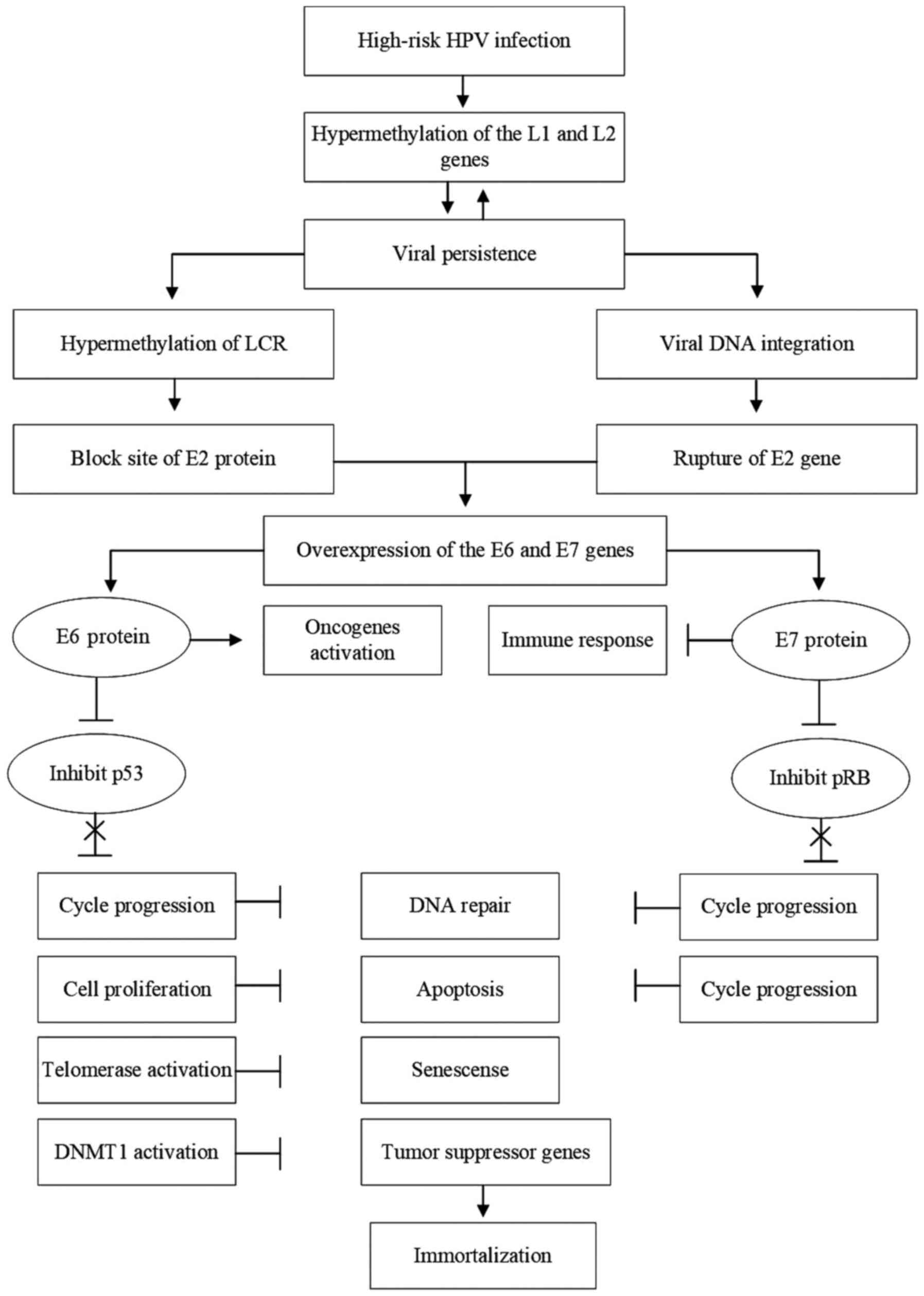
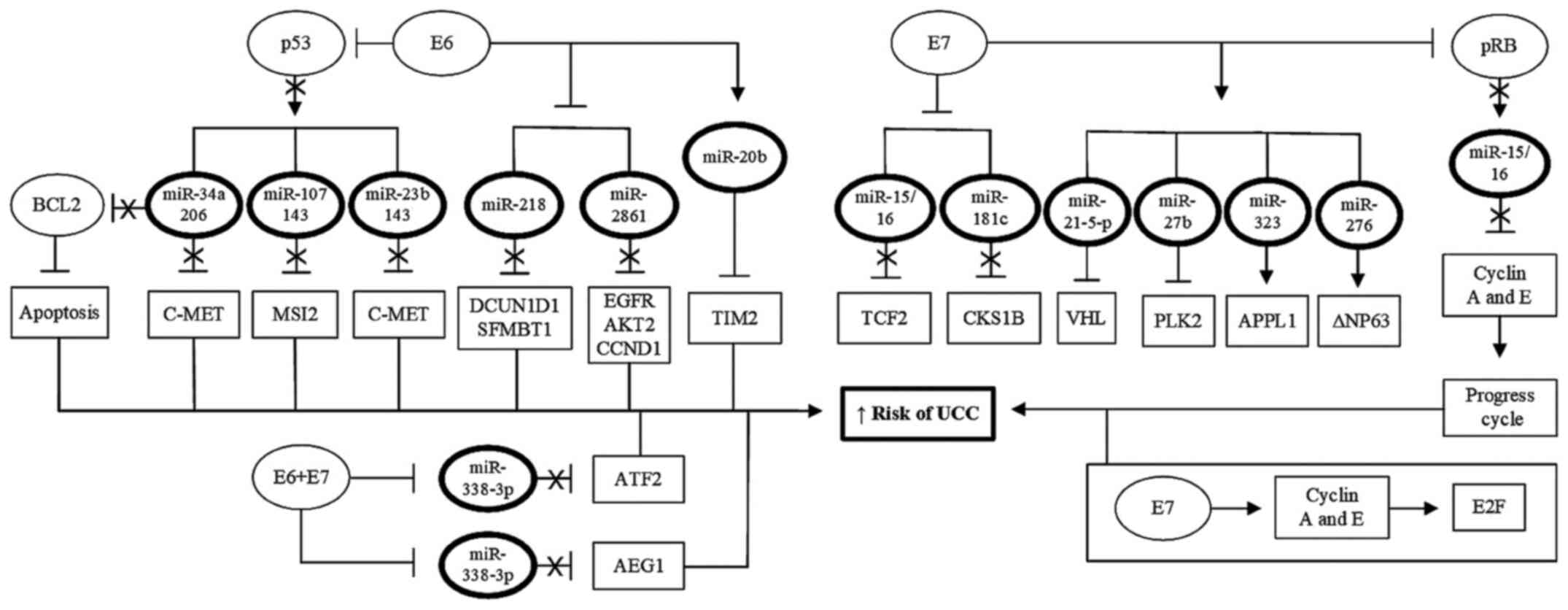
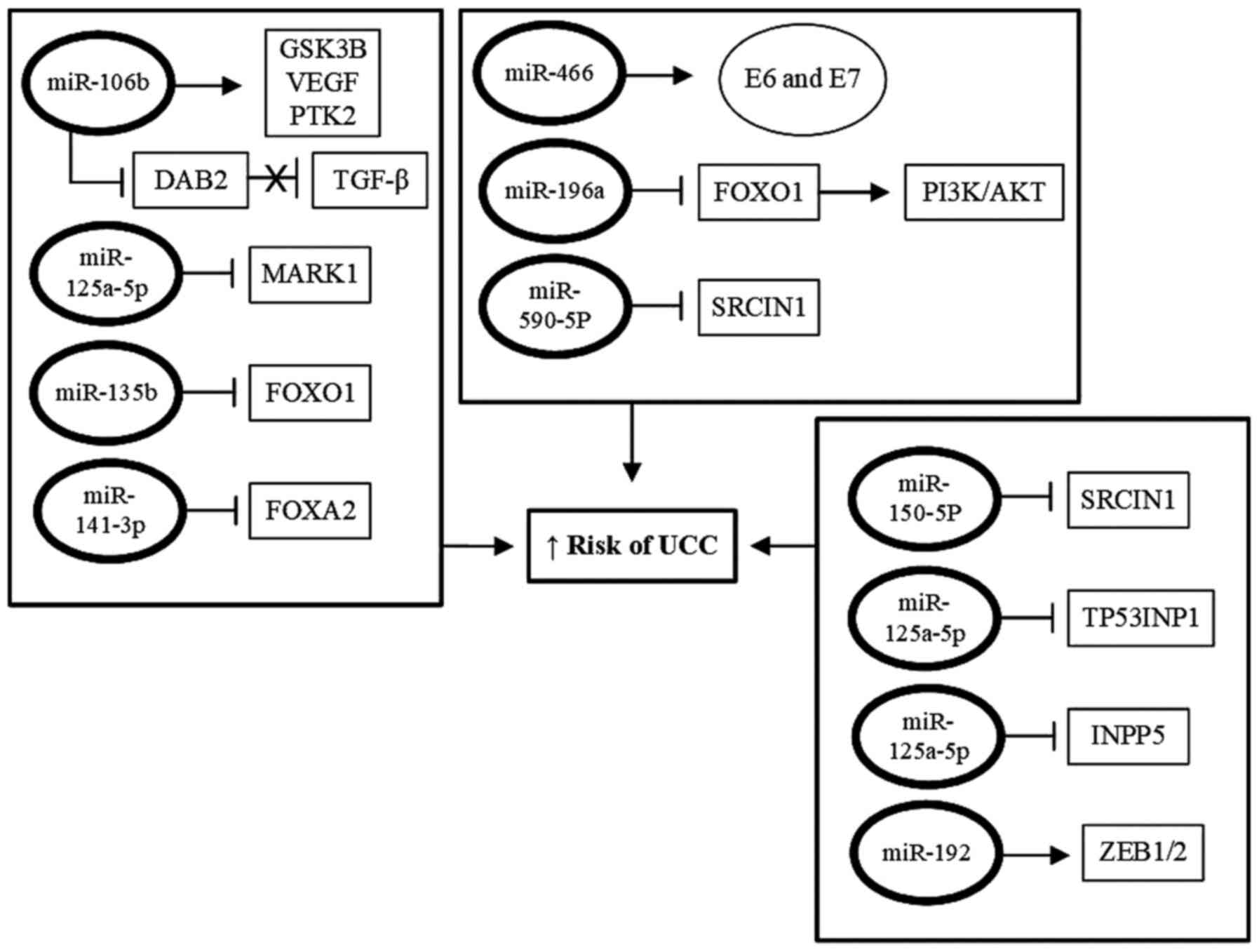
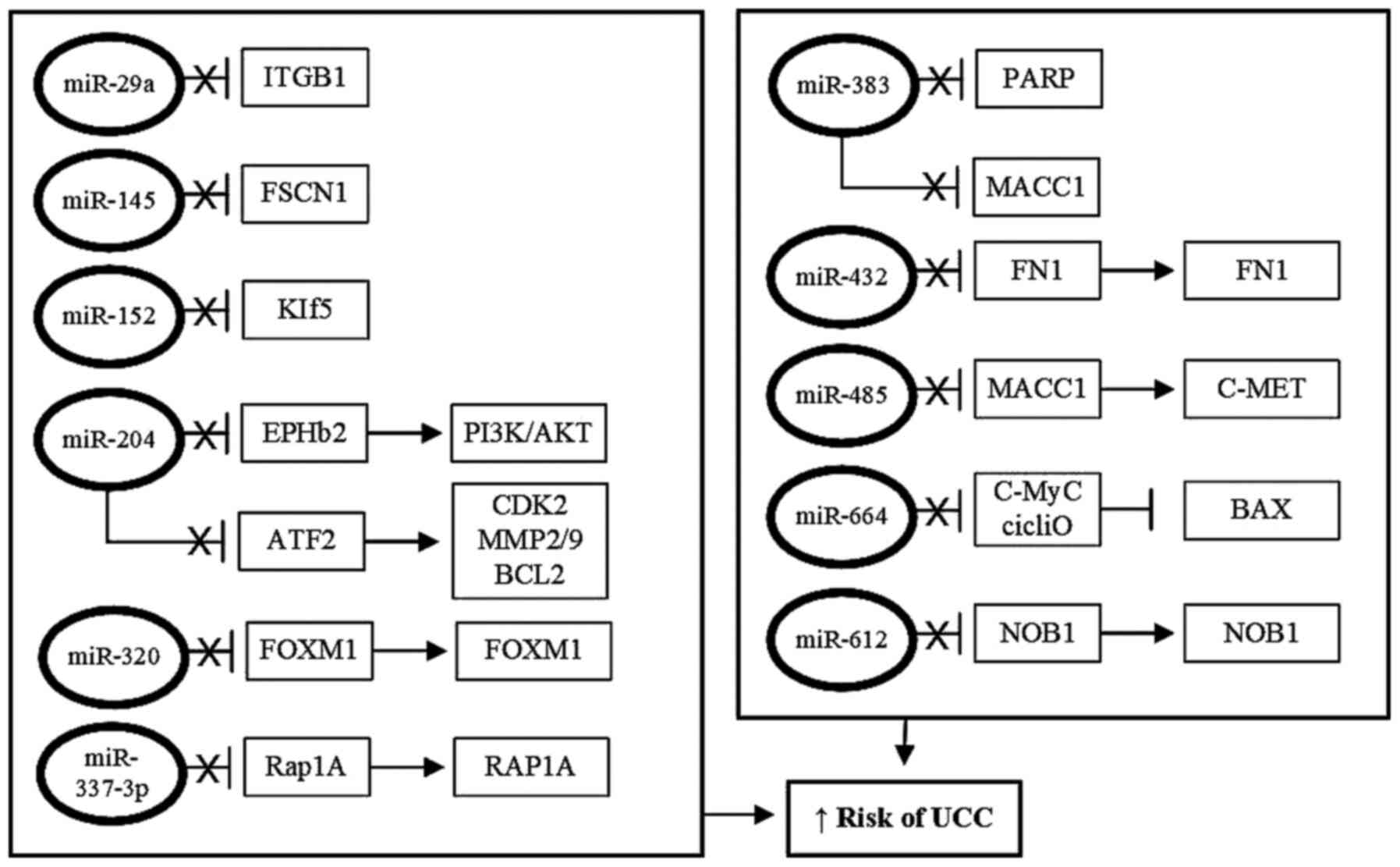
|
1
|
Liang J, Zhang S, Wang W, Xu Y, Kawuli A,
Lu J and Xiu X: Long non-coding RNA DSCAM-AS1 contributes to the
tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3
axis. Biosci Rep. 40(BSR20192061)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
INCA: Estimativa 2018: Incidência de
câncer no Brasil/Instituto Nacional de Câncer Jose Alencar Gomes da
Silva. Coordenação de Prevenção e Vigilância, Rio de Janeiro,
2017.
|
|
5
|
Chen L, Qiu X, Zhang N, Wang Y, Wang M, Li
D, Wang L and Du Y: APOBEC-mediated genomic alterations link
immunity and viral infection during human papillomavirus-driven
cervical carcinogenesis. Biosci Trends. 11:383–388. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sen P, Ganguly P and Ganguly N: Modulation
of DNA methylation by human papillomavirus E6 and E7 oncoproteins
in cervical cancer. Oncol Lett. 15:11–22. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Senapati R, Senapati NN and Dwibedi B:
Molecular mechanisms of HPV mediated neoplastic progression. Infect
Agent Cancer. 11(59)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Steenbergen RD, Snijders PJ, Heideman DA
and Meijer CJ: Clinical implications of (epi)genetic changes in
HPV-induced cervical precancerous lesions. Nat Rev Cancer.
14:395–405. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Morel A, Baguet A, Perrard J, Demeret C,
Jacquin E, Guenat D, Mougin C and Prétet JL: 5azadC treatment
upregulates miR-375 level and represses HPV16 E6 expression.
