
Di Rocco G, Boni L, Dionigi G and Rausei S: N staging system:
Tumor-node-metastasis and future perspectives. Transl Gastroenterol
Hepatol. 2(4)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pauler FM, Koerner MV and Barlow DP:
Silencing by imprinted noncoding RNAs: Is transcription the answer?
Trends Genet. 23:284–292. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu
H, Tong N, Chen J, Zhang Z and Wang M: Genome-wide analysis of long
noncoding RNA signature in human colorectal cancer. Gene.
556:227–234. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep1.
35:479–487. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dou J, Ni Y, He X, Wu D, Li M, Wu S, Zhang
R, Guo M and Zhao F: Decreasing lncRNA HOTAIR expression inhibits
human colorectal cancer stem cells. Am J Transl Res. 8:98–108.
2016.PubMed/NCBI
|
|
28
|
Lu X, Liu Z, Ning X, Huang L and Jiang B:
The long noncoding RNA HOTAIR promotes colorectal cancer
progression by sponging miR-197. Oncol Res. 26:473–481.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang X and Lu S: MicroR-545 mediates
colorectal cancer cells proliferation through up-regulating
epidermal growth factor receptor expression in HOTAIR long
non-coding RNA dependent. Mol Cell Biochem. 431:45–54.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515.
2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhong M, Wang J, Gong YB, Li JC, Zhang B
and Hou L: Expression of HOXC13 in ameloblastoma. Zhonghua Kou
Qiang Yi Xue Za Zhi. 42:43–46. 2007.(In Chinese). PubMed/NCBI
|
|
33
|
Hong YS, Wang J, Liu J, Zhang B, Hou L and
Zhong M: Expressionof HOX C13 in odontogenic tumors. Shanghai Kou
Qiang Yi Xue. 16:587–591. 2007.(In Chinese). PubMed/NCBI
|
|
34
|
Cantile M, Scognamiglio G, Anniciello A,
Farina M, Gentilcore G, Santonastaso C, Fulciniti F, Cillo C,
Franco R, Ascierto PA and Botti G: Increased HOXC13 expression in
metastatic melanoma progression. J Transl Med.
10(91)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cantile M, Galletta F, Franco R, Aquino G,
Scognamiglio G, Marra L, Cerrone M, Malzone G, Manna A, Apice G, et
al: Hyperexpression of HOXC13, located in the 12q13 chromosomal
region, in welldifferentiated and dedifferentiated human
liposarcomas. Oncol Rep. 30:2579–2586. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Luo J, Wang Z, Huang J, Yao Y, Sun Q, Wang
J, Shen Y, Xu L and Ren B: HOXC13 promotes proliferation of
esophageal squamous cell carcinoma via repressing transcription of
CASP3. Cancer Sci. 109:317–329. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M,
Lin X, Qian Q, Zhang Y, Cao L, et al: HOXC13 promotes proliferation
oflung adenocarcinoma via modulation of CCND1 and CCNE1. Am J
Cancer Res. 7:1820–183. 2017.PubMed/NCBI
|
|
38
|
Tatangelo F, Di Mauro A, Scognamiglio G,
Aquino G, Lettiero A, Delrio P, Avallone A, Cantile M and Botti G:
Posterior HOX genesand HOTAIR expression in the proximal and distal
colon cancer pathogenesis. J Transl Med. 16(350)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gao C, Lu W, Lou W, Wang L and Xu Q: Long
noncoding RNA HOXC13-AS positively affects cell proliferation and
invasion in nasopharyngeal carcinoma via modulating
miR-383-3p/HMGA2 axis. J Cell Physiol. 234:12809–12820.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Feng X, Li T, Liu Z, Shi Y and Peng Y:
HOXC10 up-regulation contributes to human thyroid cancer and
indicates poor survival outcome. Mol Biosyst. 11:2946–2954.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ansari KI, Hussain I, Kasiri S and Mandal
SS: HOXC10 is overexpressed in breast cancer and transcriptionally
regulated by estrogen via involvement of histone methylases MLL3
and MLL4. J Mol Endocrinol. 48:61–75. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sadik H, Korangath P, Nguyen NK, Gyorffy
B, Kumar R, Hedayati M, Teo WW, Park S, Panday H, Munoz TG, et al:
HOXC10 expression supports the development of chemotherapy
resistance by fine tuning DNA repair in breast cancer cells. Cancer
Res. 76:4443–4456. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tang XL, Ding BX, Hua Y, Chen H, Wu T,
Chen ZQ and Yuan CH: Hoxc10 promotes the metastasis of human lung
adenocarcinoma and indicates poor survival outcome. Front Physiol.
8(557)2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kim J, Bae DH, Kim JH, Song KS, Kim YS and
Kim SY: HOXC10 overexpression promotes cell proliferation and
migration in gastric cancer. Oncol Rep. 42:202–212. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhang E, He X, Zhang C, Su J, Lu X, Si X,
Chen J, Yin D Han L and De W: A novel long noncoding RNA HOXC-AS3
mediates tumorigenesis of gastric cancer by binding to YBX1. Genome
Biol. 19(154)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bhatlekar S, Fields JZ and Boman BM: HOX
genes and their role in the development of human cancers. J Mol
Med. 92:811–823. 2014.PubMed/NCBI View Article : Google Scholar
|