Introduction
Cervical cancer (CC) is the fourth most common type
of female malignancy with a high mortality rate worldwide, and
almost all cases of CC are caused by human papillomavirus (HPV)
infection (1). In China, HPV
infection is the most common etiological factor underlying CC
(2). Our previous study showed that
the prevalence of HPV in the Jiangxi Province is 22.49% (3). However, although HPV infection is a
prerequisite of CC, it is not sufficient to induce CC alone
(4). The specific molecular
mechanisms between the persistent high-risk HPV infection and the
pathological process of CC are still controversial (5). Increasing evidence has shown that the
abnormal expression of multiple genes are involved in the
pathogenesis of CC (6). As
tumorigenesis is a complex pathological process involving various
genetic and epigenetic events, such as the inactivation of
suppressor genes and/or the overexpression of oncogenes (7), the identification of dysregulated genes
in cancer-associated pathways may highlight the molecular
mechanisms underlying tumorigenesis, assisting in the development
of novel strategies for treatment of CC. Therefore, it is important
to elucidate the potential molecular mechanisms underlying CC, to
provide novel therapeutic targets and prognostic biomarkers of CC
(8,9).
To improve our understanding of the molecular
mechanisms underlying CC, bioinformatics analysis has been widely
used to identify differentially expressed genes (DEGs), functional
pathways and hub genes involved in the carcinogenesis and
progression of CC. Although several genes have been identified in
predicting the clinical outcome of CC, there have been
inconsistencies between previous studies (10,11),
which may be due to small sample sizes, heterogeneous histological
subtypes, different detection platforms and various data processing
methods. In the present study, three mRNA microarray datasets were
downloaded from the Gene Expression Omnibus (GEO) database to
acquire DEGs between CC and normal tissues. Subsequently, Gene
Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis, protein-protein interaction (PPI)
network analysis and the Cytoscape plugin, CytoHubba, were used to
identify hub genes in CC. Furthermore, Kaplan-Meier plotter was
used on the cBioPortal website and the DNA topoisomerase II α
(TOP2A) gene was identified as an independent prognostic biomarker
of disease-free survival in CC. A total of 188 DEGs and 8 hub genes
were identified, which may be candidate biomarkers for CC.
Materials and methods
Microarray data information
GSE63514(12),
GSE9750(13) and GSE7410(14) were obtained from the NCBI GEO
database (ncbi.nlm.nih.gov/geo) (15,16). The
GSE63514 dataset used the GPL570 [HG-U133 Plus 2] Affymetrix Human
Genome U133 Plus 2.0 Array platform, which contained 28 CC tissue
samples and 24 normal samples. The GSE9750 dataset used the GPL96
Affymetrix Human Genome U133A Array platform and included 33 CC
tissue samples that were primarily marked by HPV16 or HPV18 and 21
normal cervical samples. GSE7410 contained 40 CC and 5 normal
cervical samples, and used the GPL1708 Agilent-012391 Whole Human
Genome Oligo Microarray G4112A platform.
Screening of DEGs in CC
The DEGs between CC and normal cervical samples were
screened using GEO2R (ncbi.nlm.nih.gov/geo/geo2r). The adjusted P-values
(adj.P) and Benjamini and Hochberg false discovery rates were used
to provide a balance between the discovery of statistically
significant genes and limit false-positives. Probe sets without
corresponding gene symbols or genes with >1 probe set were
removed or averaged, respectively. Only genes with a log
fold-change >1 and adj.P-value of <0.01 were regarded as
statistically significant DEGs.
Functional annotation and pathway
enrichment analysis
The screened DEGs were submitted to the Database for
Annotation, Visualization and Integrated Discovery (DAVID;
david.abcc.ncifcrf.gov/; version 6.7)
(17), an online tool for functional
annotation. KEGG is a database for understanding higher-level
functions and biological information generated by high throughput
experimental technologies (18). GO
analysis is a useful bioinformatics tool for annotating genes and
analyzing their biological processes (19). The significant enrichment analysis of
DEGs was evaluated using GO and KEGG, with P<0.05 as the cutoff.
If there were >5 terms enriched in the category, the top 10 were
selected based on the P-value.
PPI network construction and module
analysis
The identified DEGs were imported into the Search
Tool for the Retrieval of Interacting Genes (STRING; string-db.org; version 10.0) (20) database to evaluate the functional
interactions among them. Subsequently, Cytoscape (cytoscape.org/; version 3.6.1) was used to screen the
modules from the PPI network (21),
as a molecular interaction network tool. The plugins Cytoscape and
CytoHubba were used to predict and explore the important nodes and
subnetworks in the network with 12 topological algorithms, which
included degree, edge percolated component, maximum neighborhood
component and density of maximum neighborhood component (22). CytoHubba was used to rank nodes in a
network by their network features and select the overlapping top 15
hub genes by 6 ranked methods. Finally, all identified hub genes in
this module were mapped to DAVID to perform KEGG and GO enrichment
analysis.
Key gene selection and analysis
Overall and disease-free survival analysis of hub
genes using Kaplan-Meier survival curves, as well as correlation
analysis of TOP2A and co-expression genes, were performed in
cBioPortal (cbioportal.org; version 2.4.3) (23,24). The
expression profile of TOP2A was analyzed and displayed using the
online database UALCAN (ualcan.path.uab.edu). The relationship between TOP2A
and expression patterns and HPV viral infection status were
analyzed using the 201291_s_at and 201292_probes in the Zhai Cervix
dataset available from Oncomine (oncomine.com)
(13,14,25). The
heat maps of TOP2A gene expression in clinical CC samples vs.
normal tissues are respectively: Cervical Squamous Cell Carcinoma
vs. Normal Biewenga Cervix, Gynecol Oncol, 2008(14); Cancer Type: CC Bittner Multi-cancer,
unpublished data, 2006; Cervical Squamous Cell Carcinoma vs. Normal
Scotto Cervix 2, Genes Chromosomes Cancer, 2008(15); and Cervical Squamous Cell Carcinoma
Epithelia vs. Normal Zhai Cervix, Cancer Res, 2007(25).
