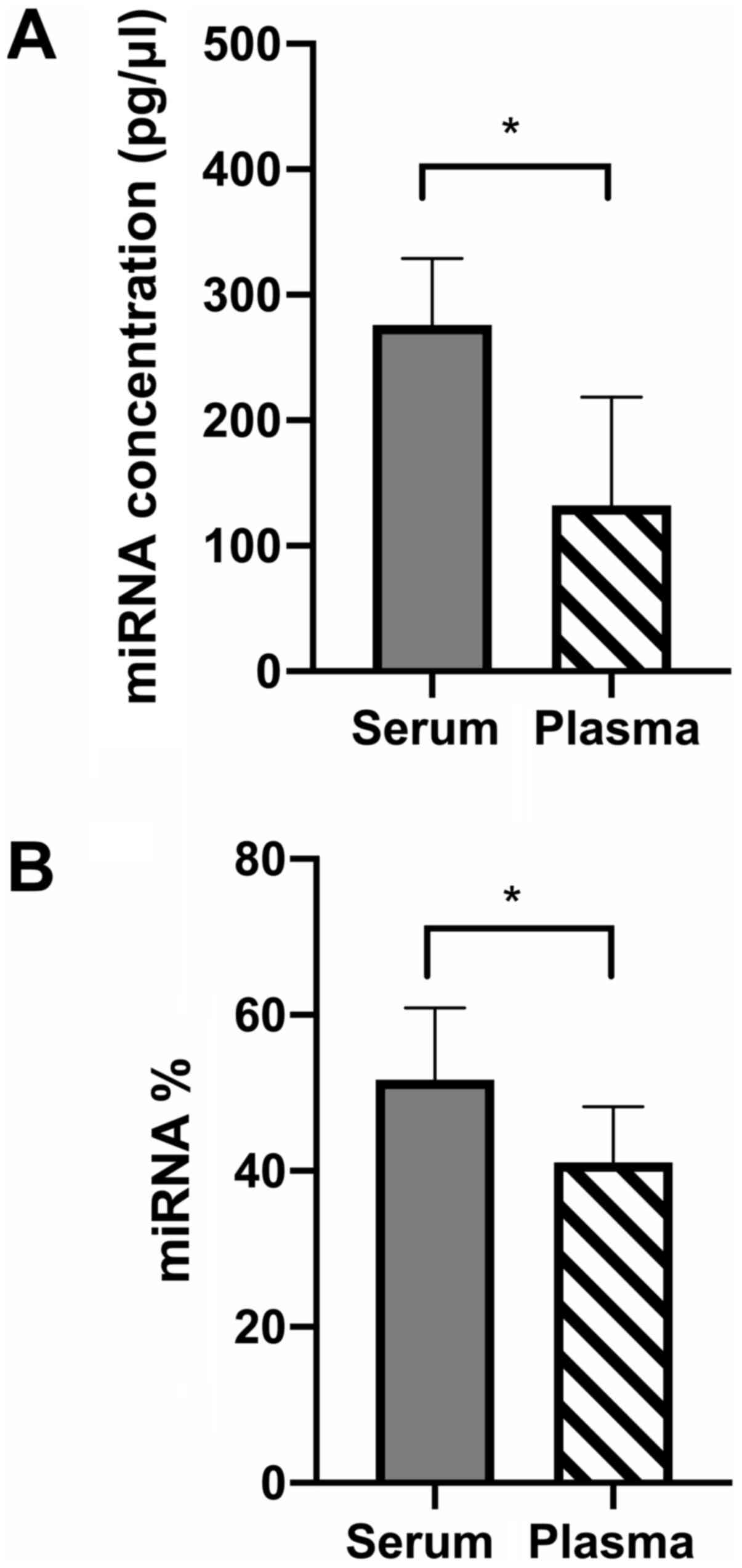
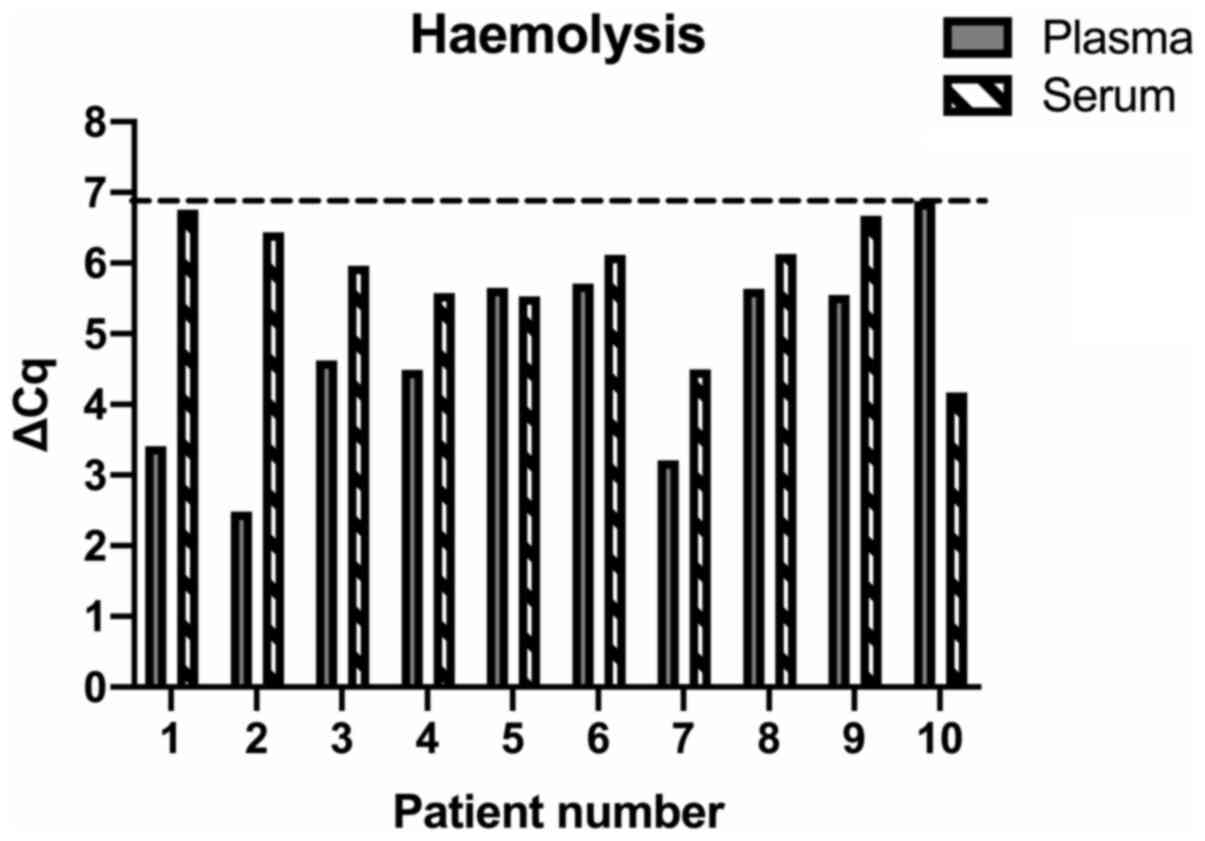
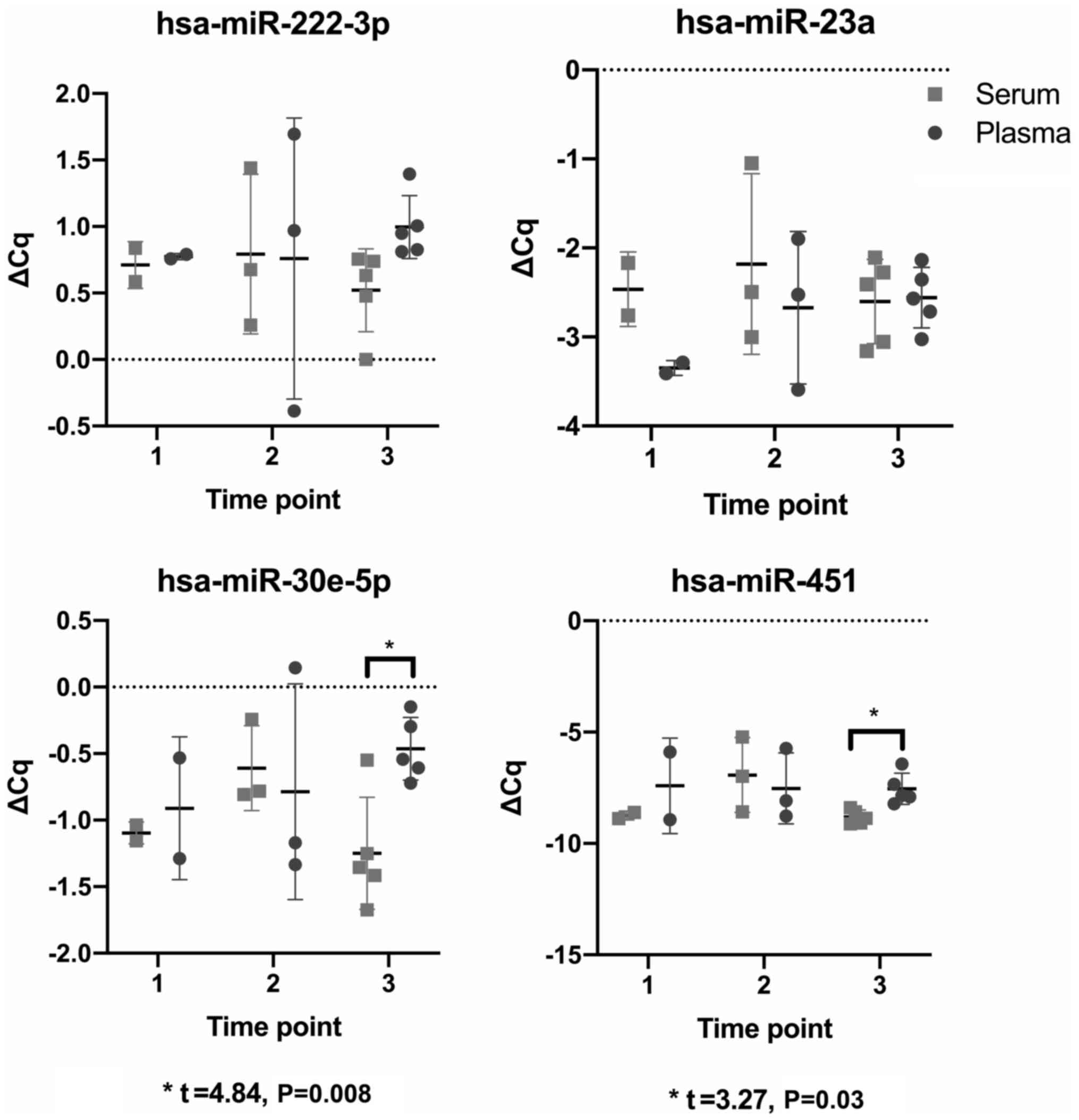
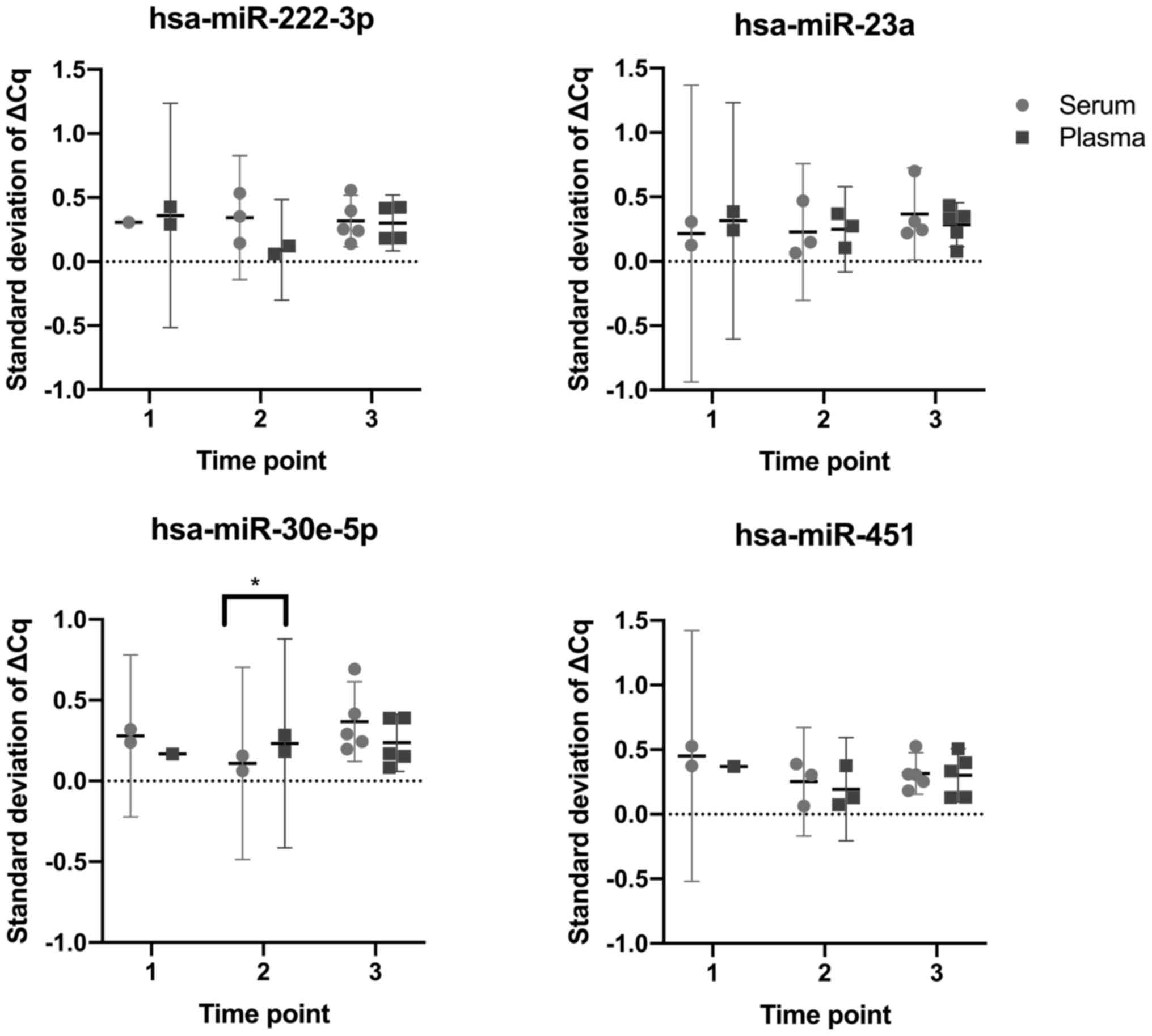
|
1
|
Mishra PJ: MicroRNAs as promising
biomarkers in cancer diagnostics. Biomark Res. 2(19)2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nair VS, Pritchard CC, Tewari M and
Ioannidis JP: Design and analysis for studying micrornas in human
disease: A primer on-omic technologies. Am J Epidemiol.
180:140–152. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen X, Wang L, Qu J, Guan NN and Li JQ:
