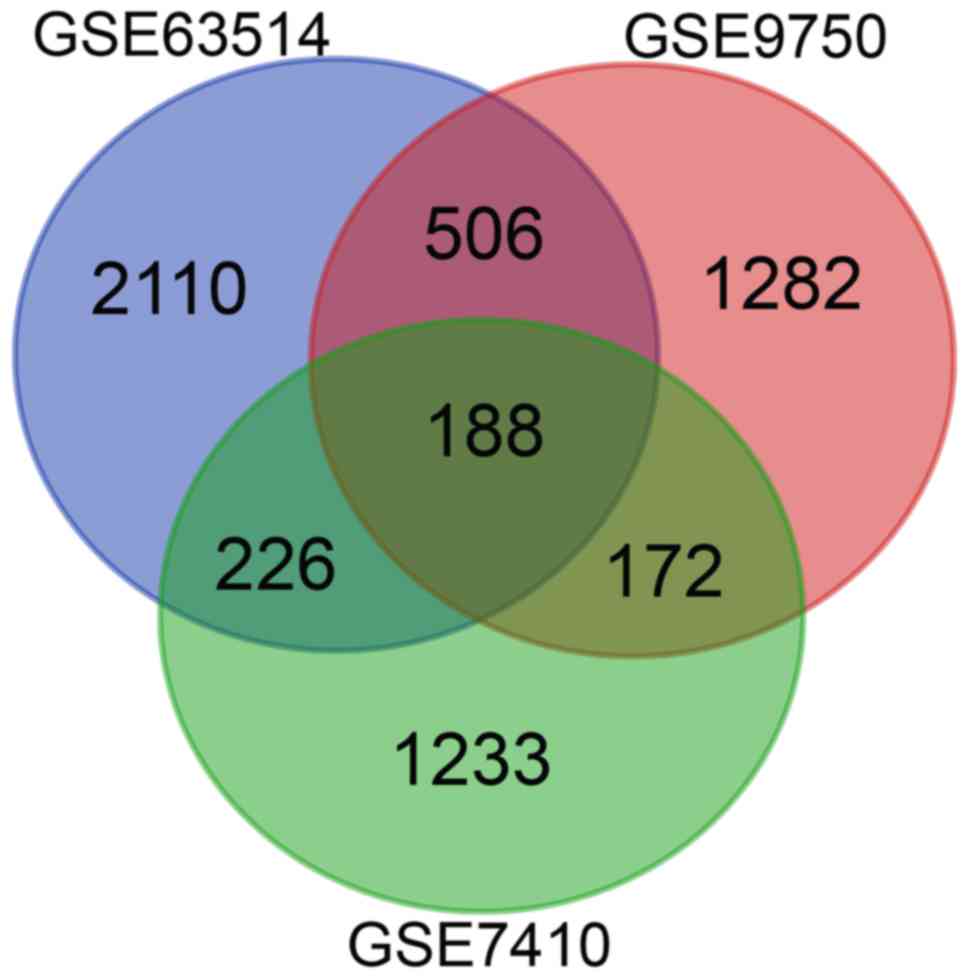
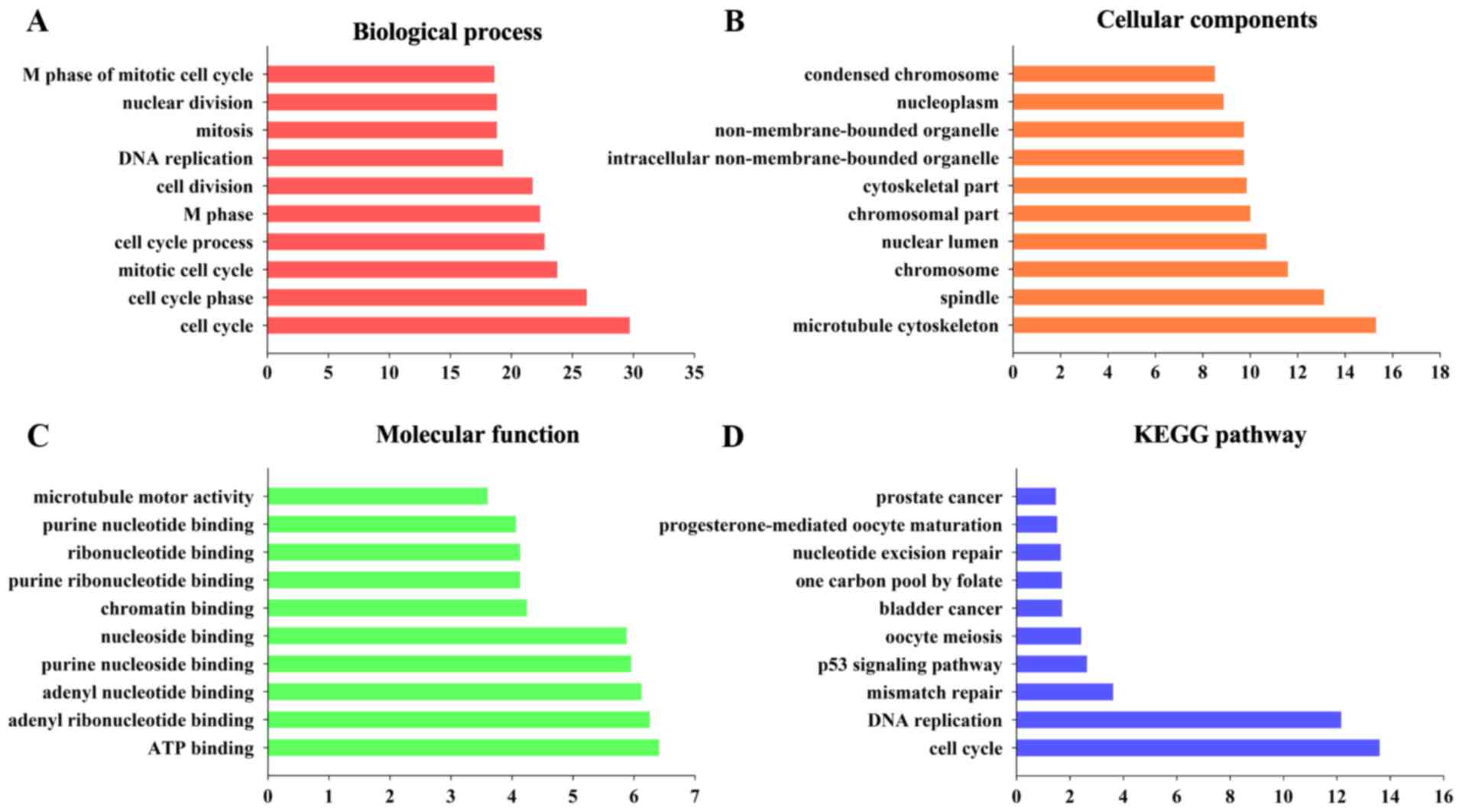
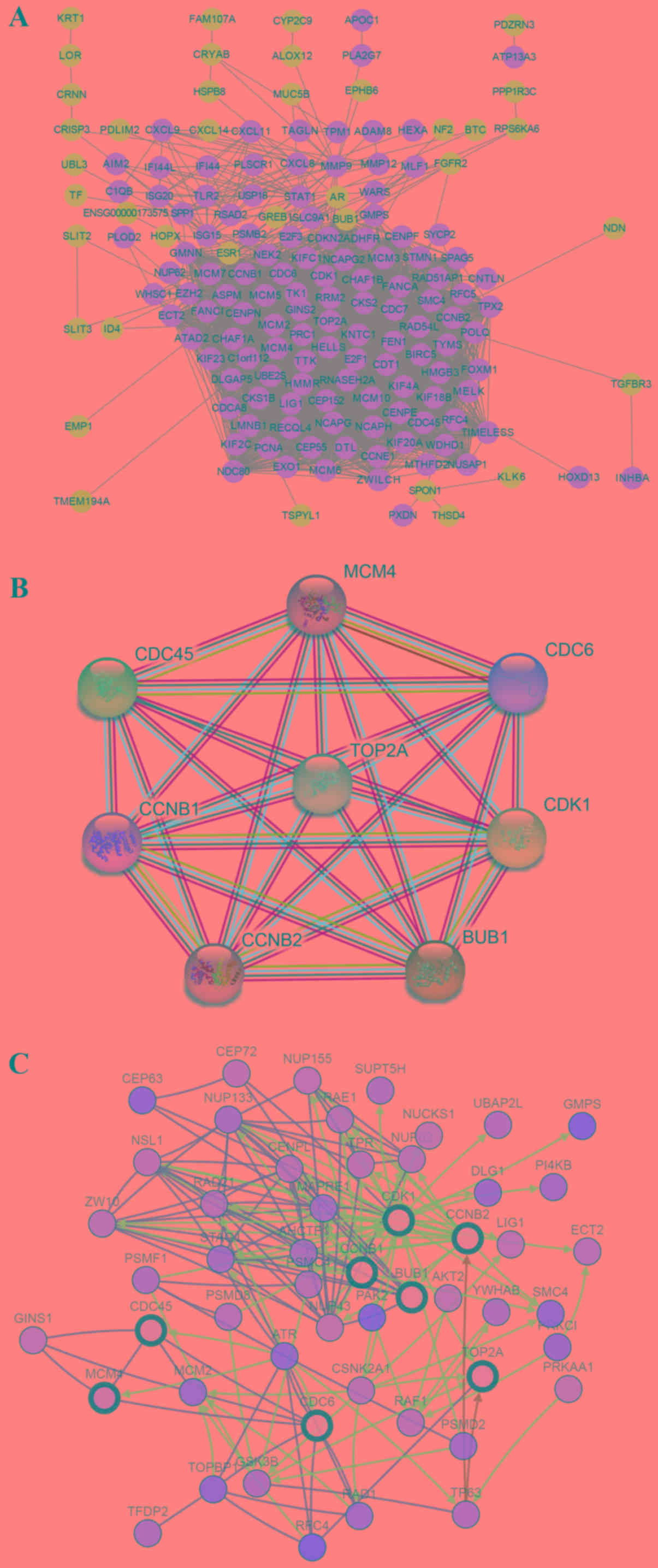
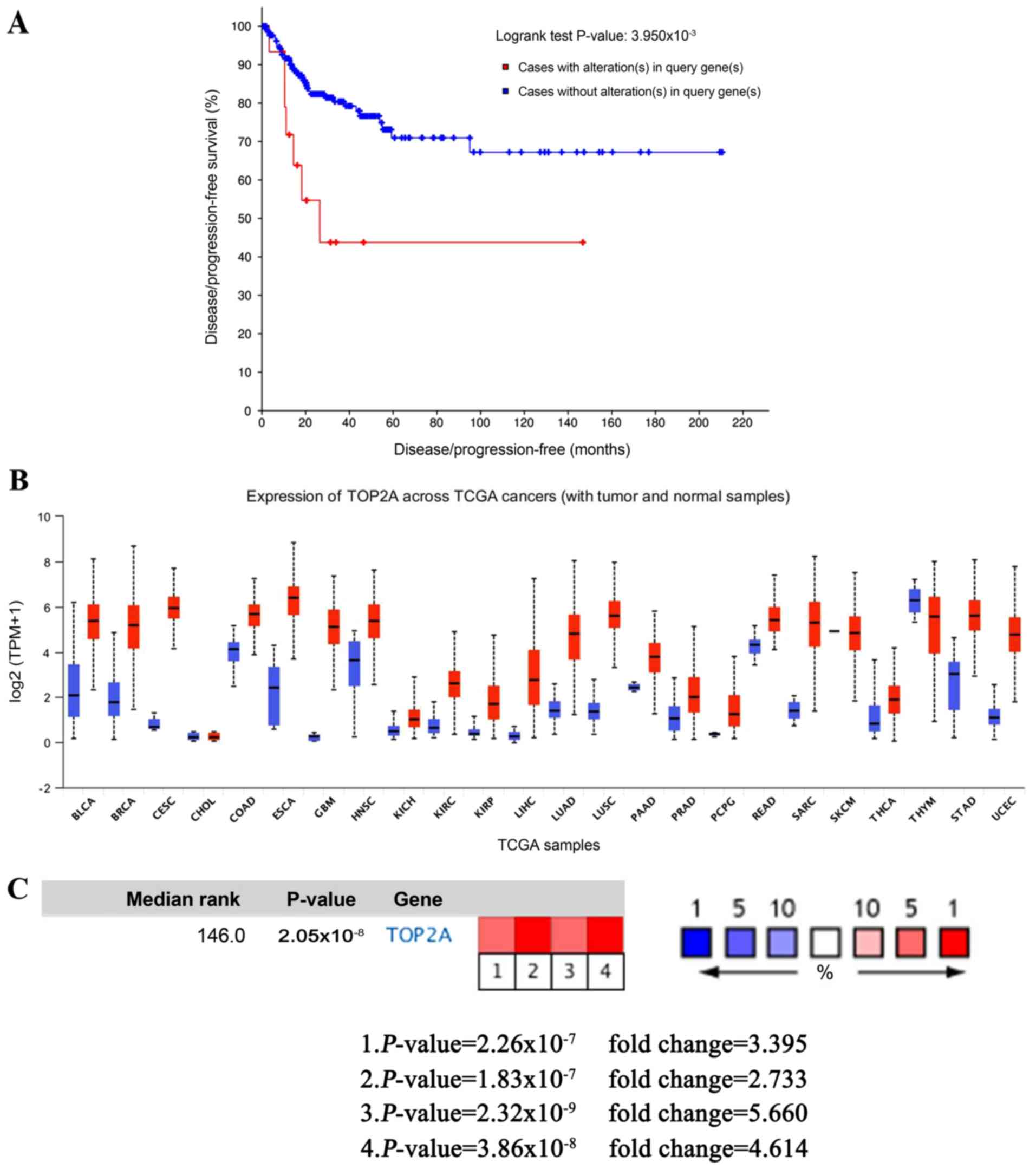
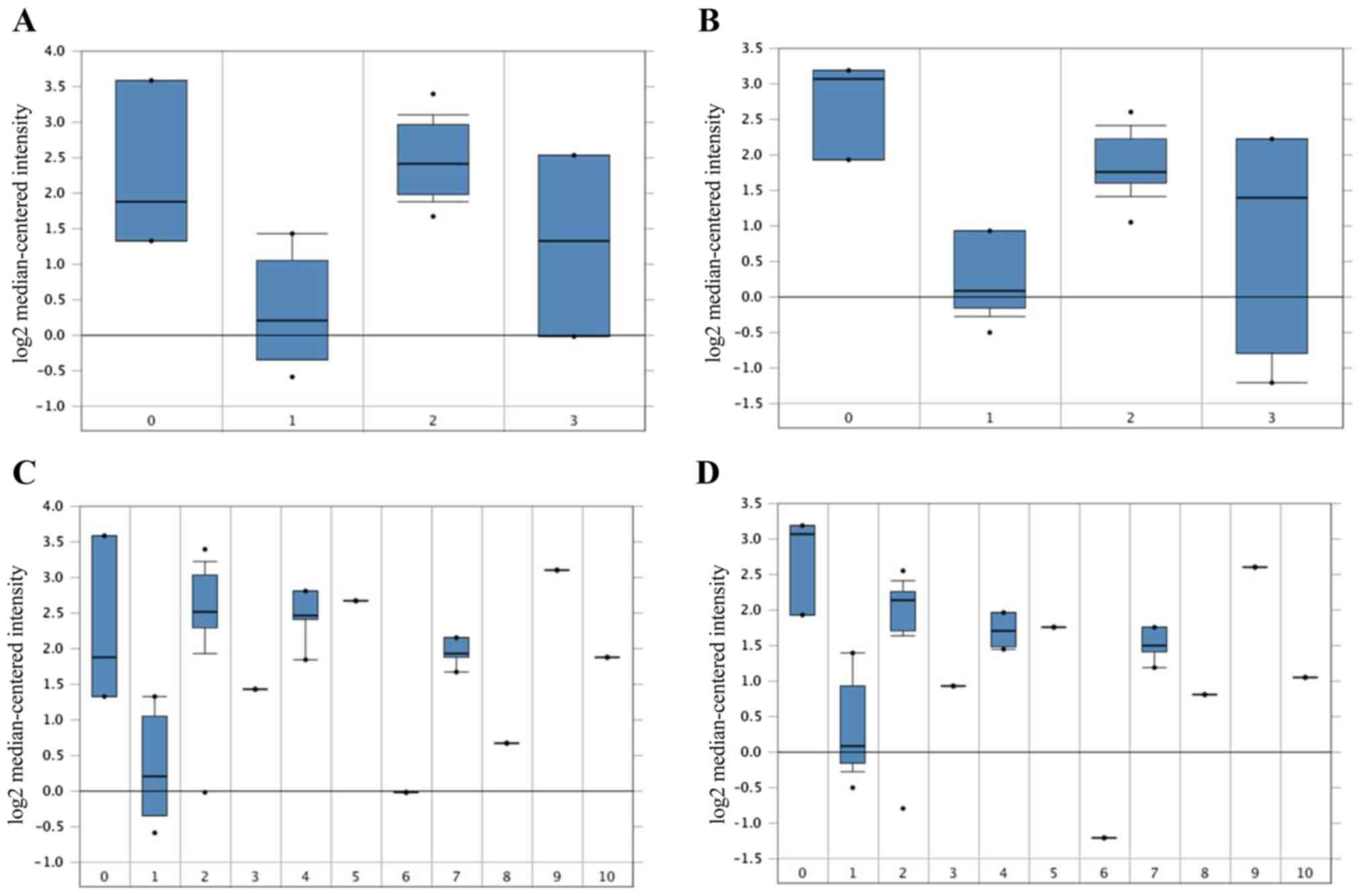
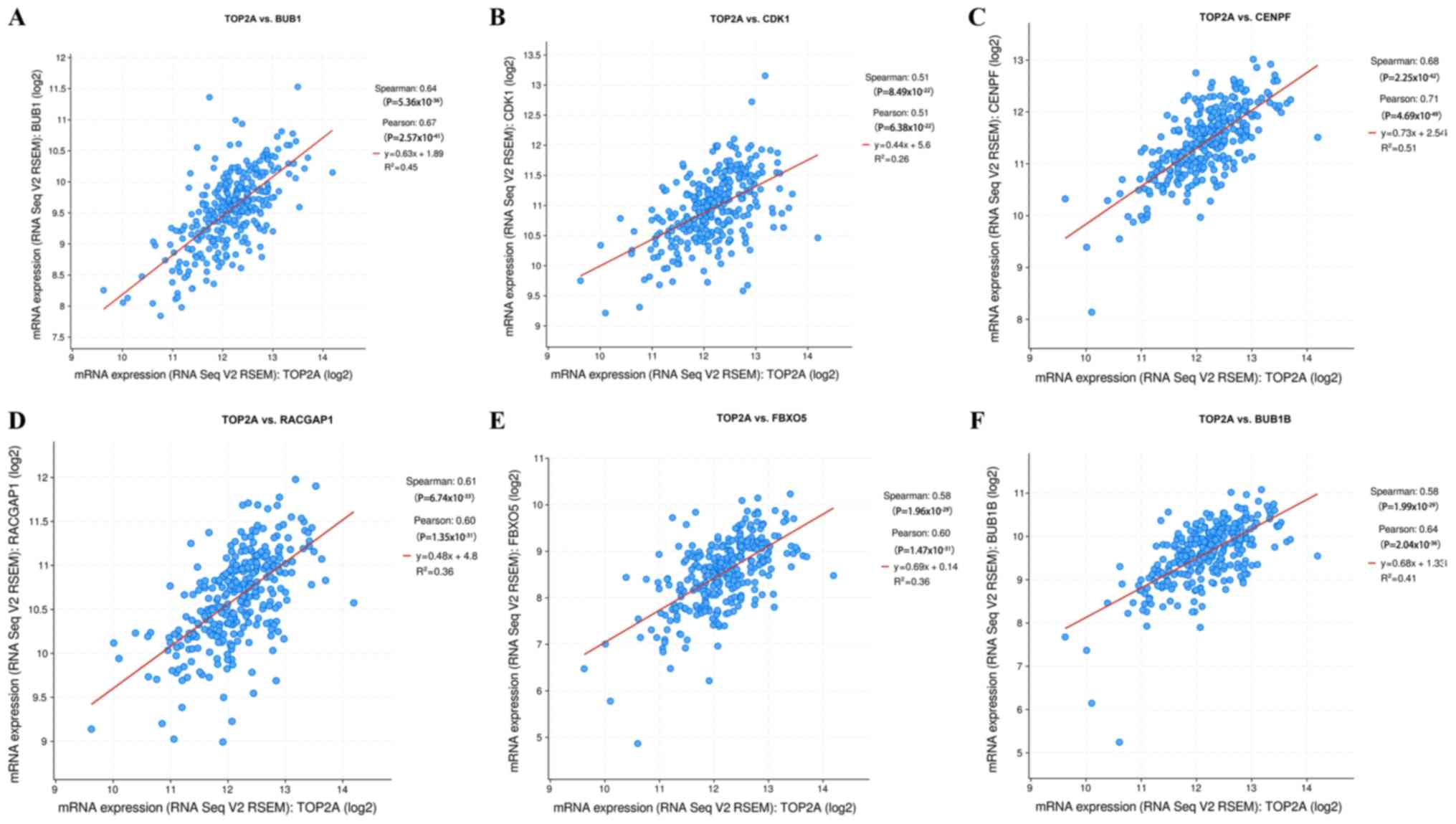
|
1
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9(248)2017.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Zhong TY, Zhou JC, Hu R, Fan XN, Xie XY,
Liu ZX, Lin M, Chen YG, Hu XM, Wang WH, et al: Prevalence of human
papillomavirus infection among 71,435 women in Jiangxi Province,
China. J Infect Public Health. 10:783–788. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: Unresolved issues. Nat
Rev Cancer. 7:11–22. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &
Research Institute at Christiana Care Health Services; HudsonAlpha
Institute for Biotechnology et al. Integrated genomic and
molecular characterization of cervical cancer. Nature. 543:378–384.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
He H, Liu X, Liu Y, Zhang M, Lai Y, Hao Y,
Wang Q, Shi D, Wang N, Luo XG, et al: Human papillomavirus E6/E7
and long noncoding RNA TMPOP2 mutually upregulated gene expression
in cervical cancer cells. J Virol. 93:e01808–e01818.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Zhang J, Yao T, Lin Z and Gao Y: Aberrant
methylation of MEG3 functions as a potential plasma-based biomarker
for cervical cancer. Sci Rep. 7(6271)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen AH, Qin YE, Tang WF, Tao J, Song HM
and Zuo M: MiR-34a and miR-206 act as novel prognostic and therapy
biomarkers in cervical cancer. Cancer Cell Int.