Statistical analysis
All statistical analyses were performed using the
aforementioned bioinformatics tools. Adj.P were corrected for in
multiple comparisons using the Benjamini and Hochberg's false
discovery rate. Co-expression and networks were calculated
according to the cBioPortal online instructions. A log-rank test
was performed to identify the significance of the Spearman's
Correlation coefficient between the mRNA expression z-Scores
(RNASeq V2 RSEM). Spearman's Correlation coefficient >0.5 and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of DEGs
Following the standardization of the microarray
results, 3,816 genes in the GSE63514 dataset, 2,625 genes in the
GSE9750 dataset and 2,093 genes in the GSE7410 dataset were
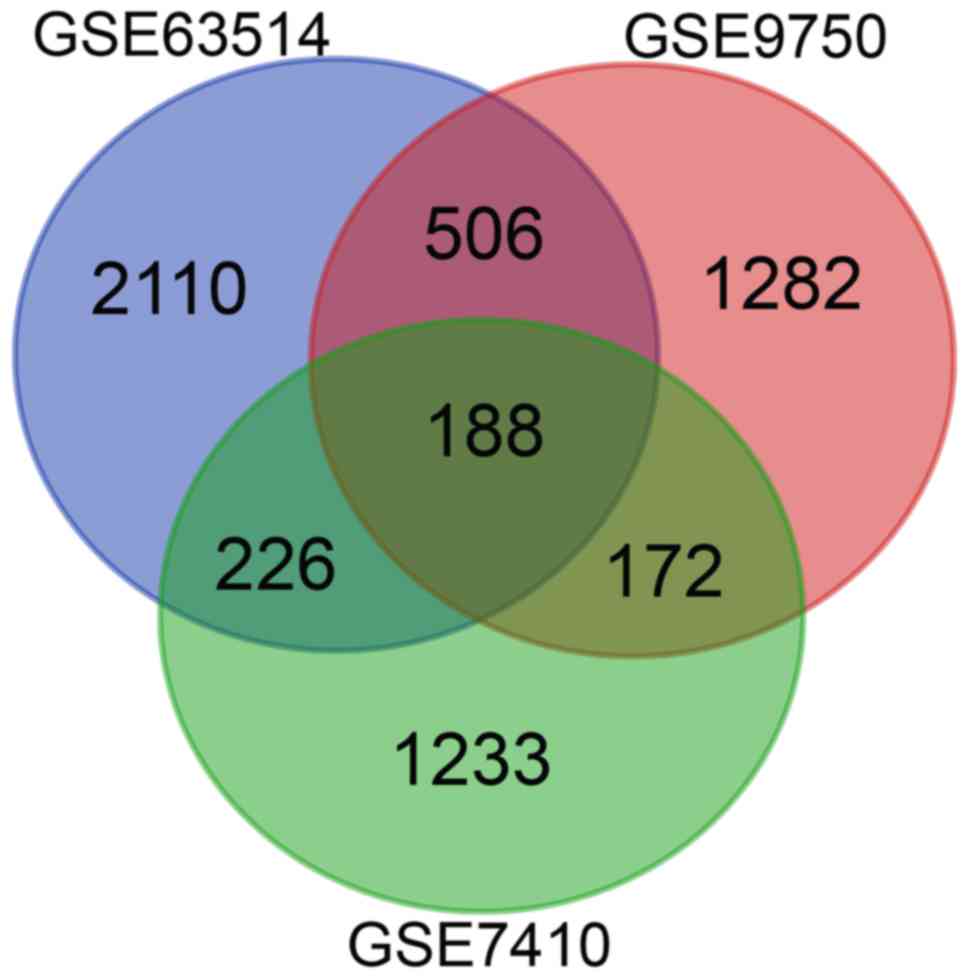
identified as DEGs. The 188 overlapping DEGs amongst the three
datasets were determined using a Venn diagram (Fig. 1), and consisted of 56 downregulated
genes and 132 upregulated genes between normal and cancerous
tissues.
GO and KEGG enrichment analysis of
DEGs
To determine the biological significance of the 188
DEGs, functional and pathway enrichment analyses were performed
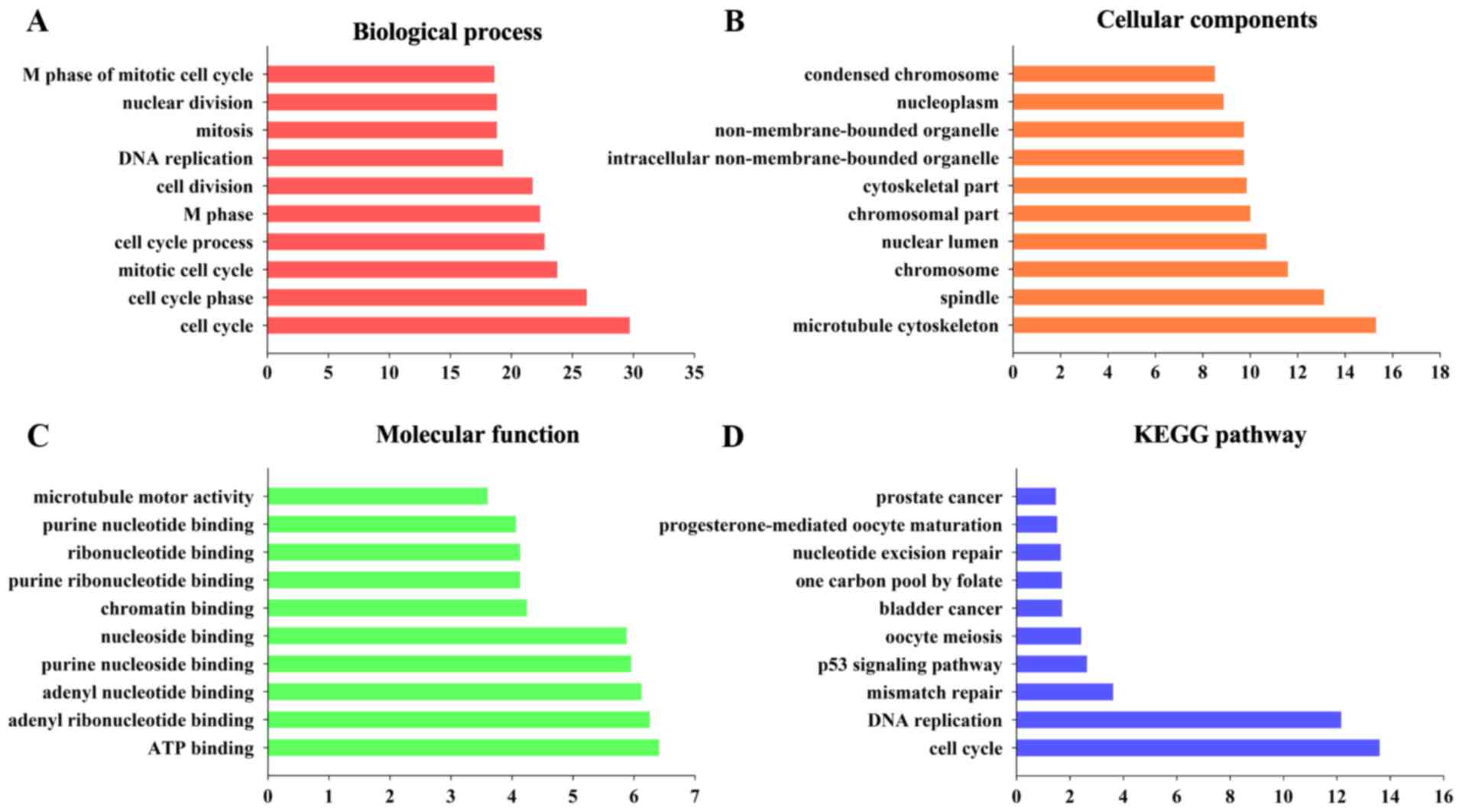
using DAVID. As shown by GO analysis, the DEGs were significantly
enriched in cell cycle, cell cycle phase, mitotic cell cycle, cell
cycle process and M phase (Fig. 2A).