Predicting miRNA-disease association based on inductive matrix
completion. Bioinformatics. 34:4256–4265. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Papadaki C, Stratigos M, Markakis G,
Spiliotaki M, Mastrostamatis G, Nikolaou C, Mavroudis D and Agelaki
S: Circulating microRNAs in the early prediction of disease
recurrence in primary breast cancer. Breast Cancer Res.
20(72)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rapado-Gonzalez O, Alvarez-Castro A,
Lopez-Lopez R, Iglesias-Canle J, Suarez-Cunqueiro MM and
Muinelo-Romay L: Circulating microRNAs as promising biomarkers in
colorectal cancer. Cancers (Basel). 11(898)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
van Schooneveld E, Wouters MC, Van der
Auwera I, Peeters DJ, Wildiers H, Van Dam PA, Vergote I, Vermeulen
PB, Dirix LY and Van Laere SJ: Expression profiling of cancerous
and normal breast tissues identifies microRNAs that are
differentially expressed in serum from patients with (metastatic)
breast cancer and healthy volunteers. Breast Cancer Res.
14(R34)2012.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang H, Peng R, Wang J, Qin Z and Xue L:
Circulating microRNAs as potential cancer biomarkers: The advantage
and disadvantage. Clin Epigenetics. 10(59)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ghasabi M, Mansoori B, Mohammadi A, Duijf
PH, Shomali N, Shirafkan N, Mokhtarzadeh A and Baradaran B:
MicroRNAs in cancer drug resistance: Basic evidence and clinical
applications. J Cell Physiol. 234:2152–2168. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gomes BC, Rueff J and Rodrigues AS:
MicroRNAs and cancer drug resistance. Methods Mol Biol.
