Programmed death-ligand 1 (PD-L1), with the gene
name CD274, was first discovered in interleukin (IL)-3-deprived
LyD9 (murine hematopoietic progenitor) and 2B4-11 (murine T cell
hybridoma) cell lines in 1992(1) and
was described as B7-H1 by Dong et al (2) in 1999. PD-L1 is the third member of the
B7 family that does not bind CD28, cytotoxic T-lymphocyte A4 or
inducible co-stimulator, and has 10-25% homology with B7.1 and B7.2
proteins (2). PD-L1 is encoded by
the PDCDL1 gene, which was discovered at p24.1 on human chromosome
9. The amino acid sequence of PD-L1 is encoded by 7 exons, which
form a protein of ~40 kDa. PD-L1 is a type I transmembrane protein,
is part of the immunoglobulin (Ig) superfamily and is composed of
IgV-like and IgC-like extracellular domains, a hydrophobic
transmembrane domain and a short cytoplasmic tail composed of 30
amino acids. The signal transduction mechanisms of PD-L1 remain
unclear (3). The most important role
of PD-L1 is binding with programmed death-1 (PD-1; CD279), a type I
transmembrane receptor that is 288 amino acids long and was first
found on T cells (4). The engagement
of PD-L1 and PD-1 on cancer cells activates Src homology region 2
domain-containing phosphatases, which inhibit the T cell receptor
(TCR) pathway. Inhibition of the TCR pathway leads to inhibition of
T cell activities, including proliferation, survival and cytokine
production, such as that of IL-2, tumour necrosis factor α (TNF-α)
and interferon γ (IFN-γ) (5), as
well as the inhibition of B7-1 and T cell tolerance (6,7). To the
best of our knowledge, the present review will discuss for the
first time how autophagy, a protein degradation pathway that
regulates homeostasis of cells, also regulates PD-L1 expression on
cancer cells.
Considering the role of PD-L1 in suppressing the
activation of T cells, it has been an outstanding target for
targeted tumour therapy during the past few years (8). To be specific, obstructing the
PD-L1/PD-1 signalling pathway by antibodies can reactivate
exhausted T cells in the tumour microenvironment, thus making
tumour cells vulnerable to attack by cytotoxic T cells (6). Currently, the efficacy and safety of
drugs targeting the PD-L1/PD-1 axis have been identified in
>1,000 clinical trials, and these drugs have been authorised for
the treatment of various cancer types, including melanoma (9), Hodgkin's lymphoma (10), non-small cell lung cancer (NSCLC)
(11), microsatellite
instability-high renal cell carcinoma (12), deficient mismatch repair cancer
(13), urothelial carcinoma
(14), Merkel cell carcinoma
(15), hepatocellular carcinoma
(16), gastric cancer (17) and head and neck squamous cell
carcinoma (18). Even so, in some
tumour types, such as non-MSI (microsatellite instability)
colorectal cancer, prostate cancer, ovarian cancer and breast
cancer, the clinical efficacy of the drugs targeting PD-1/PD-L1
when used alone was limited (19).
The factors that influenced the effective response of PD-1/PD-L1
include tumour-infiltrating lymphocyte (TIL) infiltration and
localisation at an early stage (20), TIL activation level (21) and the effect of mutations in tumour
cells (22). The expression level of
PD-L1 is the first factor notably associated with prognosis and
clinical effects of the drugs targeted to PD-1/PD-L1 among various
tumours such as melanoma, gastric cancer and NSCLC (23-25).
Among the 45 FDA-approved drugs across 15 tumour types, PD-L1 was
predictive in only 28.9% of cases, and was either not predictive
(53.3%) or not tested (17.8%) in the remaining cases. This
indicates that PD-L1 has some limitations as a predictive
biomarker; however, it still serves a notable role as a biomarker
in bladder, NSCLC, triple-negative breast and cervical cancer
(26). The expression of PD-L1 was
found to be increased among different solid tumours and was
associated with worse survival, prognosis and treatment responses
in multiple malignancies such as esophageal, gastric, urothelial
and colorectal cancer and hepatocellular carcinoma (27,28),
mostly due to the upregulation of PD-L1, which created a favourable
environment for tumour progression by inhibiting antitumour
immunity (29,30). The mechanisms that regulate the
expression of PD-L1 remain unclear. Hence, it is imperative to
broaden understanding of the regulation of PD-L1 expression in
order to improve the therapeutic efficacy of current immune
checkpoint blockade drugs, such as pembrolizumab and nivolumab. In
addition, greater understanding will improve tumour immunotherapy,
which will lead to an improved prognosis in patients with
cancer.