In terms of cellular components, the DEGs were primarily enriched
in microtubule cytoskeleton, spindle, chromosome, nuclear lumen and
chromosomal part (Fig. 2B). The
molecular function terms were primarily enriched in ATP binding,
adenyl ribonucleotide binding, adenyl nucleotide binding, purine
nucleoside binding and nucleoside binding (Fig. 2C). These significantly enriched terms
may assist in improving our understanding of the roles of the DEGs
in the development and progression of CC. In the KEGG analysis, the
DEGs were primarily enriched in cell cycle, DNA replication and
mismatch repair (Fig. 2D).
PPI network construction and
interaction network analysis of the hub genes
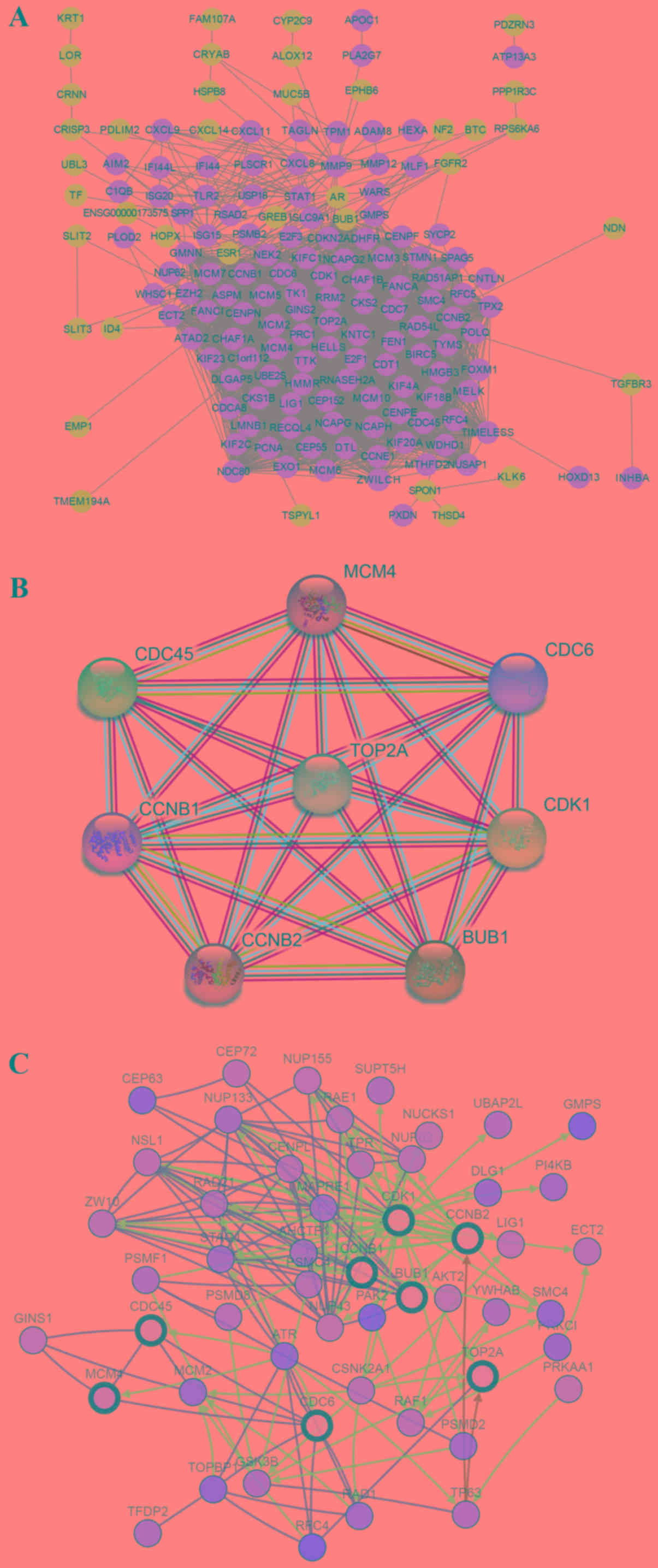
The screened DEGs were uploaded to the STRING
website to reconstruct the PPI network. The resulting network
contained 188 nodes and 2,854 edges (Fig. 3A). Subsequently, hub genes were
filtered using CytoHubba. Cell division cycle 6 (CDC6),
cyclin-dependent kinase 1 (CDK1), cell division control protein 45
(CDC45), budding uninhibited by benzimidazoles 1 (BUB1), TOP2A,
minichromosome maintenance complex component 4 (MCM4), CCNB2 and
CCNB1 were identified as the 8 hub genes, all of which were
imported into STRING to reconstruct the PPI network of 8 nodes and
28 edges (Fig. 3B). A network of the
hub genes and their co-expression genes was then analyzed using the
cBioPortal online platform, which shows the gene-gene interaction
network among the hub genes and the most frequently altered
neighboring co-expression genes (Fig.
3C). These results suggest that the above hub genes may serve a
critical role in the development of CC.