Oncotarget. 8:46163–46176. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fernandes JV, de Medeiros Fernandes TA, de
Azevedo JC, Cobucci RN, de Carvalho MG, Andrade VS and de Araújo
JM: Link between chronic inflammation and human
papillomavirus-induced carcinogenesis (Review). Oncol Lett.
9:1015–1026. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9(248)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Durzynska J, Lesniewicz K and Poreba E:
Human papillomaviruses in epigenetic regulations. Mutat Res Rev
Mutat Res. 772:36–50. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Amaro-Filho SM, Pereira Chaves CB, Felix
SP, Basto DL, de Almeida LM and Moreira MAM: HPV DNA methylation at
the early promoter and E1/E2 integrity: A comparison between HPV16,
HPV18 and HPV45 in cervical cancer. Papillomavirus Res. 5:172–179.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jacquin E, Baraquin A, Ramanah R,
Carcopino X, Morel A, Valmary-Degano S, Bravo IG, de Sanjosé S,
Riethmuller D, Mougin C and Prétet JL: Methylation of human
papillomavirus type 16 CpG sites at E2-binding site 1 (E2BS1),
E2BS2, and the Sp1-binding site in cervical cancer samples as
determined by high-resolution melting analysis-PCR. J Clin
Microbiol. 51:3207–3215. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Loscalzo J and Handy DE: Epigenetic
modifications: Basic mechanisms and role in cardiovascular disease
(2013 Grover Conference series). Pulm Circ. 4:169–174.
2014.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Shamsi MB, Firoz AS, Imam SN, Alzaman N
and Samman MA: Epigenetics of human diseases and scope in future