Autophagy is a tightly coordinated process that
isolates misfolded or mutated proteins, damaged or aged organelles
into a double membrane vesicle called an autophagosome.
Autophagosomes fuse with lysosomes, forming autolysosomes, the
contents of which can then be used to degrade the components of the
autolysosomes (31). Nucleotides,
amino acids and other nutrients produced by the aforementioned
degradation process can be recycled by cells. The recycling
capacity of autophagy is conserved from yeast to humans and
regulates cellular homeostasis in both physiological and
pathophysiological contexts. Currently, autophagy has been
classified into 3 forms: i) Macroautophagy, ii) microautophagy and
iii) chaperone-mediated autophagy (CMA). Among these,
macroautophagy is the dominate type of autophagy, and is a common
process for the degradation of cytoplasmic components and
organelles for nutrient recovery in cell recycling. However,
microautophagy is a non-selective degradation process that directly
swallows intracellular components into tubules or lysosomes
(32). CMA is unlike the other two
aforementioned types of autophagy in that it only participates in
the degradation of soluble proteins that contain the KFERQ sequence
motif, such as perilipin2 and pirilipin3(33). After being recognised by the 70-kDa
cytoplasmic heat shock protein (Hsc70), a complex is formed by
these proteins in combination with Hsc70 and its chaperones, which
are transported to the lysosome and interact with the lysosomal
associated membrane protein-2A receptor and are degraded by acid
hydrolase in lysosomes (34).
The process of autophagy is achieved in 4 distinct
stages: i) Initiation, ii) nucleation, iii) maturation and iv)
degradation (35). First is the
process of autophagosome initiation, which is controlled by the
Unc-51-like kinase 1 (ULK)-autophagy-related gene 13 (Atg13)-family
interacting protein 200 kDa kinase complex. This complex is
negatively regulated by the mammalian target of rapamycin complex 1
(mTORC1) via activation of ULK and AMP-activated protein kinase
(AMPK) (36). Second, the phagophore
nucleation step is achieved by the phosphatidylinositol 3-kinase
(PI3K) complex, Beclin 1, Atg14L, Vps (vesicular protein sorting)
15, Vps 34, UV radiation resistance associated gene and Bax
interacting factor 1(36). The
proteins participating in the process of initiation and nucleation
can synergistically facilitate the formation of the double membrane
structure of autophagosomes (36).
The type of membranes can originate from the mitochondria, plasma
membrane or the endoplasmic reticulum (37,38).
Third, is the elongation or expansion step, in which the
Atg5-Atg12-Atg16 complex is formed to elongate the autophagosome
double membranes. Simultaneously, members of the g-aminobutyric
acid receptor-associated protein and LC3 families of proteins are
recruited to the membrane after binding to the lipid
phosphatidylethanolamine (PE). LC3 (Atg8)-I is cleaved by Atg4 and
in turn conjugated with PE by Atg3 and Atg7 to form LC3-II (also
known as MAP1LC3B). LC3-II can serve as a marker to exhibit the
quantity of autophagosomes formed at all points of the process.
Finally, autophagosomes fuse with the lysosomes to degrade the
substances in autophagosomes by the action of acid proteases in
lysosomes. Moreover, this promotes energy efficiency through ATP
generation and attenuates damage to the cell by removing
non-functional proteins and organelles (35).
T cell immunity is essential for homeostasis of the
body, as it identifies antigens and kills cells with gene mutations
or with aberrant pathology, which includes tumour cells.
Unfortunately, excessive activation of uncontrolled T cells can
also kill normal tissue cells, contributing to autoimmune diseases
such as rheumatoid arthritis (51).