TOP2A mRNA expression and gene
expression correlation analysis
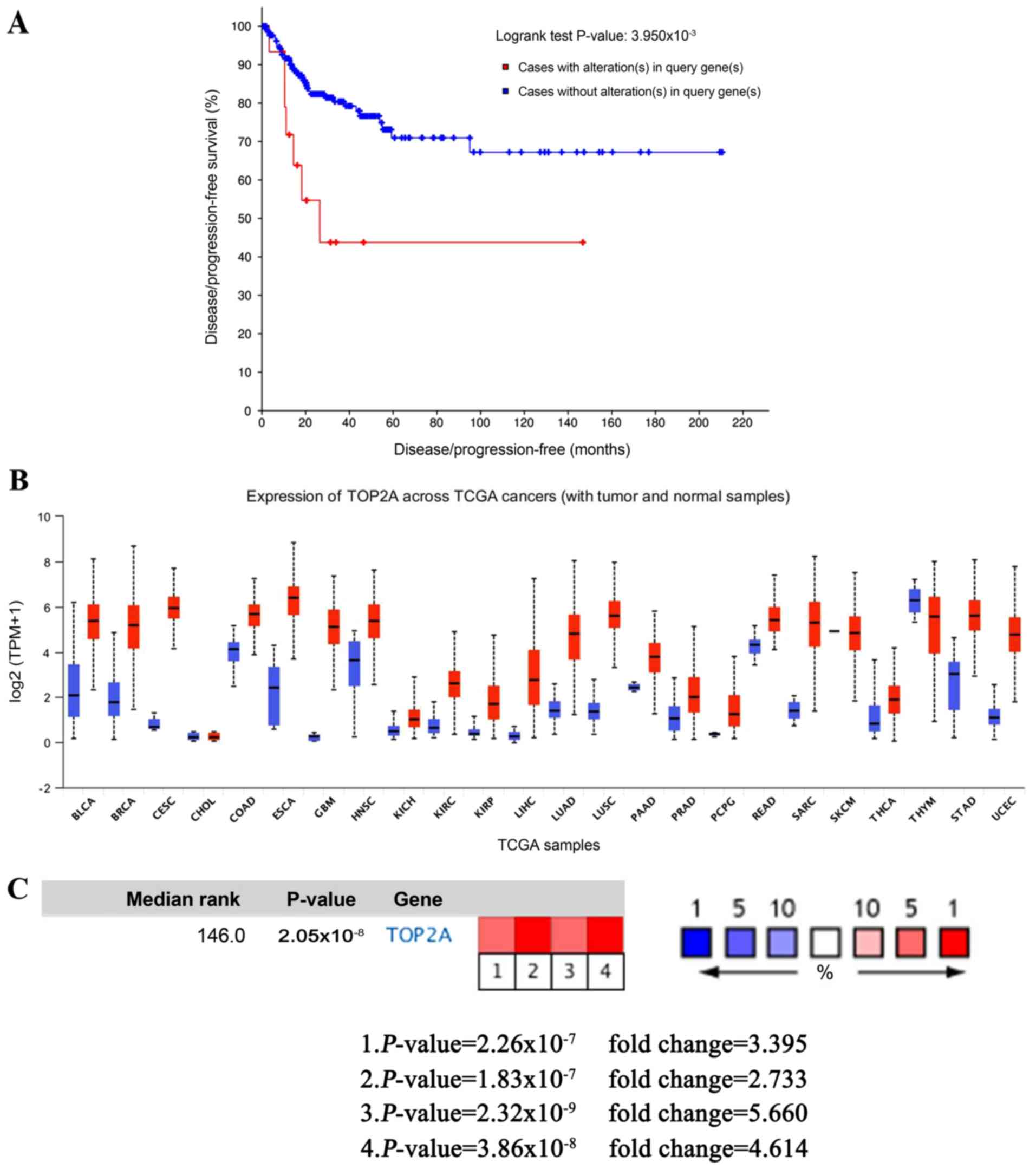
cBioPortal was used to perform the survival analysis
of these hub genes to evaluate their effects on CC. Briefly, only
TOP2A was clearly associated with the prognosis of patients
(Fig. 4A). Patients whose tissues
exhibited upregulated expression levels of TOP2A had significantly
shorter disease/progression-free survival times compared to those
with lower expression levels (P<0.01). The expression profile of
TOP2A in human tissues was displayed using UALCAN. It was found
that the TOP2A mRNA expression in bladder, breast, cervical,
esophageal, liver, kidney and lung cancer, as well as glioblastoma
multiforme, was higher compared with the respective normal tissues
(Fig. 4B). Oncomine analysis of
cancer vs. normal tissue confirmed that TOP2A expression was
significantly upregulated in CC (Fig.
4C). In the Zhai Cervix dataset, higher TOP2A mRNA expression
levels were associated with cancer type and human papillomavirus
(HPV) infection status (Fig.
5A-D).
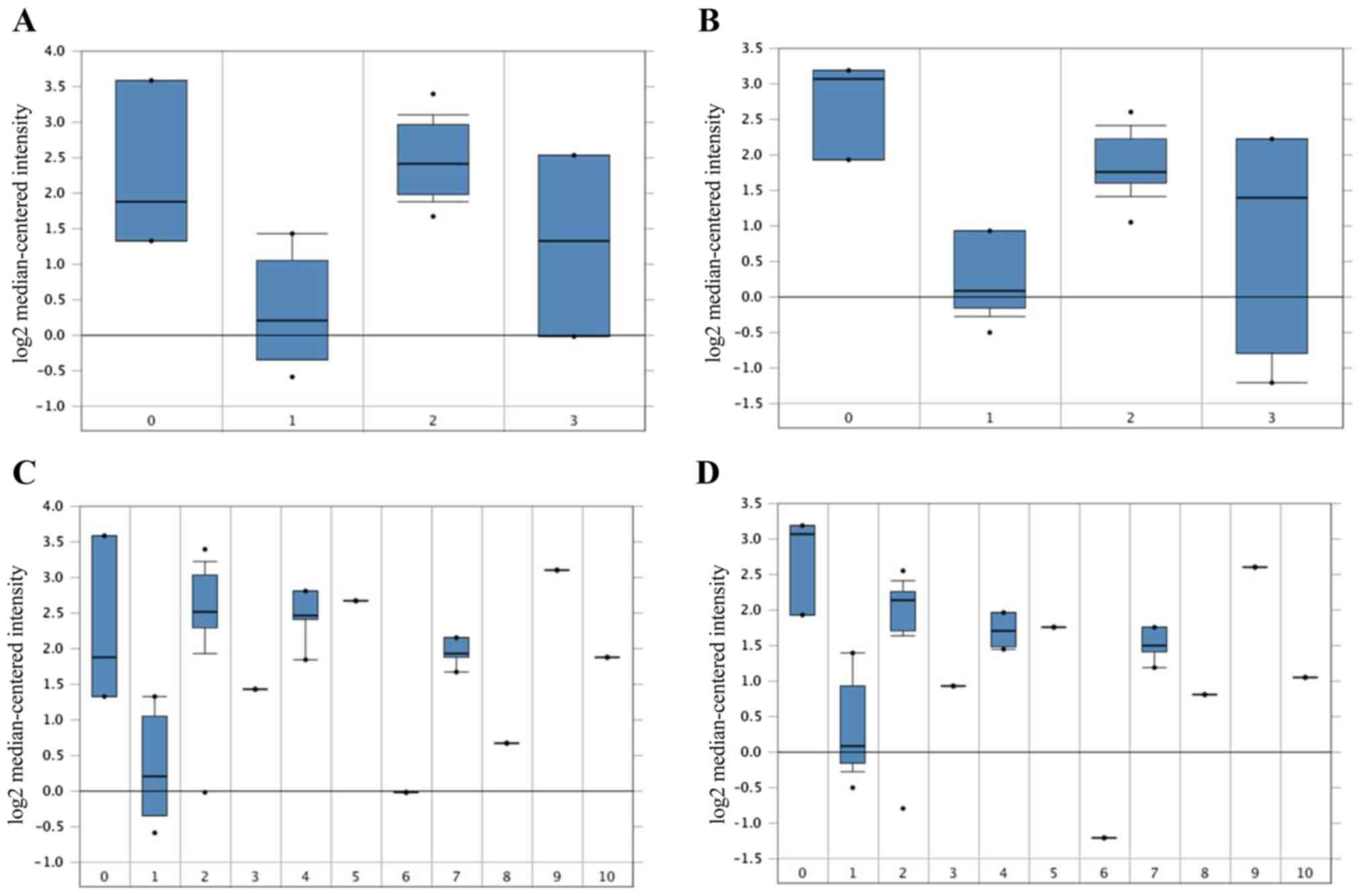 | Figure 5Association between the expression of
TOP2A, cancer type and HPV Infection Status in the 201291_s_at and
201292_at probes in the Zhai Cervix dataset. (A and B) TOP2A mRNA
expression in CC between normal CC and precursor tissues. 0, no
value (3); 1, cervix squamous
epithelium (10); 2, cervical
squamous cell carcinoma (21); 3,
high grade cervical squamous intraepithelial neoplasia (7). (C and D) TOP2A mRNA expression and HPV
Infection Status. 0, no value (3);
1, HPV negative (10); 2, HPV type
16 positive (14); 3, HPV type 18
positive (weak) (1); 4, HPV type 18
positive (4); 5, HPV type 18 and 45
positive (1); 6, HPV type 33
positive (1); 7, HPV type 33, 52 and
58 positive (4); 8, HPV type 56
positive (1); 9, HPV type 58
positive (1); 10, HPV type 59
positive (1). TOP2A, DNA
topoisomerase II α; CC, cervical cancer; HPV, human papillomavirus.