17(63)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yi YX, Fang Yan, Wu KJ, Liu YY and Zhang
W: Comprehensive gene and pathway analysis of cervical cancer
progression. Oncol Lett. 19:3316–3332. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang HJ, Xue JM, Li J, Wan LH and Zhu YX:
Identification of key genes and pathways of diagnosis and prognosis
in cervical cancer by bioinformatics analysis. Mol Genet Genomic
Med. 8(e1200)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Biewenga P, Buist MR, Moerland PD, Ver
Loren van Themaat E, van Kampen AH, ten Kate FJ and Baas F: Gene
expression in early stage cervical cancer. Gynecol Oncol.
108:520–526. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41 (Database
Issue):D991–D995. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8(R183)2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101; discussion 101-103, 119-128, 244-152.
2002.PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res. 41
(Database Issue):D808–D815. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ,
Trimble CL, Fearon ER and Cho KR: Gene expression analysis of
preinvasive and invasive cervical squamous cell carcinomas
identifies HOXC10 as a key mediator of invasion. Cancer Res.
67:10163–10172. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Medi K, Esra G and Kazim YA: Novel genomic
biomarker candidates for cervical cancer as identified by
differential co-expression network analysis. OMICS. 23:261–273.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dai FF, Chen GT, Wang YQ, Zhang L, Long
YM, Yuan MQ, Yang DY, Liu SY, Cheng YX and Zhang LP: Identification
of candidate biomarkers correlated with the diagnosis and prognosis
of cervical cancer via integrated bioinformatics analysis. Onco
Targets Ther. 12:4517–4532. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu JH, Nie SP, Gao M, Jiang Y, Wan YC, Ma
XL, Zhou SL and Cheng WJ: Identification of EPHX2 and RMI2 as two
novel key genes in cervical squamous cell carcinoma by an
integrated bioinformatic analysis. J Cell Physiol. 234:21260–21273.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
van Dam PA, Rolfo C, Ruiz R, Pauwels P,
Van Berckelaer C, Trinh XB, Ferri Gandia J, Bogers JP and Van Laere
S: Potential new biomarkers for squamous carcinoma of the uterine
cervix. ESMO Open. 3(e000352)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Iqbal J, Ejaz SA, Miliutina M, Langer P
and Saeed A: Anti-proliferative effects of chromones: Potent
derivatives affecting cell growth and apoptosis in breast,
bone-marrow and cervical cancer cells. Med Chem. 15:883–891.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao H, Li S, Wang G, Zhao W, Zhang D,
Wang F, Li W and Sun L: Study of the mechanism by which dinaciclib
induces apoptosis and cell cycle arrest of lymphoma Raji cells
through a CDK1-involved pathway. Cancer Med. 8:4348–4358.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chang WH, Forde D and Lai AG: Dual
prognostic role of 2-oxoglutarate-dependent oxygenases in ten
cancer types: Implications for cell cycle regulation and cell
adhesion maintenance. Cancer Commun (Lond). 39(23)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lee JW, Park S, Kim SY, Um SH and Moon EY:
Curcumin hampers the antitumor effect of vinblastine via the
inhibition of microtubule dynamics and mitochondrial membrane
potential in HeLa cervical cancer cells. Phytomedicine. 23:705–713.
2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Saha SK and Khuda-Bukhsh AR: Berberine
alters epigenetic modifications, disrupts microtubule network, and
modulates HPV-18 E6-E7 oncoproteins by targeting p53 in cervical
cancer cell HeLa: A mechanistic study including molecular docking.
Eur J Pharmacol. 744:132–146. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Das D, Bristol ML, Smith NW, James CD,
Wang X, Pichierri P and Morgan IM: Werner Helicase control of human
papillomavirus 16 E1-E2 DNA replication is regulated by SIRT1
deacetylation. mBio. 10(e00263-19)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lou W, Ding B, Xu L and Fan W:
Construction of potential glioblastoma multiforme-related
miRNA-mRNA regulatory network. Front Mol Neurosci.