Therefore, preventing autoimmunity by regulating activated T cells
is an important feature of immune homeostasis. The co-inhibitory
immune checkpoints, which include CTLA4-CD80, PD-1-PD-L1,
galectin-9-T cell immunoglobulin mucin-3 and TCR-lymphocyte
activation gene 3, can regulate the activity of T cells under
normal physiological conditions (52). However, upregulation of these
inhibitory checkpoints leads to the immune microenvironment
becoming immunosuppressed (53),
which can cause immune tolerance and immune escape. The PD-1-PDL1
axis has been particularly identified as the most clinically
significant, as antibodies against it have led to benefits in a
variety of cancer types such as NSCLC, melanoma and gastric cancer
(9,11,17).
Inhibition of PD-L1 expression on tumour cells heightens
immunosurveillance and decreases the immune checkpoint function
derived from PD-L1(5). Hence, it is
critical to explore the mechanisms that regulate the expression of
PD-L1.
In the past ten years, the mechanisms that regulate
PD-L1 expression via different pathways have been explored. First,
the genomic alternation/rearrangements in chromosome 9p24.1, on
which CD274 is located, have been identified to upregulate PD-L1
expression (54-58).
It has been reported in the literature that amplification and
mutations in the Janus kinase (JAK) family promote the upregulation
of PD-L1 expression by inducing its mRNA expression (55,59). The
increase in activity of the JAK2/STAT signalling pathway resulting
from gene mutations also increases PD-L1 expression (55,59). DNA
double-strand breaks also upregulate PD-L1 expression by activating
the STAT signalling pathway via the kinases ATM/ATM and
Rad3-related/checkpoint kinase 1 (60,61). The
expression of PD-L1 was induced by disrupting the CD274 3'UTR as
well, by using CRISPR technology/Cas9 or miRNAs, such as miR-200,
miR-34a, miR-152 and miR-424 (62-65).
Epigenetic regulation, histone acetylation and methylation boost
PD-L1 expression on melanoma and pancreatic cancer cells (66-68).
In addition, oncogenic transcription factors, for example MYC, can
combine with the PD-L1 promoter to enhance PD-L1 expression in
hepatocellular carcinoma, human melanoma and NSCLC cell lines
(69). Anaplastic lymphoma kinase
can promote PD-L1 expression via STAT3(70). Besides MYC and ALK, the mutated and
amplified HIF1/2α (hypoxia-inducible factor 1/2-α), NF-κB,
phosphatase and tensin homologue/PI3K, mitogen-activated protein
kinase and epidermal growth factor receptor oncogenic pathways can
upregulate PD-L1 mRNA expression on melanoma, ovarian cancers and
lung squamous carcinoma cells (71-76).
In addition, the IFN-γ/JAK/STAT1 inflammatory pathway is used by
cancer cells to enhance PD-L1 mRNA expression (77,78).
Several inflammatory cytokines, such as Toll-like receptor 3,
TNF-α, transforming growth factor β, IFN-α/β and IL-4/6/17/27 have
been demonstrated to upregulate PD-L1 mRNA expression on tumour
cells or tumour-associated stromal cells (78-85).
Post-transcriptional modifications, such as N-linked glycosylation
(86), serine/threonine and tyrosine
phosphorylation (87),
polyubiquitination (88) and
palmitoylation (89) have been found
to serve significant roles in protein stability, as well as PD-L1
translocation regulation.