therapeutics. J Taibah Univ Med Sci. 212:205–211. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vogt G: Facilitation of environmental
adaptation and evolution by epigenetic phenotype variation:
Insights from clonal, invasive, polyploid, and domesticated
animals. Environ Epigenet. 3(dvx002)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Marsit CJ: Influence of environmental
exposure on human epigenetic regulation. J Exp Biology. 218:71–79.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li Y and Tollefsbol TO: Age-related
epigenetic drift and phenotypic plasticity loss: Implications in
prevention of age-related human diseases. Epigenomics. 8:1637–1651.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zannas AS and Chrousos GP: Epigenetic
programming by stress and glucocorticoids along the human lifespan.
Mol Psychiatry. 22:640–646. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Osborne A: The role of epigenetics in
human evolution. Biosc Horizons. 10(hzx007)2017.
|
|
22
|
Stefansson OA and Esteller M: Epigenetic
modifications in breast cancer and their role in personalized
medicine. Am J Pathol. 183:1052–1063. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Llinàs-Arias P and Esteller M: Epigenetic
inactivation of tumor suppressor coding and non-coding genes in
human cancer: An update. Open Biol. 7(170152)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ramassone A, Pagotto S, Veronese A and
Visone R: Epigenetics and microRNAs in cancer. Int J Mol Sci.
19(459)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kurdyukov S and Bullock M: DNA methylation
analysis: Choosing the right method. Biology (Basel).