12(66)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Brown CA, Bogers J, Sahebali S, Depuydt
CE, De Prins F and Malinowski DP: Role of protein biomarkers in the
detection of high-grade disease in cervical cancer screening
programs. J Oncol. 2012(289315)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Peres AL, Paz E Silva KM, de Araújo RF, de
Lima Filho JL, de Melo Júnior MR, Martins DB and de Pontes Filho
NT: Immunocytochemical study of TOP2A and Ki-67 in cervical smears
from women under routine gynecological care. J Biomed Sci.
23(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Smrkolj S, Erzen M and Rakar S: Prognostic
significance of topoisomerase II alpha and collagen IV
immunoexpression in cervical cancer. Eur J Gynaecol Oncol.
31:380–385. 2010.PubMed/NCBI
|
|
41
|
Korkolopoulou P and Vassilakopoulos TP:
Topoisomerase IIalpha as a prognostic factor in mantle cell
lymphoma. Leukemia. 18:1347–1349. 2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fritz P, Cabrera CM, Dippon J, Gerteis A,
Simon W, Aulitzky WE and van der Kuip H: c-erbB2 and topoisomerase
IIalpha protein expression independently predict poor survival in
primary human breast cancer: A retrospective study. Breast Cancer
Res. 7:R374–R384. 2005.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients survival and chemoresistance. Int J Cancer.
124:644–652. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Del Pino M, Svanholm-Barrie C, Torné A,
Marimon L, Gaber J, Sagasta A, Persing DH and Ordi J: mRNA
biomarker detection in liquid-based cytology: A new approach in the
prevention of cervical cancer. Mod. Pathol. 28:312–320.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Scapulatempo-Neto C, Veo C, Fregnani JHTG,
Lorenzi A, Mafra A, Melani AGF, Loaiza EAA, Rosa LAR, de Oliveira
CM, Levi JE and Longatto-Filho A: Characterization of topoisomerase
II α and minichromosome maintenance protein 2 expression in anal
carcinoma. Oncol Lett. 13:1891–1898. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu D, Huang CL, Kameyama K, Hayashi E,
Yamauchi A, Sumitomo S and Yokomise H: Topoisomerase IIalpha gene
expression is regulated by the p53 tumor suppressor gene in
nonsmall cell lung carcinoma patients. Cancer. 94:2239–2247.
2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Schaefer-Klein JL, Murphy SJ, Johnson SH,
Vasmatzis G and Kovtun IV: Topoisomerase 2 alpha cooperates with
androgen receptor to contribute to prostate cancer progression.
PLoS One. 10(e0142327)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hua W, Sa KD, Zhang X, Jia LT, Zhao J,
Yang AG, Zhang R, Fan J and Bian K: MicroRNA-139 suppresses
proliferation in luminal type breast cancer cells by targeting
Topoisomerase II alpha. Biochem Biophys Res Commun. 463:1077–1083.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015(381602)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Erriquez J, Becco P, Olivero M, Ponzone R,
Maggiorotto F, Ferrero A, Scalzo MS, Canuto EM, Sapino A, Verdun di
Cantogno L, et al: TOP2A gene copy gain predicts response of
epithelial ovarian cancers to pegylated liposomal doxorubic in:
TOP2A as marker of response to PLD in ovarian cancer. Gynecol
Oncol. 138:627–633. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Deguchi S, Katsushima K, Hatanaka A,
Shinjo K, Ohka F, Wakabayashi T, Zong H, Natsume A and Kondo Y:
Oncogenic effects of evolutionarily conserved noncoding RNA
ECONEXIN on gliomagenesis. Oncogene. 36:4629–4640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Santin AD, Zhan F, Bignotti E, Siegel ER,
Cané S, Bellone S, Palmieri M, Anfossi S, Thomas M, Burnett A, et
al: Gene expression profiles of primary HPV16- and HPV18-infected
early stage cervical cancers and normal cervical epithelium:
Identification of novel candidate molecular markers for cervical
cancer diagnosis and therapy. Virology. 331:269–291.
2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jain M, Zhang L, He M, Zhang YQ, Shen M
and Kebebew E: TOP2A is overexpressed and is a therapeutic target
for adrenocortical carcinoma. Endocr Relat Cancer. 20:361–370.
2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang H, Yang B, Geng T, Li B, Dai P and
Chen C: Tissue-specific selection of optimal reference genes for
expression analysis of anti-cancer drug-related genes in tumor
samples using quantitative real-time RT-PCR. Exp Mol Pathol.