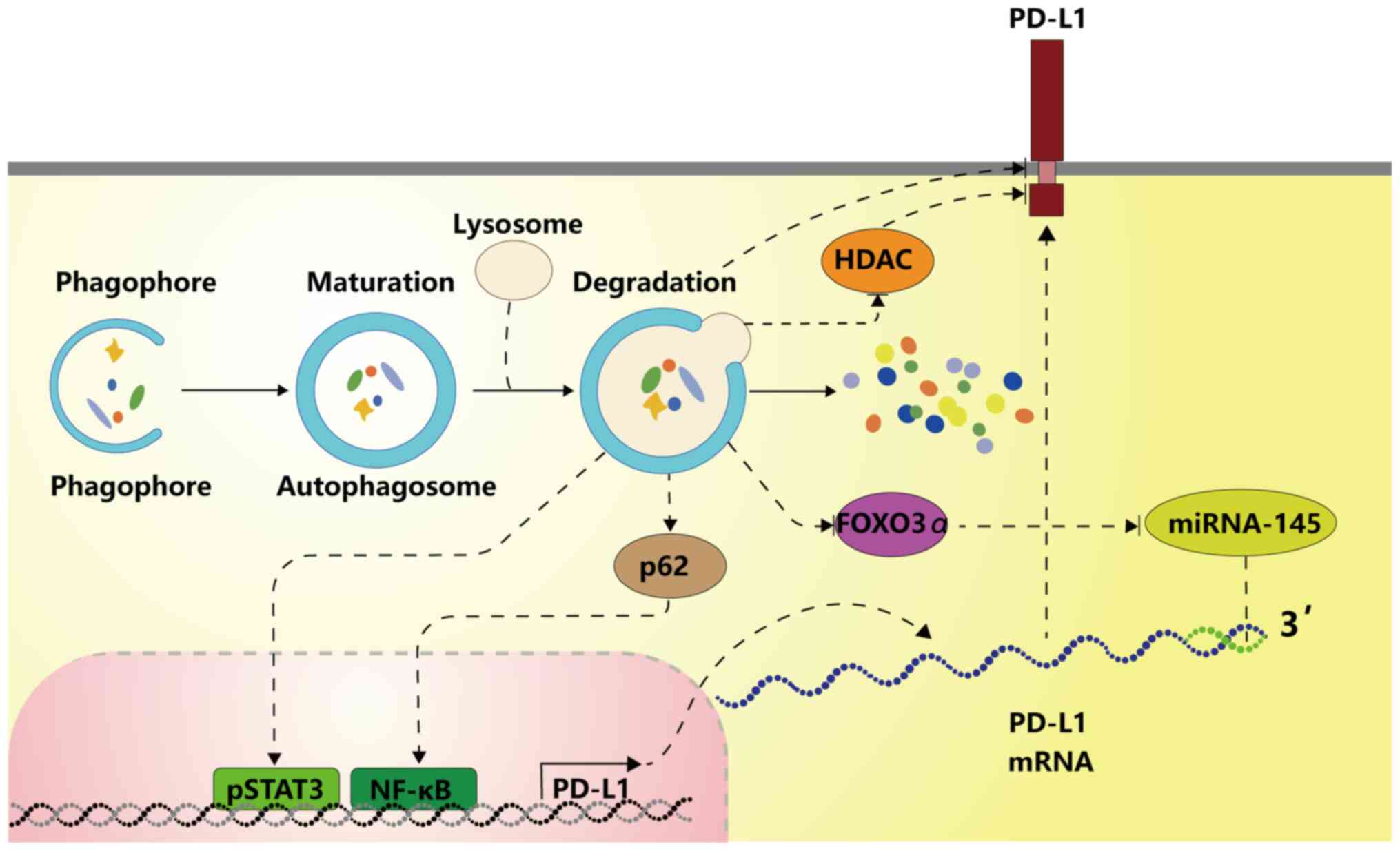
The present study demonstrated that autophagy can be
induced by various molecules to downregulate PD-L1 through NF-kB,
STAT3, HDAC6 or the degradation of autophagic flux. Whether
autophagy can regulate PD-L1 expression through MYC, ALK, HIF1/2,
several inflammatory cytokines, or other mechanisms remains to be
investigated. Oncogenic pathways, such as PI3K/AKT and
Ras/Raf/MEK/ERK, can induce autophagy and also regulate PD-L1
expression, which suggests that these pathways may regulate PD-L1
expression via autophagy.
In addition, the MEK-ERK signalling pathway has been
identified frequently to be activated in a variety of cancer types
such as hepatocellular carcinoma and colon cancer (98,99).