5(3)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang X, Hu M, Lyu X, Li C, Thannickal VJ
and Sanders YY: DNA methylation regulated gene expression in organ
fibrosis. Biochim Biophys Acta Mol Basis Dis. 1863:2389–2397.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yang X, Han H, de Carvalho DD, Lay FD,
Jones PA and Liang G: Gene body methylation can alter gene
expression and is a therapeutic target in cancer. Cancer Cell.
26:577–590. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jin Z and Liu Y: DNA methylation in human
diseases. Genes Dis. 5:1–8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Moore LD, Le T and Fan G: DNA Methylation
and its basic function. Neuropsychopharmacology. 38:23–38.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yang HJ: Aberrant DNA methylation in
cervical carcinogenesis. Chin J Cancer. 32:42–48. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lu Q, Ma D and Zhao S: DNA methylation
changes in cervical cancers. Methods Mol Biol. 863:155–176.
2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu G: CDH1 promoter methylation in
patients with cervical carcinoma: A systematic meta-analysis with
trial sequential analysis. Future Oncol. 14:51–63. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Snoek BC, Splunter AP, Bleeker MC, Ruiten
MC, Heideman DA, Rurup WF, Verlaat W, Schotman H, Gent MV, Trommel
NE and Steenbergen RD: Cervical cancer detection by DNA methylation
analysis in urine. Sci Rep. 9(3088)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
McCormick TM, Canedo NH, Furtado YL,
Silveira FA, de Lima RJ, Rosman AD, Almeida Filho GL and Carvalho
Mda G: Association between human papillomavirus and Epstein-Barr
virus DNA and gene promoter methylation of RB1 and CDH1 in the
cervical lesions: A transversal study. Diagn Pathol.
10(59)2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cardoso MF, Castelletti CH, Lima-Filho JL,
Martins DB and Teixeira JA: Putative biomarkers for cervical
cancer: SNVs, methylation and expression profiles. Mutat Res.
773:161–173. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fernandes JV, Araújo JM and Fernandes TA:
Biology and natural history of human papillomavirus infection. Open
Access J Clin Trials. 2013(5)2013.
|
|
38
|
Bashaw AA, Leggatt GR, Chandra J, Tuong ZK
and Frazer IH: Modulation of antigen presenting cell functions
during chronic HPV infection. Papillomavirus Res. 4:58–65.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang M, Wang M, Li X, Xie Y, Xia X, Tian
J, Zhang K and Tang A: Wnt signaling in cervical cancer? J Cancer.
9:1277–1286. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ayala-Calvillo A, Mojica-Vázquer LH,
García-Carrancá A and González-Maya L: Wnt/β-catenin pathway
activation and silencing of the APC gene in HPV-positive human
cervical cancer-derived cells. Mol Med Rep. 17:200–208.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lee J and Kim SS: The function of p27 KIP1
during tumor development. Exp Mol Med. 41:765–771. 2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Qi Q, Ling Y, Zhu M, Zhou L, Wan M, Bao Y
and Liu Y: Promoter region methylation and loss of protein
expression of PTEN and significance in cervical cancer. Biomed Rep.
2:653–658. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li JY, Huang T, Zhang C, Jiang DJ, Hong
QX, Ji HH, Ye M and Duan SW: Association between RASSF1A promoter
hypermethylation and oncogenic HPV infection status in invasive
cervical cancer: A meta-analysis. Asian Pac J Cancer Prev.
16:5749–754. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sherr CJ and Bartek J: Cell cycle-targeted
cancer therapies. Ann Rev Cancer Biol. 1:41–57. 2017.
|
|
45
|
Li X, Tao L, Tan Q, Dong Y, Pan X, Pang L,
Qi Y, Zou H, Liang W, Liu W, et al: CpG island methylation of the
CADM1 gene correlates with cervical carcinogenesis in the Uighur
and Han populations of Xinjiang, China. Int J Clin Exp Pathol.
9:6977–6987. 2016.
|
|
46
|
Holubeková V, Mendelová A, Grendár M,
Meršaková S, Kapustová I, Jašek K, Vaňochová A, Danko J and
Lasabová Z: Methylation pattern of CDH1 promoter and its
association with CDH1 gene expression in cytological cervical
specimens. Oncol Lett. 12:2613–2621. 2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Siegel EM, Riggs BM, Delmas AL, Koch A,
Hakam A and Brown KD: Quantitative DNA methylation analysis of
candidate genes in cervical cancer. PLoS One.
10(e0122495)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lorincz AT: Virtues and weaknesses of DNA
methylation as a test for cervical cancer prevention. Acta Cytol.
60:501–512. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kim MK, Lee IH, Lee KH, Lee YK, So KA,
Hong SR, Hwang CS, Kee MK, Rhee JE, Kang C, et al: DNA methylation
in human papillomavirus-infected cervical cells is elevated in
high-grade squamous intraepithelial lesions and cancer. J Gynecol
Oncol. 27(e14)2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chang CC, Huang RL, Wang HC, Liao YP, Yu
MH and Lai HC: High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR,
SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol
Cancer. 24:201–209. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu L, Xu J, Hu Z, Yang B, Wang L, Lin X,
Xia Z, Zhang Z and Zhu Y: Quantitative DNA methylation analysis of
paired box gene 1 and LIM homeobox transcription factor 1α genes in
cervical cancer. Oncol Lett. 15:4477–4484. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Flamini MI, Gauna GV, Sottile ML, Nadin
BS, Sanchez AM and Vargas-Roig LM: Retinoic acid reduces migration
of human breast cancer cells: Role of retinoic acid receptor beta.
J Cell Mol Med. 18:1113–1123. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Yin FF, Wang N, Bi XN, Yu X, Xu XH, Wang
YL, Zhao CQ, Luo B and Wang YK: Serine/threonine kinases 31(STK31)
may be a novel cellular target gene for the HPV16 oncogene E7 with
potential as a DNA hypomethylation biomarker in cervical cancer.
Virol J. 13(60)2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Molano M, Moreno-Acosta P, Morales N,
Burgos M, Buitrago L, Gamboa O, Alvarez R, Garland SM, Tabrizi SN,
Steenbergen RD and Mejía JC: Association between type-specific HPV
infections and hTERT DNA methylation in patients with invasive
cervical cancer. Cancer Genomics Proteomics. 13:483–491.