1395:137–162. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11(25)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu
Y, Chen D, Xu J, Huo R, Dai J, et al: Early second-trimester serum
miRNA profiling predicts gestational diabetes mellitus. PLoS One.
6(e23925)2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhu Y, Tian F, Li H, Zhou Y, Lu J and Ge
Q: Profiling maternal plasma microRNA expression in early pregnancy
to predict gestational diabetes mellitus. Int J Gynaecol Obstet.
130:49–53. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hromadnikova I, Kotlabova K, Ivankova K,
Vedmetskaya Y and Krofta L: Profiling of cardiovascular and
cerebrovascular disease associated microRNA expression in umbilical
cord blood in gestational hypertension, preeclampsia and fetal
growth restriction. Int J Cardiol. 249:402–409. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hromadnikova I, Dvorakova L, Kotlabova K
and Krofta L: The Prediction of gestational hypertension,
preeclampsia and fetal growth restriction via the first trimester
screening of plasma exosomal C19MC microRNAs. Int J Mol Sci.
20(2972)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sheikh AM, Small HY, Currie G and Delles
C: Systematic review of Micro-RNA expression in Pre-eclampsia
identifies a number of common pathways associated with the disease.
PLoS One. 11(e0160808)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Skalis G, Katsi V, Miliou A, Georgiopoulos
G, Papazachou O, Vamvakou G, Nihoyannopoulos P, Tousoulis D and
Makris T: MicroRNAs in preeclampsia. Microrna. 8:28–35.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cai M, Kolluru GK and Ahmed A: Small
molecule, big prospects: MicroRNA in pregnancy and its
complications. J Pregnancy. 2017(6972732)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L
and Guo X: Potential role of maternal serum microRNAs as a
biomarker for fetal congenital heart defects. Med Hypotheses.
76:424–426. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Duenas A, Exposito A, Aranega A and Franco
D: The role of non-coding RNA in congenital heart diseases. J
Cardiovasc Dev Dis. 6(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tay JW, James I, Hughes QW, Tiao JY and
Baker RI: Identification of reference miRNAs in plasma useful for
the study of oestrogen-responsive miRNAs associated with acquired
Protein S deficiency in pregnancy. BMC Res Notes.
10(312)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ge Q, Shen Y, Tian F, Lu J, Bai Y and Lu
Z: Profiling circulating microRNAs in maternal serum and plasma.
Mol Med Rep. 12:3323–3330. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Blondal T, Jensby Nielsen S, Baker A,
Andreasen D, Mouritzen P, Wrang Teilum M and Dahlsveen IK:
Assessing sample and miRNA profile quality in serum and plasma or
other biofluids. Methods. 59 (Suppl):S1–S6. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang K, Yuan Y, Cho JH, McClarty S, Baxter
D and Galas DJ: Comparing the MicroRNA spectrum between serum and
plasma. PLoS One. 7(e41561)2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
He S, Zeng S, Zhou ZW, He ZX and Zhou SF:
Hsa-microRNA-181a is a regulator of a number of cancer genes and a
biomarker for endometrial carcinoma in patients: A bioinformatic
and clinical study and the therapeutic implication. Drug Des Devel
Ther. 9:1103–1175. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable Blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Gilad S, Meiri E, Yogev Y, Benjamin S,
Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed
N, et al: Serum microRNAs are promising novel biomarkers. PLoS One.
3(e3148)2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kontomanolis EN, Kalagasidou S and
Fasoulakis Z: MicroRNAs as potential serum biomarkers for early
detection of ectopic pregnancy. Cureus. 10(e2344)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Exiqon: Biofluids Guideline: Analysing
microRNAs in liquid biopsies. urihttps://www.gene-quantification.de/exiqon-biofluids-guidelines-2016.pdfsimplehttps://www.gene-quantification.de/exiqon-biofluids-guidelines-2016.pdf.
|
|
34
|
Rounge TB, Lauritzen M, Langseth H, Enerly
E, Lyle R and Gislefoss RE: microRNA biomarker discovery and
high-throughput DNA sequencing are possible using long-term
archived serum samples. Cancer Epidemiol Biomarkers Prev.