The numbers in the brackets refers to the number of samples. |
TOP2A gene expression correlation analysis results
from the cBioPortal online platform showed that, of the hub genes,
BUB1 (P<0.0001, rs=0.635) and CDK1 (P<0.0001, rs=0.511) had a
close association with TOP2A (Fig.
6A and B). In addition, strong
positive correlations were observed between TOP2A mRNA expression
levels and centromere protein F (CENPF; P<0.0001, rs=0.677), Rac
GTPase activating protein 1 (RACGAP1; P<0.0001, rs=0.612), F-box
protein 5 (FBXO5; P<0.0001, rs=0.585) and BUB1 mitotic
checkpoint serine/threonine kinase B (P<0.0001, rs=0.584;
Fig. 6C-F).
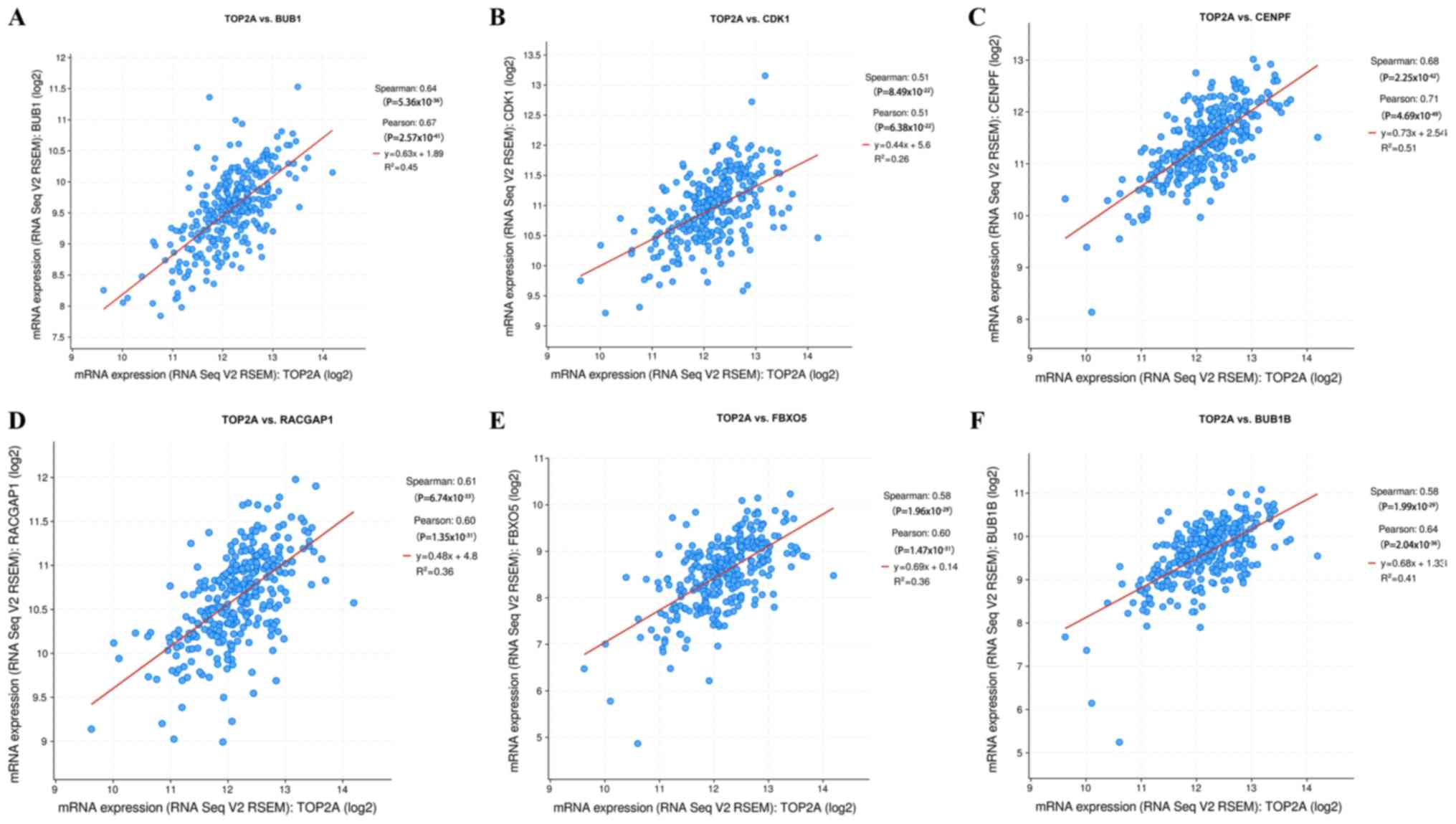 | Figure 6Gene expression correlation analysis
for BUB1, CDK1, CENPF, RACGAP1, FBXO5 and BUB1B with the TOP2A gene
using cBioPortal. The scatter plot shows the Spearman's correlation
of TOP2A expression with the expression of (A) BUB1, (B) CDK1, (C)
CENPF, (D) RACGAP1, (E) FBXO5 and (F) BUB1B. BUB1, budding
uninhibited by benzimidazoles 1; CDK1, cyclin-dependent kinase 1;
CENPF, centromere protein F; RACGAP1, Rac GTPase activating protein
1; FBXO5, F-box protein 5; BUB1B, BUB1 mitotic checkpoint
serine/threonine kinase B; TOP2A, DNA topoisomerase II α. |
Discussion
In the present study the key words ‘cervical cancer’
were used to search the GEO database and screen out the datasets
which contained Homo sapiens mRNA expression profiles of
both CC tissues and normal tissues. A total of three datasets
(GSE63514, GSE9750 and GSE7410) were selected for further
investigation. Among these, GSE63514 and GSE9750 have been analyzed
together (26,27). Other studies have also mined the
GSE63514(28) and GSE9750(29) datasets; only the GSE7410 dataset had
not yet been thoroughly examined previously, to the best of our
knowledge. Therefore, the GEO2R online tool was used to perform the
analyses of the three datasets to determine any potentially
relevant factors.
In the present study, 188 DEGs were identified in
CC, including 132 upregulated and 56 downregulated genes. As shown
by the results of KEGG and GO enrichment analysis, the 188 DEGs
were primarily enriched in cell cycle. Previous studies have
reported that the abnormal regulation of cell cycle process serves
a vital role in the tumorigenesis and progression of several types
of cancer (30-32).