2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Johannsen E and Lambert PF: Epigenetics of
human papillomaviruses. Virology. 445:205–212. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Simanaviciene V, Popendikyte V,
Gudleviciene Z and Zvirbliene A: Different DNA methylation pattern
of HPV16, HPV18 and HPV51 genomes in asymptomatic HPV infection as
compared to cervical neoplasia. Virology. 484:227–233.
2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang W, Sun Z, Liu J, Wang G, Lu Z, Zhou
W, Qi T and Ruan Q: Increased methylation of human papillomavirus
type 16 DNA is associated with the severity of cervical lesions in
infected females from northeast China. Oncol Lett. 13:3809–3816.
2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
McBride AA and Warburton A: The role of
integration in oncogenic progression of HPV-associated cancers.
PLoS Pathog. 13(e1006211)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Vinokurova S and von Knebel Doeberitz M:
Differential methylation of the HPV 16 upstream regulatory region
during epithelial differentiation and neoplastic transformation.
PLoS One. 6(e24451)2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
McBride AA: The papillomavirus E2
proteins. Virology. 445:57–79. 2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Filho SM, Bertoni N, Brant AC, Vidal JP,
Felix SP, Cavalcanti SM, Carestiato FN, Martins LF, Almeida LM and
Moreira MA: Methylation at 3'LCR of HPV16 can be affected by
patient age and disruption of E1 or E2 genes. Virus Res. 232:48–53.
2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Fertey J, Hagmann J, Ruscheweyh HJ, Munk
C, Kjaer S, Huson D, Haedicke-Jarboui J, Stubenrauch F and Iftner
T: Methylation of CpG 5962 in L1 of the human papillomavirus 16
genome as a potential predictive marker for viral persistence: A
prospective large cohort study using cervical swab samples. Cancer
Med. 9:1058–1068. 2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Niyazi M, Sui S, Zhu K, Wang L, Jiao Z and
Lu P: Correlation between methylation of human papillomavirus-16 L1
gene and cervical carcinoma in Uyghur women. Gynecol Obstet Invest.
82:22–29. 2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Huang J, Tan ZR, Yu J, Li H, Lv QL, Shao
YY and Zhou HH: DNA hypermethylated status and gene expression of
PAX1/SOX1 in patients with colorectal carcinoma. Onco Targets Ther.
10:4739–4751. 2017.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Luan T, Hua Q, Liu X, Xu P, Gu Y, Qian H,
Yan L, Xu X, Geng R, Zeng X and Li P: PAX1 Methylation as a
potential biomarker to predict the progression of cervical
intraepithelial neoplasia: A Meta-analysis of related studies. Int
J Gynecol Cancer. 27:1480–1488. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lin YW, Tsao CM, Yu PN, Shih YL, Lin CH
and Yan MD: SOX1 suppresses cell growth and invasion in cervical
cancer. Gynecol Oncol. 131:174–181. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Bowden SJ, Kalliala I, Veroniki AA, Arbyn
M, Mitra A, Lathouras K, Mirabello L, Chadeau-Hyam M, Paraskevaidis
E, Flanagan JM and Kyrgiou M: The use of human papillomavirus DNA
methylation in cervical intraepithelial neoplasia: A systematic
review and meta-analysis. EBioMedicine. 50:246–59. 2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Torres-Rojas FI, Alarcón-Romero LC,
Leyva-Vázquez MA, Ortiz-Ortiz J, Mendoza-Catalán MÁ,
Hernández-Sotelo D, Moral-Hernández OD, Rodríguez-Ruiz HA,
Leyva-Illades D, Flores-Alfaro E and Illades-Aguiar B: Methylation
of the L1 gene and integration of human papillomavirus 16 and 18 in
cervical carcinoma and premalignant lesions. Oncol Lett.
15:2278–2286. 2018.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Clarke MA, Gradissimo A, Schiffman M, Lam
J, Sollecito CC, Fetterman B, Lorey T, Poitras N, Raine-Bennett TR,
Castle PE, et al: Human papillomavirus DNA methylation as a
biomarker for cervical precancer: Consistency across 12 genotypes
and potential impact on management of HPV-positive women. Clin
Cancer Res. 24:2194–2202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Shestakova EA and Nakatani Y:
Characterization of histone predeposition complexes from different
cellular compartments. J Investig Genomics. 2:25–28. 2015.
|
|
71
|
Bornelöv S, Reynolds N, Xenophontos M,
Gharbi S, Johnstone E, Floyd R, Ralser M, Signolet J, Loos R,
Dietmann S, et al: The nucleosome remodeling and deacetylation
complex modulates chromatin structure at sites of active
transcription to fine-tune gene expression. Mol Cell. 71:56–72.e4.
2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Feng T, Wang H, Su H, Lu H, Yu L, Zhang X,
Sun H and You Q: Novel N-hydroxyfurylacrylamide-based histone
deacetylase (HDAC) inhibitors with branched CAP group (Part 2).
Bioorg Med Chem. 21:5339–5354. 2013.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Li B, Carey M and Workman JL: The role of
chromatin during transcription. Cell. 128:707–719. 2007.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhang H, Dai X, Qi Y, He Y, Du W and Pang
JJ: Histone deacetylases inhibitors in the treatment of retinal
degenerative diseases: Overview and perspectives. J Ophthalmol.
2015(250812)2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Song C, Zhu S, Wu C and Kang J: Histone
deacetylase (HDAC) 10 suppresses cervical cancer metastasis through
inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J
Biol Chem. 288:28021–2833. 2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Kondo Y, Shen L, Cheng AS, Ahmed S,
Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B,
et al: Gene silencing in cancer by histone H3 lysine 27
trimethylation independent of promoter DNA methylation. Nat Genet.
40:741–750. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
77
|
Hiragami-Hamada K, Xie SQ, Saveliev A,
Uribe-Lewis S, Pombo A and Festenstein R: The molecular basis for
stability of heterochromatin-mediated silencing in mammals.
Epigenetics Chromatin. 2(14)2009.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Scarpini CG, Groves IJ, Pett MR, Ward D
and Coleman N: Virus transcript levels and cell growth rates after
naturally occurring HPV16 integration events in basal cervical
keratinocytes. J Pathol. 233:281–293. 2014.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yeo-Teh NSL, Ito Y and Jha S: High-Risk
human papillomaviral oncogenes E6 and E7 target key cellular
pathways to achieve oncogenesis. Int J Mol Sci.
19(1706)2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Uchida C: Roles of pRB in the regulation
of nucleosome and chromatin structures. Biomed Res Int.