24:1381–1387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kreth S, Hubner M and Hinske LC: MicroRNAs
as clinical biomarkers and therapeutic tools in perioperative
medicine. Anesth Analg. 126:670–681. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Morales-Prieto DM, Ospina-Prieto S,
Chaiwangyen W, Schoenleben M and Markert UR: Pregnancy-associated
miRNA-clusters. J Reprod Immunol. 97:51–61. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bidarimath M, Khalaj K, Wessels JM and
Tayade C: MicroRNAs, immune cells and pregnancy. Cell Mol Immunol.
11:538–547. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Luo SS, Ishibashi O, Ishikawa G, Ishikawa
T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A,
et al: Human villous trophoblasts express and secrete
placenta-specific microRNAs into maternal circulation via exosomes.
Biol Reprod. 81:717–729. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mouillet JF, Ouyang Y, Coyne CB and
Sadovsky Y: MicroRNAs in placental health and disease. Am J Obstet
Gynecol. 213 (4 Suppl):S163–S172. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Moreno-Moya JM, Vilella F and Simon C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Qiagen: miRNeasy Serum/Plasma-Handbook for
miRNeasy Serum/Plasma Kit. Journal.
|
|
44
|
Promega: Maxwell RSC miRNA Plasma and
Serum Kit: Instructions for use of product AS1680. urihttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocolssimplehttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols.
|
|
45
|
Exiqon: Profiling of microRNA in blood
(serum/plasma). Guidelines for the miRCURY LNA Universal RT
microRNA PCR system. urihttps://www.gene-quantification.de/microRNA-serum-plasma-guidelines-exiqon.pdfsimplehttps://www.gene-quantification.de/microRNA-serum-plasma-guidelines-exiqon.pdf.
|
|
46
|
Zou P, Luo L, Zhao C, Chen Z, Dong R, Li
N, Wang Y, Wang J, Wang T, Chen M, et al: The serum microRNA
profile of intrahepatic cholestasis of pregnancy: Identification of
novel noninvasive biomarkers. Cell Physiol Biochem. 51:1480–1488.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Foye C, Yan IK, David W, Shukla N,
Habboush Y, Chase L, Ryland K, Kesari V and Patel T: Comparison of
miRNA quantitation by Nanostring in serum and plasma samples. PLoS
One. 12(e0189165)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Max KEA, Bertram K, Akat KM, Bogardus KA,
Li J, Morozov P, Ben-Dov IZ, Li X, Weiss ZR, Azizian A, et al:
Human plasma and serum extracellular small RNA reference profiles
and their clinical utility. Proc Natl Acad Sci USA.
115:E5334–E5343. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Glinge C, Clauss S, Boddum K, Jabbari R,
Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kääb S, Wakili R,
et al: Stability of circulating Blood-based MicroRNAs-Pre-analytic
methodological considerations. PLoS One.
12(e0167969)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Dufourd T, Robil N, Mallet D, Carcenac C,
Boulet S, Brishoual S, Rabois E, Houeto JL, de la Grange P and
Carnicella S: Plasma or serum? A qualitative study on rodents and
humans using high-throughput microRNA sequencing for circulating
biomarkers. Biol Methods Protoc. 4(bpz006)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
The Early Detection Research Network
(EDRN): Standard Operating Procedure (SOP) for collection of EDTA
plasma. urihttps://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/plasma-sop.pdfsimplehttps://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/plasma-sop.pdf.
|
|
52
|
Qiagen: miRCURY LNA miRNA PCR-Exosomes,
serum/plasma and other biofluid samples handbook. urihttps://www.qiagen.com/fi/resources/resourcedetail?id=7ab5f614-f5d6-4bdc-b22b-246ec3601588&lang=ensimplehttps://www.qiagen.com/fi/resources/resourcedetail?id=7ab5f614-f5d6-4bdc-b22b-246ec3601588&lang=en.
|
|
53
|
Shah JS, Soon PS and Marsh DJ: Comparison
of methodologies to detect low levels of hemolysis in serum for
accurate assessment of serum microRNAs. PLoS One.