In addition, with regard to GO terms, the DEGs were primarily
enriched in cell cycle, mitotic cell cycle, microtubule
cytoskeleton, spindle, ATP binding and adenyl ribonucleotide
binding, whereas changes in the KEGG pathway were largely enriched
in cell cycle, DNA replication and mismatch repair. Previous
studies have shown the antitumor effect of disrupting the
microtubule cytoskeleton (33,34).
Furthermore, HPV infection facilitating DNA replication and
stimulating DNA damage response are potential important causes of a
range of diseases, including CC (35). Therefore, these potential mechanisms
support the results of the present study. The results of the
present study may improve our understanding of the underlying
mechanisms by which the identified DEGs may promote development or
progression of CC. The other enriched functions and pathways may
also be involved in CC carcinogenesis, and thus should be further
studied.
Cytoscape is an important open source bioinformatics
software platform for visualizing molecular interaction networks.
The CytoHubba plugin was used to screen out core genes for
subsequent prognosis analysis similar to previous studies (36,37).
These hub genes may serve an important role in the development of
CC. Through the PPI network, CDC6, CDK1, CDC45, BUB1, TOP2A, MCM4,
CCNB2 and CCNB1 were further selected as hub genes and these hub
gens were considered important. Subsequently, the hub genes were
chosen for further survival analysis. Only TOP2A was associated
with patient prognosis, and TOP2A dysregulation was significantly
associated with reduced disease-free survival, but not overall
survival (P<0.01). TOP2A is an essential nuclear enzyme for
chromosome condensation, chromatid separation and the relief of
supercoiled DNA during mitosis, and it is crucial for the
segregation of daughter chromosomes at the end of cell division
(38). Upregulated expression of
TOP2A is significantly associated with increased CC cell division
(39) and reduced survival periods
(40-43).
In addition, TOP2A may be used as an immunohistochemical biomarker
for CC (39). Several studies have
confirmed the use of TOP2A as a sensitive biomarker for the
screening of carcinoma tissues (39,44,45).
TOP2A was also frequently overexpressed in several types of cancer,
including lung (46), prostate
(47), breast (42,48),
hepatic (49) and ovarian cancer
(50), and glioma (51). Upregulated expression of TOP2A is
significantly associated with the progression from cervical
intraepithelial neoplasia grade 2 to more advanced cervical lesion
division (38), and the Oncomine
analysis performed in the present study confirmed these results. In
addition, due to HPV infection being the primary etiological factor
of CC, the data in the Zhai Cervix dataset suggested that high-risk
HPV infection resulted in increased TOP2A expression, compared with
no HPV infection. Santin et al (52) showed that TOP2A was coordinately
dysregulated in primary HPV16 and HPV18-infected stage-IB-IIA CC,
potentially representing a common signaling pathway initiated by
HPV transformation. Thus, the encoding enzyme gene is regarded as
the target of several anticancer agents (53-55)
and a variety of mutations in this gene have been associated with
the development of drug resistance (56-58).
To examine the underlying molecular mechanisms in
CC, gene expression correlation analysis was performed in
cBioPortal. CENPF, RACGAP1 FBXO5 and BUB1B had a strong positive
correlation with TOP2A expression, as well as the hub genes BUB1
and CDK1 in CC. Nevertheless, previous studies have not shown
interactions between TOP2A and co-expression genes BUB1, CDK1,
CENPF, RACGAP1, FBXO5 and BUB1B in CC. However, Guo et al
(59) suggested that BUB1, CDK1,
RACGAP1 and TOP2A may be involved in the tumorigenesis of
adrenocortical carcinoma. Li et al (60) showed that lower expression levels of
TOP2A and CENPF were associated with improved overall survival in
patients with bladder cancer. BUB1 is a mitotic checkpoint
serine/threonine kinase that is crucial for physiological
chromosomal segregation and is correlated with cancer stem cell
potential; it may also be a target for anti-breast cancer stem cell
therapies (61). CDK1 is a member of
the Ser/Thr protein kinase family, which is associated with
proliferation and has been demonstrated to eb a prognostic factor
in the development of ovarian cancer (62,63).
Schwermer et al (64)
confirmed a pivotal role of the CDK1/CCNB1 complex in tumor cell
survival. CENPF is a centromere-kinetochore complex and chromosomal
segregation-associated protein during mitosis (65). Preclinical analysis of mouse models
showed that FOXM1 and CENPF were master drug treatment-responsive
genes in prostate cancer (65).
RACGAP1 serves a key role in metastasis, regulating cell
morphology, motility and establishment of cell polarity (66). Furthermore, RACGAP1 has been shown to
function as a potential biomarker, due to its significant
association with poor disease-free and overall survival in
colorectal cancer (67). FBXO5, also
known as EMI1, is an inhibitor of the anaphase promoting
complex/cyclosome (APC/C) and is upregulated in HPV16 E7
oncoprotein-expressing mitotic cells, where it interferes with the
degradation of APC/C substrates in CC (68). BUB1B encoded a kinase involved in
spindle checkpoint function, and also serves a role in inhibiting
the APC/C; the spindle checkpoint function has been shown to be
impaired in several types of cancer (69-71).
Further studies are required to confirm whether TOP2A also
participates in any biological processes associated with cancer
development or progression, due to its strong positive correlation
with FBXO5 and BUB1B.
The present study has some limitations. Firstly,
although upregulated TOP2A mRNA expression levels was a biomarker
for the prognosis of CC, all the data in the present study are
based on online databases; further studies with larger sample sizes
are required to validate these results and to determine potential
targets for diagnosis and treatment of CC. Gene Set Enrichment
Analysis was not performed, which is a more powerful strategy for
functional and pathway enrichment analysis, to assess the
significant enrichment analysis of DEGs; thus, future studies are
required to identify the significant molecular signaling pathways
in CC. Finally, due to the lack of sufficient clinical sample
information, multivariate Cox regression analysis could not be
performed to further elucidate the significance of TOP2A. In future
studies, the underlying mechanisms of TOP2A in CC should be
clarified.
In conclusion, TOP2A may be used a biomarker which
predicts poor prognosis and may serve as a therapeutic target for
treatment of CC. Further studies are required to explore and
demonstrate the potential use of the identified hub genes for the
diagnosis, prognosis and treatment of CC.
Acknowledgements
The authors are grateful for the help provided by Dr
Rui Hou (Harry Perkins Institute of Medical Research, The Western
University of Australia).
Funding
This study was partially supported by the National
Natural Science Foundation of China (grant no. 81702580) and the
Natural Science Foundation of Jiangxi Province (grant nos.