2016(5959721)2016.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Fischer M: Census and evaluation of p53
target genes. Oncogene. 36:3943–3956. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Zhang Y, Dakic A, Chen R, Dai Y, Schlegel
R and Liu X: Direct HPV E6/Myc interactions induce histone
modifications, Pol II phosphorylation, and hTERT promoter
activation. Oncotarget. 8:96323–96339. 2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Johansson C, Jamal Fattah T, Yu H, Nygren
J, Mossberg AK and Schwartz S: Acetylation of intragenic histones
on HPV16 correlates with enhanced HPV16 gene expression. Virology.
482:244–259. 2015.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Zhang L, Yuan C, Wang Y and Zhao S:
Histone deacetylases 3 (HDAC3) is highly expressed in cervical
cancer and inhibited by siRNA. Int J Clin Exp Pathol. 9:3600–3605.
2016.
|
|
85
|
Chen X, Loo JX, Shi X, Xiong W, Guo Y, Ke
H, Yang M, Jiang Y, Xia S, Zhao M, et al: E6 protein expressed by
high-risk HPV activates super-enhancers of the EGFR and c-MET
oncogenes by destabilizing the histone demethylase KDM5C. Cancer.
78:1418–1430. 2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang WT, Han C, Sun YM, Chen TQ and Chen
YQ: Noncoding RNAs in cancer therapy resistance and targeted drug
development. J Hematol Oncol. 12(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Sadri Nahand J, Moghoofei M, Salmaninejad
A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh
MH, Bokharaei-Salim F, Mirzaei H and Hamblin MR: Pathogenic role of
exosomes and microRNAs in HPV-mediated inflammation and cervical
cancer: A review. Int J Cancer. 146:305–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Łaniewski P, Cui H, Roe DJ, Barnes D,
Goulder A, Monk BJ, Greenspan DL, Chase DM and Herbst-Kralovetz MM:
Features of the cervicovaginal microenvironment drive cancer
biomarker signatures in patients across cervical carcinogenesis.
Sci Rep. 9(7333)2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Gupta SM and Mania-Pramanik J: Retraction
note: Molecular mechanisms in progression of HPV-associated
cervical carcinogenesis. J Biomed Sci. 26(50)2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Zheng ZM and Wang X: Regulation of
cellular miRNA expression by human papillomaviruses. Biochim
Biophys Acta. 26:668–677. 2011.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Liu Z, Luo S, Wu M, Huang C, Shi H and
Song X: LncRNA GHET1 promotes cervical cancer progression through
regulating AKT/mTOR and Wnt/β-catenin signaling pathways. Biosci
Rep. 40(BSR20191265)2020.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Fouad YA and Aanei C: Revisiting the
hallmarks of cancer. Am J Cancer Res. 7:1016–1036. 2017.PubMed/NCBI
|
|
93
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Zamani S, Sohrabi A, Hosseini SM,
Rahnamaye-Farzami M and Akbari A: Deregulation of miR-21 and
miR-29a in cervical cancer related to HPV infection. Microrna.
8:110–115. 2019.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Vaschetto LM: miRNA activation is an
endogenous gene expression pathway. RNA Biol. 156:826–828.
2018.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Zhou K, Liu M and Cao Y: New insight into
microRNA fnctions in cancer: Oncogene-microRNA-tumor suppressor
gene network. Front Mol Biosci. 4(46)2017.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Yeung CLA, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 suppresses microRNA-23b expression
in human cervical cancer cells through DNA methylation of the host
gene C9orf3. Oncotarget. 8:12158–12173. 2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Chen AH, Qin YE, Tang WF, Tao J, Song HM
and Zuo M: miR-34a and miR-206 act as novel prognostic and therapy
biomarkers in cervical cancer. Cancer Cell Int.
17(63)2017.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Dong P, Xiong Y, Hanley SJB, Yue J and
Watari H: Musashi-2, a novel oncoprotein promoting cervical cancer
cell growth and invasion, is negatively regulated by p53-induced
miR-143 and miR-107 activation. J Exp Clin Cancer Res.
36(150)2017.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Ofir M, Hacohen D and Ginsberg D: miR-15
and miR-16 are direct transcriptional targets of E2F1 that limit
E2F-induced proliferation by targeting cyclin E. Mol Cancer Res.
9:440–447. 2011.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Chen X, Cao R, Liu H, Zhang T, Yuan X and
Xu S: MicroRNA-15a-5p-targeting oncogene YAP1 inhibits cell
viability and induces cell apoptosis in cervical cancer cells. Int
J Mol Med. 46:1301–1310. 2020.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Zhou M, Chen X, Wu J, He X and Ren R:
MicroRNA-143 regulates cell migration and invasion by targeting
GOLM1 in cervical cancer. Oncol Lett. 16:6393–6400. 2018.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Sannigrahi MK, Sharma R, Singh V, Panda
NK, Rattan V and Khullar M: Role of host miRNA Hsa-miR-139-3p in
HPV-16-induced carcinomas. Clin Cancer Res. 23:3884–3895.
2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Jiang Z, Song Q, Zeng R, Li J, Li J, Lin
X, Chen X, Zhang J and Zheng Y: MicroRNA-218 inhibits EMT,
migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical
cancer. Oncotarget. 7:45622–45636. 2016.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zhu L, Tu H, Liang Y and Tang D: miR-218
produces anti-tumor effects on cervical cancer cells in vitro.
World J Surg Oncol. 16(204)2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
McLaughlin-Drubin ME and Münger K: The
human papillomavirus E7 oncoprotein. Virology. 384:335–344.
2009.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Myklebust MP, Bruland O, Fluge Ø,
Skarstein A, Balteskard L and Dahl O: MicroRNA-15b is induced with
E2F-controlled genes in HPV-related cancer. Br J Cancer.
105:1719–1725. 2011.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Liu Z, Wu M, Shi H, Huang C, Luo S and
Song X: DDN-AS1-miR-15a/16-TCF3 feedback loop regulates tumor
progression in cervical cancer. J Cell Biochem. 120:10228–10238.
2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Cheng Y, Geng L, Zhao L, Zuo P and Wang J:
Human papillomavirus E6-regulated microRNA-20b promotes invasion in
cervical cancer by targeting tissue inhibitor of metalloproteinase
2. Mol Med Rep. 16:5464–5470. 2017.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Kong Q, Wang W and Li P: Regulator role of
HPV E7 protein on miR-21 expression in cervical carcinoma cells and
its functional implication. Int J Clin Exp Pathol. 8:15808–15813.