11(e0153200)2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Kirschner MB, Edelman JB, Kao SCH, Vallely
MP, van Zandwijk N and Reid G: The impact of hemolysis on Cell-free
microRNA biomarkers. Front Genet. 4(94)2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
GmbH T: TechNote TN-01: The impact of
sample type (serum and EDTA-plasma) and platelet contamination on
osteomiR detection.
|
|
56
|
The Early Detection Research Network
(EDRN): Standard Operating Procedure (SOP) For collection of serum.
urihttps://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/serum-sop.pdfsimplehttps://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/serum-sop.pdf.
|
|
57
|
Promega: Maxwell® RSC miRNA
Plasma and Serum Kit: Instructions for use of product AS1680.
urihttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocolssimplehttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols.
|
|
58
|
Qiagen: RNA Spike-In Kit, For RT handbook.
urihttps://www.qiagen.com/us/products/discovery-and-translational-research/pcr-qpcr-dpcr/qpcr-assays-and-instruments/mirna-qpcr-assay-and-panels/rna-spike-in-kit-for-rt/?clear=true#resourcessimplehttps://www.qiagen.com/us/products/discovery-and-translational-research/pcr-qpcr-dpcr/qpcr-assays-and-instruments/mirna-qpcr-assay-and-panels/rna-spike-in-kit-for-rt/?clear=true#resources.
|
|
59
|
Promega: Maxwell® RSC
Instrument, AS4500. urihttps://www.promega.co.uk/products/lab-automation/maxwell-instruments/maxwell-rsc-instrument/?catNum=AS4500simplehttps://www.promega.co.uk/products/lab-automation/maxwell-instruments/maxwell-rsc-instrument/?catNum=AS4500.
|
|
60
|
Promega: Maxwell® RSC miRNA
Tissue Kit: Instructions for use of product. urihttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocolssimplehttps://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols.
|
|
61
|
Exiqon: miRCURY microRNA QC PCR Panel
Instruction Manual 203887-203892. urihttp://www.exiqon.com/ls/Documents/Scientific/QC-PCR-Panel-Manual.pdfsimplehttp://www.exiqon.com/ls/Documents/Scientific/QC-PCR-Panel-Manual.pdf.
|
|
62
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Yoon H, Belmonte KC, Kasten T, Bateman R
and Kim J: Intra- and Inter-individual Variability of microRNA
levels in human cerebrospinal fluid: Critical implications for
biomarker discovery. Sci Rep. 7(12720)2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Agilent: Bioanalyzer Small RNA Analysis.
urihttps://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-rna-kits-reagents/bioanalyzer-small-rna-analysis-228257#supportsimplehttps://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-rna-kits-reagents/bioanalyzer-small-rna-analysis-228257#support.
|
|
65
|
Masotti A, Caputo V, Prudente S and
Bottazzo GF: Analysis of small RNAs with the Agilent 2100
Bioanalyzer. Application note. Agilent Technologies, 2006.
urihttps://www.agilent.com/cs/library/applications/5989-5215EN.pdfsimplehttps://www.agilent.com/cs/library/applications/5989-5215EN.pdf.
|
|
66
|
Magee R, Telonis AG, Cherlin T, Rigoutsos
I and Londin E: Assessment of isomiR discrimination using
commercial qPCR methods. Noncoding RNA Vol. 3(18)2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Kuchenbauer F, Morin RD, Argiropoulos B,
Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu
AL, et al: In-depth characterization of the microRNA transcriptome
in a leukemia progression model. Genome Res. 18:1787–1797.
2008.PubMed/NCBI View Article : Google Scholar
|
|
68
|
University of Manchester: miRBase: the
microRNA database. urihttp://www.mirbase.org/simplehttp://www.mirbase.org/.
|
|
69
|
Hussing C, Kampmann ML, Mogensen HS,
Børsting C and Morling N: Quantification of massively parallel
sequencing libraries-a comparative study of eight methods. Sci Rep.
8(1110)2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Garcia-Elias A, Alloza L, Puigdecanet E,
Nonell L, Tajes M, Curado J, Enjuanes C, Díaz O, Bruguera J,
Martí-Almor J, et al: Defining quantification methods and
optimizing protocols for microarray hybridization of circulating
microRNAs. Sci Rep. 8(1110)2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
de Ronde MWJ, Ruijter JM, Lanfear D,
Bayes-Genis A, Kok MGM, Creemers EE, Pinto YM and Pinto-Sietsma SJ:
Practical data handling pipeline improves performance of qPCR-based
circulating miRNA measurements. RNA. 23:811–821. 2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Hardikar AA, Farr RJ and Joglekar MV:
Circulating microRNAs: Understanding the limits for quantitative
measurement by real-time PCR. J Am Heart Assoc.
3(e000792)2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010.PubMed/NCBI View Article : Google Scholar
|