2017ACB21066 and 20171BBG70051).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ, HL and TZ designed the study. QZ, LZ, SH, XX,
and TZ analyzed the data and prepared the figures. QZ and YL
revised the manuscript. HL, YL and JL conceived the study and
interpreted the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9(248)2017.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Zhong TY, Zhou JC, Hu R, Fan XN, Xie XY,
Liu ZX, Lin M, Chen YG, Hu XM, Wang WH, et al: Prevalence of human
papillomavirus infection among 71,435 women in Jiangxi Province,
China. J Infect Public Health. 10:783–788. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: Unresolved issues. Nat
Rev Cancer. 7:11–22. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &
Research Institute at Christiana Care Health Services; HudsonAlpha
Institute for Biotechnology et al. Integrated genomic and
molecular characterization of cervical cancer. Nature. 543:378–384.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
He H, Liu X, Liu Y, Zhang M, Lai Y, Hao Y,
Wang Q, Shi D, Wang N, Luo XG, et al: Human papillomavirus E6/E7
and long noncoding RNA TMPOP2 mutually upregulated gene expression
in cervical cancer cells. J Virol. 93:e01808–e01818.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Zhang J, Yao T, Lin Z and Gao Y: Aberrant
methylation of MEG3 functions as a potential plasma-based biomarker
for cervical cancer. Sci Rep. 7(6271)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen AH, Qin YE, Tang WF, Tao J, Song HM
and Zuo M: MiR-34a and miR-206 act as novel prognostic and therapy
biomarkers in cervical cancer. Cancer Cell Int.
17(63)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yi YX, Fang Yan, Wu KJ, Liu YY and Zhang
W: Comprehensive gene and pathway analysis of cervical cancer
progression. Oncol Lett. 19:3316–3332. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang HJ, Xue JM, Li J, Wan LH and Zhu YX:
Identification of key genes and pathways of diagnosis and prognosis
in cervical cancer by bioinformatics analysis. Mol Genet Genomic
Med. 8(e1200)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Biewenga P, Buist MR, Moerland PD, Ver
Loren van Themaat E, van Kampen AH, ten Kate FJ and Baas F: Gene
expression in early stage cervical cancer. Gynecol Oncol.
108:520–526. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41 (Database
Issue):D991–D995. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8(R183)2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101; discussion 101-103, 119-128, 244-152.
2002.PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res. 41
(Database Issue):D808–D815. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ,
Trimble CL, Fearon ER and Cho KR: Gene expression analysis of
preinvasive and invasive cervical squamous cell carcinomas
identifies HOXC10 as a key mediator of invasion. Cancer Res.
67:10163–10172. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Medi K, Esra G and Kazim YA: Novel genomic
biomarker candidates for cervical cancer as identified by
differential co-expression network analysis. OMICS. 23:261–273.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dai FF, Chen GT, Wang YQ, Zhang L, Long
YM, Yuan MQ, Yang DY, Liu SY, Cheng YX and Zhang LP: Identification
of candidate biomarkers correlated with the diagnosis and prognosis
of cervical cancer via integrated bioinformatics analysis. Onco
Targets Ther. 12:4517–4532. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu JH, Nie SP, Gao M, Jiang Y, Wan YC, Ma
XL, Zhou SL and Cheng WJ: Identification of EPHX2 and RMI2 as two
novel key genes in cervical squamous cell carcinoma by an
integrated bioinformatic analysis. J Cell Physiol. 234:21260–21273.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
van Dam PA, Rolfo C, Ruiz R, Pauwels P,
Van Berckelaer C, Trinh XB, Ferri Gandia J, Bogers JP and Van Laere
S: Potential new biomarkers for squamous carcinoma of the uterine
cervix. ESMO Open. 3(e000352)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Iqbal J, Ejaz SA, Miliutina M, Langer P
and Saeed A: Anti-proliferative effects of chromones: Potent
derivatives affecting cell growth and apoptosis in breast,
bone-marrow and cervical cancer cells. Med Chem. 15:883–891.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao H, Li S, Wang G, Zhao W, Zhang D,
Wang F, Li W and Sun L: Study of the mechanism by which dinaciclib
induces apoptosis and cell cycle arrest of lymphoma Raji cells
through a CDK1-involved pathway. Cancer Med. 8:4348–4358.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chang WH, Forde D and Lai AG: Dual
prognostic role of 2-oxoglutarate-dependent oxygenases in ten
cancer types: Implications for cell cycle regulation and cell
adhesion maintenance. Cancer Commun (Lond). 39(23)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lee JW, Park S, Kim SY, Um SH and Moon EY:
Curcumin hampers the antitumor effect of vinblastine via the
inhibition of microtubule dynamics and mitochondrial membrane
potential in HeLa cervical cancer cells. Phytomedicine. 23:705–713.
2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Saha SK and Khuda-Bukhsh AR: Berberine
alters epigenetic modifications, disrupts microtubule network, and
modulates HPV-18 E6-E7 oncoproteins by targeting p53 in cervical
cancer cell HeLa: A mechanistic study including molecular docking.
Eur J Pharmacol. 744:132–146. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Das D, Bristol ML, Smith NW, James CD,
Wang X, Pichierri P and Morgan IM: Werner Helicase control of human
papillomavirus 16 E1-E2 DNA replication is regulated by SIRT1
deacetylation. mBio. 10(e00263-19)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lou W, Ding B, Xu L and Fan W:
Construction of potential glioblastoma multiforme-related
miRNA-mRNA regulatory network. Front Mol Neurosci.
12(66)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Brown CA, Bogers J, Sahebali S, Depuydt
CE, De Prins F and Malinowski DP: Role of protein biomarkers in the
detection of high-grade disease in cervical cancer screening
programs. J Oncol. 2012(289315)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Peres AL, Paz E Silva KM, de Araújo RF, de
Lima Filho JL, de Melo Júnior MR, Martins DB and de Pontes Filho
NT: Immunocytochemical study of TOP2A and Ki-67 in cervical smears
from women under routine gynecological care. J Biomed Sci.
23(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Smrkolj S, Erzen M and Rakar S: Prognostic
significance of topoisomerase II alpha and collagen IV
immunoexpression in cervical cancer. Eur J Gynaecol Oncol.
31:380–385. 2010.PubMed/NCBI
|
|
41
|
Korkolopoulou P and Vassilakopoulos TP:
Topoisomerase IIalpha as a prognostic factor in mantle cell
lymphoma. Leukemia. 18:1347–1349. 2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fritz P, Cabrera CM, Dippon J, Gerteis A,
Simon W, Aulitzky WE and van der Kuip H: c-erbB2 and topoisomerase
IIalpha protein expression independently predict poor survival in
primary human breast cancer: A retrospective study. Breast Cancer
Res. 7:R374–R384. 2005.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients survival and chemoresistance. Int J Cancer.
124:644–652. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Del Pino M, Svanholm-Barrie C, Torné A,
Marimon L, Gaber J, Sagasta A, Persing DH and Ordi J: mRNA
biomarker detection in liquid-based cytology: A new approach in the
prevention of cervical cancer. Mod. Pathol. 28:312–320.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Scapulatempo-Neto C, Veo C, Fregnani JHTG,
Lorenzi A, Mafra A, Melani AGF, Loaiza EAA, Rosa LAR, de Oliveira
CM, Levi JE and Longatto-Filho A: Characterization of topoisomerase
II α and minichromosome maintenance protein 2 expression in anal
carcinoma. Oncol Lett. 13:1891–1898. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu D, Huang CL, Kameyama K, Hayashi E,
Yamauchi A, Sumitomo S and Yokomise H: Topoisomerase IIalpha gene
expression is regulated by the p53 tumor suppressor gene in
nonsmall cell lung carcinoma patients. Cancer. 94:2239–2247.