2015.PubMed/NCBI
|
|
111
|
Cai L, Wang W, Li X, Dong T, Zhang Q, Zhu
B, Zhao H and Wu S: MicroRNA-21-5p induces the metastatic phenotype
of human cervical carcinoma cells in vitro by targeting the
von Hippel-Lindau tumor suppressor. Oncol Lett. 15:5213–5219.
2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Xu L, Xu Q, Li X and Zhang X: MicroRNA-21
regulates the proliferation and apoptosis of cervical cancer cells
via tumor necrosis factor-α. Mol Med Rep. 16:4659–4663.
2017.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Liu F, Zhang S, Zhao Z, Mao X, Huang J, Wu
Z, Zheng L and Wang Q: MicroRNA-27b up-regulated by human
papillomavirus 16 E7 promotes proliferation and suppresses
apoptosis by targeting polo-like kinase2 in cervical cancer.
Oncotarget. 7:19666–19679. 2016.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Ding H, Wu YL, Wang YX and Zhu FF:
Characterization of the microRNA expression profile of cervical
squamous cell carcinoma metastases. Asian Pac J Cancer Prev.
15:1675–1679. 2014.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Park S, Lee S, Kim J, Kim G, Park KH, Kim
TU, Chung D and Lee H: ΔNp63 to TAp63 expression ratio as a
potential molecular marker for cervical cancer prognosis. PLoS One.
14(e0214867)2019.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Coimbra EC, Da Conceição Gomes Leitão M,
Júnior MR, De Oliveira TH, Da Costa Silva Neto J and De Freitas AC:
Expression profile of microRNA-203 and its ΔNp63 target in cervical
carcinogenesis: Prospects for cervical cancer screening. Anticancer
Res. 36:3939–3946. 2016.PubMed/NCBI
|
|
117
|
Mandal P, Saha SS, Sen S, Bhattacharya A,
Bhattacharya NP, Bucha S, Sinha M, Chowdhury RR, Mondal NR,
Chakravarty B, et al: Cervical cancer subtypes harbouring
integrated and/or episomal HPV16 portray distinct molecular
phenotypes based on transcriptome profiling of mRNAs and miRNAs.
Cell Death Discov. 5(81)2019.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Balasubramaniam SD, Balakrishnan V, Oon CE
and Kaur G: Key molecular events in cervical cancer development.
Medicina (Kaunas). 55(384)2019.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Yi Y, Liu Y, Wu W, Wu K and Zhang W: The
role of miR-106p-5p in cervical cancer: From expression to
molecular mechanism. Cell Death Discov. 4(36)2018.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Cheng Y, Guo Y, Zhang Y, You K, Li Z and
Geng L: MicroRNA-106b is involved in transforming growth factor
β1-induced cell migration by targeting disabled homolog 2 in
cervical carcinoma. J Exp Clin Cancer Re. 35(11)2016.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Natalia MA, Alejandro GT, Virginia TJ and
Alvarez-Salas LM: MARK1 is a novel target for miR-125a-5p:
Implications for cell migration in cervical tumor cells. Microrna.
7:54–61. 2018.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Xu Y, Zhao S, Cui M and Wang Q:
Down-regulation of microRNA-135b inhibited growth of cervical
cancer cells by targeting FOXO1. Int J Clin Exp Pathol.
8:10294–10304. 2015.PubMed/NCBI
|
|
123
|
Li JH, Zhang Z, Du MZ, Guan YC, Yao JN, Yu
HY, Wang BJ, Wang XL, Wu SL and Li Z: MicroRNA-141-3p fosters the
growth, invasion, and tumorigenesis of cervical cancer cells by
targeting FOXA2. Arch Biochem Biophys. 657:23–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Zhu J and Han S: miR-150-5p promotes the
proliferation and epithelial-mesenchymal transition of cervical
carcinoma cells via targeting SRCIN1. Pathol Res Pract.
215:738–747. 2019.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Li N, Cui T, Guo W, Wang D and Mao L:
miR-155-5p accelerates the metastasis of cervical cancer cell via
targeting TP53INP1. Onco Targets Ther. 12:3181–3196.
2019.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Yang M, Zhai X, Ge T, Yang C and Lou G:
miR-181a-5p promotes proliferation and invasion and inhibits
apoptosis of cervical cancer cells via regulating inositol
polyphosphate-5-phosphatase A (INPP5A). Oncol Res. 26:703–712.
2018.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Farzanehpour M, Mozhgani SH, Jalilvand S,
Faghihloo E, Akhavan S, Salimi V and Azad TM: Serum and tissue
miRNAs: Potential biomarkers for the diagnosis of cervical cancer.
Virol J. 16(116)2019.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Hou T, Ou J, Zhao X, Huang X, Huang Y and
Zhang Y: MicroRNA-196a promotes cervical cancer proliferation
through the regulation of FOXO1 and p27Kip1. Br J Cancer.
110:1260–1268. 2014.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Varghese VK, Shukla V, Jishnu PV,
Kabekkodu SP, Pandey D, Sharan K and Satyamoorthy K: Characterizing
methylation regulated miRNA in carcinoma of the human uterine
cervix. Life Sci. 232(116668)2019.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Zhou LL, Shen Y, Gong JM, Sun P and Sheng
JH: MicroRNA-466 with tumor markers for cervical cancer screening.
Oncotarget. 8:70821–70827. 2017.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Chu Y, Ouyang Y, Wang F, Zheng A, Bai L,
Han L, Chen Y and Wang H: MicroRNA-590 promotes cervical cancer
cell growth and invasion by targeting CHL1. J Cell Biochem.
115:847–853. 2014.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Guerrero-Gómez AO and Guerrero-Florez M:
MicroRNAs asociados al Cáncer de Cuello Uterino y sus lesiones
precursoras: Una revisión sistemática. Univ Salud. 18:345–363.
2016.
|
|
133
|
Zhang H, Lu Y, Wang S, Sheng X and Zhang
S: MicroRNA-152 acts as tumor suppressor microRNA by inhibiting
Krüppel-like factor 5 in human cervical cancer. Oncol Res.
27:335–340. 2019.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Wang S, Gao B, Yang H, Liu X, Wu X and
Wang W: MicroRNA-432 is downregulated in cervical cancer and
directly targets FN1 to inhibit cell proliferation and invasion.