98:375–381. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani
ME, Ow SG, Nandi S, Lim CT and Thiery JP: Short-term expansion of
breast circulating cancer cells predicts response to anti-cancer
therapy. Oncotarget. 6:15578–15593. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Thougaard AV, Langer SW, Hainau B,
Grauslund M, Juhl BR, Jensen PB and Sehested M: A murine
experimental anthracycline extravasation model: Pathology and study
of the involvement of topoisomerase II alpha and iron in the
mechanism of tissue damage. Toxicology. 269:67–72. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Melaiu O, Cristaudo A, Melissari E, Di
Russo M, Bonotti A, Bruno R, Foddis R, Gemignani F, Pellegrini S
and Landi S: A review of transcriptome studies combined with data
mining reveals novel potential markers of malignant pleural
mesothelioma. Mutat Res. 750:132–140. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Meagher NS, Schuster K, Voss A, Budden T,
Pang CNI, de Fazio A, Ramus SJ and Friedlander ML: Does the primary
site really matter? Profiling mucinous ovarian cancers of uncertain
primary origin (MO-CUP) to personalise treatment and inform the
design of clinical trials. Gynecol Oncol. 150:527–533.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Guo J, Gu Y, Ma X, Zhang L, Li H, Yan Z,
Han Y, Xie L and Guo X: Identification of hub genes and pathways in
adrenocortical carcinoma by integrated bioinformatic analysis. J
Cell Mol Med. 8:4428–4438. 2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li S, Liu X, Liu T, Meng X, Yin X, Fang C,
Huang D, Cao Y, Weng H, Zeng X and Wang X: Identification of
biomarkers correlated with the TNM staging and overall survival of
patients with bladder cancer. Front Physiol. 8(947)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Han JY, Han YK, Park GY, Kim SD, Kim JS,
Jo WS and Lee CG: Corrigendum: Bub1 is required for maintaining
cancer stem cells in breast cancer cell lines. Sci Rep.
6(17984)2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Xi Q, Huang M, Wang Y, Zhong J, Liu R, Xu
G, Jiang L, Wang J, Fang Z and Yang S: The expression of CDK1 is
associated with proliferation and can be a prognostic factor in
epithelial ovarian cancer. Tumour Biol. 36:4939–4948.
2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumorigenesis and progression. Cancer Lett. 362:122–130.
2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Schwermer M, Lee S, Köster J, van Maerken
T, Stephan H, Eggert A, Morik K, Schulte JH and Schramm A:
Sensitivity to cdk1-inhibition is modulated by p53 status in
preclinical models of embryonal tumors. Oncotarget. 6:15425–15435.
2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Mitrofanova A, Aytes A, Zou M, Shen MM,
Abate-Shen C and Califano A: Predicting drug response in human
prostate cancer from preclinical analysis of in vivo mouse models.
Cell Rep. 12:2060–2071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Grewal S, Carver JG, Ridley AJ and Mardon
HJ: Implantation of the human embryo requires Rac1-dependent
endometrial stromal cell migration. Proc Natl Acad Sci USA.
105:16189–16194. 2008.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Imaoka H, Toiyama Y, Saigusa S, Kawamura
M, Kawamoto A, Okugawa Y, Hiro J, Tanaka K, Inoue Y, Mohri Y and
Kusunoki M: RacGAP1 expression, increasing tumor malignant
potential, as a predictive biomarker for lymph node metastasis and
poor prognosis in colorectal cancer. Carcinogenesis. 36:346–354.
2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yu Y and Munger K: Human papillomavirus
type 16 E7 oncoprotein inhibits the anaphase promoting
complex/cyclosome activity by dysregulating EMI1 expression in
mitosis. Virology. 446:251–259. 2013.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Yost S, de Wolf B, Hanks S, Zachariou A,
Marcozzi C, Clarke M, de Voer R, Etemad B, Uijttewaal E, Ramsay E,
et al: Biallelic TRIP13 mutations predispose to Wilms tumor and
chromosome missegregation. Nat Genet. 49:1148–1151. 2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Tan CL, Teissier S, Gunaratne J, Quek LS
and Bellanger S: Stranglehold on the spindle assembly checkpoint:
The human papillomavirus E2 protein provokes BUBR1-dependent
aneuploidy. Cell Cycle. 14:1459–1470. 2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Suematsu T, Li Y, Kojima H, Nakajima K,
Oshimura M and Inoue T: Deacetylation of the mitotic checkpoint
protein BubR1 at lysine 250 by SIRT2 and subsequent effects on
BubR1 degradation during the prometaphase/anaphase transition.
Biochem Biophys Res Commun. 453:588–594. 2014.PubMed/NCBI View Article : Google Scholar
|