2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Schaefer-Klein JL, Murphy SJ, Johnson SH,
Vasmatzis G and Kovtun IV: Topoisomerase 2 alpha cooperates with
androgen receptor to contribute to prostate cancer progression.
PLoS One. 10(e0142327)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hua W, Sa KD, Zhang X, Jia LT, Zhao J,
Yang AG, Zhang R, Fan J and Bian K: MicroRNA-139 suppresses
proliferation in luminal type breast cancer cells by targeting
Topoisomerase II alpha. Biochem Biophys Res Commun. 463:1077–1083.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015(381602)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Erriquez J, Becco P, Olivero M, Ponzone R,
Maggiorotto F, Ferrero A, Scalzo MS, Canuto EM, Sapino A, Verdun di
Cantogno L, et al: TOP2A gene copy gain predicts response of
epithelial ovarian cancers to pegylated liposomal doxorubic in:
TOP2A as marker of response to PLD in ovarian cancer. Gynecol
Oncol. 138:627–633. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Deguchi S, Katsushima K, Hatanaka A,
Shinjo K, Ohka F, Wakabayashi T, Zong H, Natsume A and Kondo Y:
Oncogenic effects of evolutionarily conserved noncoding RNA
ECONEXIN on gliomagenesis. Oncogene. 36:4629–4640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Santin AD, Zhan F, Bignotti E, Siegel ER,
Cané S, Bellone S, Palmieri M, Anfossi S, Thomas M, Burnett A, et
al: Gene expression profiles of primary HPV16- and HPV18-infected
early stage cervical cancers and normal cervical epithelium:
Identification of novel candidate molecular markers for cervical
cancer diagnosis and therapy. Virology. 331:269–291.
2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jain M, Zhang L, He M, Zhang YQ, Shen M
and Kebebew E: TOP2A is overexpressed and is a therapeutic target
for adrenocortical carcinoma. Endocr Relat Cancer. 20:361–370.
2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang H, Yang B, Geng T, Li B, Dai P and
Chen C: Tissue-specific selection of optimal reference genes for
expression analysis of anti-cancer drug-related genes in tumor
samples using quantitative real-time RT-PCR. Exp Mol Pathol.
98:375–381. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani
ME, Ow SG, Nandi S, Lim CT and Thiery JP: Short-term expansion of
breast circulating cancer cells predicts response to anti-cancer
therapy. Oncotarget. 6:15578–15593. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Thougaard AV, Langer SW, Hainau B,
Grauslund M, Juhl BR, Jensen PB and Sehested M: A murine
experimental anthracycline extravasation model: Pathology and study
of the involvement of topoisomerase II alpha and iron in the
mechanism of tissue damage. Toxicology. 269:67–72. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Melaiu O, Cristaudo A, Melissari E, Di
Russo M, Bonotti A, Bruno R, Foddis R, Gemignani F, Pellegrini S
and Landi S: A review of transcriptome studies combined with data
mining reveals novel potential markers of malignant pleural
mesothelioma. Mutat Res. 750:132–140. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Meagher NS, Schuster K, Voss A, Budden T,
Pang CNI, de Fazio A, Ramus SJ and Friedlander ML: Does the primary
site really matter? Profiling mucinous ovarian cancers of uncertain
primary origin (MO-CUP) to personalise treatment and inform the
design of clinical trials. Gynecol Oncol. 150:527–533.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Guo J, Gu Y, Ma X, Zhang L, Li H, Yan Z,
Han Y, Xie L and Guo X: Identification of hub genes and pathways in
adrenocortical carcinoma by integrated bioinformatic analysis. J
Cell Mol Med. 8:4428–4438. 2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li S, Liu X, Liu T, Meng X, Yin X, Fang C,
Huang D, Cao Y, Weng H, Zeng X and Wang X: Identification of
biomarkers correlated with the TNM staging and overall survival of
patients with bladder cancer. Front Physiol. 8(947)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Han JY, Han YK, Park GY, Kim SD, Kim JS,
Jo WS and Lee CG: Corrigendum: Bub1 is required for maintaining
cancer stem cells in breast cancer cell lines. Sci Rep.
6(17984)2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Xi Q, Huang M, Wang Y, Zhong J, Liu R, Xu
G, Jiang L, Wang J, Fang Z and Yang S: The expression of CDK1 is
associated with proliferation and can be a prognostic factor in
epithelial ovarian cancer. Tumour Biol. 36:4939–4948.
2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumorigenesis and progression. Cancer Lett. 362:122–130.
2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Schwermer M, Lee S, Köster J, van Maerken
T, Stephan H, Eggert A, Morik K, Schulte JH and Schramm A:
Sensitivity to cdk1-inhibition is modulated by p53 status in
preclinical models of embryonal tumors. Oncotarget. 6:15425–15435.
2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Mitrofanova A, Aytes A, Zou M, Shen MM,
Abate-Shen C and Califano A: Predicting drug response in human
prostate cancer from preclinical analysis of in vivo mouse models.
Cell Rep. 12:2060–2071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Grewal S, Carver JG, Ridley AJ and Mardon
HJ: Implantation of the human embryo requires Rac1-dependent
endometrial stromal cell migration. Proc Natl Acad Sci USA.
105:16189–16194. 2008.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Imaoka H, Toiyama Y, Saigusa S, Kawamura
M, Kawamoto A, Okugawa Y, Hiro J, Tanaka K, Inoue Y, Mohri Y and
Kusunoki M: RacGAP1 expression, increasing tumor malignant
potential, as a predictive biomarker for lymph node metastasis and
poor prognosis in colorectal cancer. Carcinogenesis. 36:346–354.
2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yu Y and Munger K: Human papillomavirus
type 16 E7 oncoprotein inhibits the anaphase promoting
complex/cyclosome activity by dysregulating EMI1 expression in
mitosis. Virology. 446:251–259. 2013.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Yost S, de Wolf B, Hanks S, Zachariou A,
Marcozzi C, Clarke M, de Voer R, Etemad B, Uijttewaal E, Ramsay E,
et al: Biallelic TRIP13 mutations predispose to Wilms tumor and
chromosome missegregation. Nat Genet. 49:1148–1151. 2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Tan CL, Teissier S, Gunaratne J, Quek LS
and Bellanger S: Stranglehold on the spindle assembly checkpoint:
The human papillomavirus E2 protein provokes BUBR1-dependent
aneuploidy. Cell Cycle. 14:1459–1470. 2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Suematsu T, Li Y, Kojima H, Nakajima K,
Oshimura M and Inoue T: Deacetylation of the mitotic checkpoint
protein BubR1 at lysine 250 by SIRT2 and subsequent effects on
BubR1 degradation during the prometaphase/anaphase transition.
Biochem Biophys Res Commun. 453:588–594. 2014.PubMed/NCBI View Article : Google Scholar
|