Oncol Lett. 18:1475–1482. 2019.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Lu Y, Hu J, Sun W, Li S, Deng S and Li M:
miR-29c inhibits cell growth, invasion, and migration of pancreatic
cancer by targeting ITGB1. Onco Targets Ther. 9:99–109.
2015.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Ma L and Li LL: miR-145 contributes to the
progression of cervical carcinoma by directly regulating FSCN1.
Cell Transplant. 28:1299–1305. 2019.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Duan S, Wu A, Chen Z, Yang Y, Liu L and
Shu Q: miR-204 regulates cell proliferation and invasion by
targeting EphB2 in human cervical cancer. Oncol Res. 26:713–723.
2018.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Shu L, Zhang Z and Cai Y: microRNA-204
inhibits cell migration and invasion in human cervical cancer by
regulating transcription factor 12. Oncol Lett. 15:161–166.
2018.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Li N, Guo X, Liu L, Wang L and Cheng R:
Molecular mechanism of miR-204 regulates proliferation, apoptosis
and autophagy of cervical cancer cells by targeting ATF2. Artif
Cells Nanomed Biotechnol. 47:2529–2535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Shi C and Zhang Z: MicroRNA-320 suppresses
cervical cancer cell viability, migration and invasion via directly
targeting FOXM1. Oncol Lett. 14:3809–3816. 2017.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Hong H, Zhu H, Zhao S, Wang K, Zhang N,
Tian Y, Li Y, Wang Y, Lv X, Wei T, et al: The novel
circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression.
Cell Death Dis. 10(950)2019.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Cao XM: Role of miR-337-3p and its target
Rap1A in modulating proliferation, invasion, migration and
apoptosis of cervical cancer cells. Cancer Biomark. 24:257–267.
2019.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Hua FF, Liu SS, Zhu LH, Wang YH, Liang X,
Ma N and Shi HR: miRNA-338-3p regulates cervical cancer cells
proliferation by targeting MACC1 through MAPK signaling pathway.
Eur Rev Med Pharmacol Sci. 21:5342–5352. 2017.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Lu R, Yang Z, Xu G and Yu S: miR-338
modulates proliferation and autophagy by PI3K/AKT/mTOR signaling
pathway in cervical cancer. Biomed Pharmacother. 105:633–644.
2018.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Jayamohan S, Kannan M, Moorthy RK,
Rajasekaran N, Jung HS, Shin YK and Arockiam AJ: Dysregulation of
miR-375/AEG-1 axis by human papillomavirus 16/18-E6/E7 promotes
cellular proliferation, migration, and invasion in cervical cancer.
Front Oncol. 9(847)2019.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Shang A, Zhou C, Bian G, Chen W, Lu W,
Wang W and Li D: miR-381-3p restrains cervical cancer progression
by downregulating FGF7. J Cell Biochem. 120:778–789.
2019.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Teng P, Jiao Y, Hao M and Tang X:
microRNA-383 suppresses the PI3K-AKT-MTOR signaling pathway to
inhibit development of cervical cancer via down-regulating PARP2. J
Cell Biochem. 119:5243–5252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Wang S, Zhang Y, Yuan S and Ji X:
MicroRNA-485 targets MACC1 and inhibits cervical cancer cell
proliferation and invasion. Mol Med Rep. 18:2407–2416.
2018.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Lv M, Ou R, Zhang Q, Lin F, Li X, Wang K
and Xu Y: MicroRNA-664 suppresses the growth of cervical cancer
cells via targeting c-Kit. Drug Des Devel Ther. 13:2371–2379.
2019.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Xu J, Wan X, Chen X, Fang Y, Cheng X, Xie
X and Lu W: miR-2861 acts as a tumor suppressor via targeting
EGFR/AKT2/CCND1 pathway in cervical cancer induced by human
papillomavirus virus 16 E6. Sci Rep. 6(28968)2016.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Jin Y, Zhou X, Yao X, Zhang Z, Cui M and
Lin Y: MicroRNA-612 inhibits cervical cancer progression by
targeting NOB1. J Cell Mol Med. 24:3149–3156. 2020.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Zhang JJ, Wang DD, Du CX and Wang Y: Long
noncoding RNA ANRIL promotes cervical cancer development by acting
as a sponge of miR-186. Oncol Res. 26:345–352. 2018.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Liu S, Song L, Yao H, Zhang L, Xu D, Gao F
and Li Q: miR-375 is epigenetically downregulated by HPV-16 E6
mediated DNMT1 upregulation and modulates EMT of cervical cancer
cells by suppressing lncRNA MALAT1. PLoS One.
11(e0163460)2016.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Song H, Liu Y, Jin X, Liu Y, Yang Y, Li L,
Wang X and Li G: Long non-coding RNA LINC01535 promotes cervical
cancer progression via targeting the miR-214/EZH2 feedback loop. J
Cell Mol Med. 23:6098–6111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9(e100340)2014.PubMed/NCBI View Article : Google Scholar
|
|
156
|
He H, Liu X, Liu Y, Zhang M, Lai Y, Hao Y,
Wang Q, Shi D, Wang N, Luo XG, et al: Human papillomavirus E6/E7
and long noncoding RNA TMPOP2 mutually upregulated gene expression
in cervical cancer cells. J Virol. 93:e01808–e18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Ma Z, Shuai Y, Gao X, Wen X and Ji J:
Circular RNAs in the tumour microenvironment. Mol Cancer.
19(8)2020.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Bach DH, Lee SK and Sood AK: Circular RNAs
in Cancer. Mol Ther Nucleic Acids. 16:118–129. 2019.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Zhao J, Lee EE, Kim J, Yang R, Chamseddin
B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al: Transforming
activity of an oncoprotein-encoding circular RNA from human
papillomavirus. Nat Commun. 10(2300)2019.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X
and Ding Y: Circular RNA circSLC26A4 accelerates cervical cancer
progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids.
19:413–420. 2020.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Chen RX, Liu HL, Yang LL, Kang FH, Xin LP,
Huang LR, Guo QF and Wang YL: Circular RNA circRNA_0000285 promotes
cervical cancer development by regulating FUS. Eur Rev Med
Pharmacol Sci. 23:8771–8778. 2019.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Ding S, Huang X, Zhu J, Xu B, Xu L, Gu D
and Zhang W: ADH7, miR-3065 and LINC01133 are associated with
cervical cancer progression in different age groups. Oncol Lett.
19:2326–2338. 2020.PubMed/NCBI View Article : Google Scholar
|