MEK-ERK is located on the outer surface of autophagosomes and
promotes the production of Beclin1 protein by inducing the
lipidation of LC3-I to LC3-II, hence enhancing autophagy (100). In addition, the MEK/ERK module
promotes autophagy via the AMPK-MEK/ERK-TSC-mTOR signalling pathway
(101). Continuous activation of
the Ras/Raf/MEK/ERK pathway may enhance the autophagy flux in
cells, and mRNA levels of LC3B and SQSTM1 are also increased
(102). When the MEK-ERK pathway
was inhibited by chemical or genetic inhibitors, PD-L1
transcription induced by IFN-γ was inhibited in multiple myeloma
cells (103). In concert with this,
when the MEK-ERK signalling pathway was activated by phorbol
myristate acetate, PD-L1 expression was enhanced. By contrast, when
MEK was inhibited in tumours, PD-L1 expression was decreased in
mouse-derived breast cancer cell lines (103,104).
Inhibition of the MEK-ERK signalling pathway abrogates increased
PD-L1 expression stimulated by TLR ligands, which has been observed
in various cancer cells and antigen presenting cells, such as
myeloma, bladder cancer, lymphoma and dendritic cells (94,97,98,103,105-107).
Higher mutation rates of RAS and excessive activation of the RAS
pathway have been demonstrated to increase PD-L1 expression among
human lung and colorectal tumours (108). Besides, autophagy, as a
pro-survival tumour mechanism, can be mediated by PD-L1 to escape
the immune system. It has been reported that PD-L1 induces
autophagy via mTORC signalling to promote the proliferation of
ovarian cancer cells (109). As
aforementioned, activated autophagy may reversely upregulate the
PD-L1 expression through the ATG/autophagy/FOXO3A/miR-145 axis,
forming a positive feedback loop to create a favourable environment
for tumour progression.
As discussed above, the mechanisms that autophagy
uses to regulate PD-L1 expression were reviewed. Autophagy can both
positively and negatively regulate the PD-L1 expression on cancer
cells. However, whether oncogenic pathways, such as PI3K/AKT and
Ras/Raf/MEK/ERK, regulate PD-L1 expression via autophagy needs to
be further elucidated. In addition, the mechanism by which
autophagy effects PD-L1 expression remains to be further clarified
in future studies.
In conclusion, autophagy can regulate PD-L1
expression in a number of cancer types via various mechanisms.
Joint use of autophagy regulators and drugs targeting the
PD-1/PD-L1 axis may enhance the therapeutic effect, hence improving
the prognosis of patients with cancer.
Not applicable.
No funding was received.
Not applicable.
LG was the main contributor to writing the
manuscript and made substantial contributions to the conception and
design. YC was responsible for the modification of the manuscript
structure and language, was given final approval of the manuscript
to be published and agreed to be accountable for all aspects of the
work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved. Both authors have read and approved the final
manuscript. Data sharing is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Ishida Y, Agata Y, Shibahara K and Honjo
T: Induced expression of PD-1, a novel member of the immunoglobulin
gene superfamily, upon programmed cell death. EMBO J. 11:3887–3895.
1992.PubMed/NCBI
|
|
2
|
Dong H, Zhu G, Tamada K and Chen L: B7-H1,
a third member of the B7 family, co-stimulates T-cell proliferation
and interleukin-10 secretion. Nat Med. 5:1365–1369. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Chen J, Jiang CC, Jin L and Zhang XD:
Regulation of PD-L1: A novel role of pro-survival signalling in
cancer. Ann Oncol. 27:409–416. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Boussiotis VA, Chatterjee P and Li L:
Biochemical signaling of PD-1 on T cells and its functional
implications. Cancer J. 20:265–271. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wherry EJ: T cell exhaustion. Nat Immunol.
12:492–499. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Butte MJ, Keir ME, Phamduy TB, Sharpe AH
and Freeman GJ: Programmed death-1 ligand 1 interacts specifically
with the B7-1 costimulatory molecule to inhibit T cell responses.
Immunity. 27:111–122. 2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Latchman YE, Liang SC, Wu Y, Chernova T,
Sobel RA, Klemm M, Kuchroo VK, Freeman GJ and Sharpe AH:
PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting
cells, and host tissues negatively regulates T cells. Proc Natl
Acad Sci USA. 101:10691–10696. 2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Brahmer JR, Rizvi NA, Lutzky J, Khleif S,
Blake-Haskins A, Robbins XLB, Vasselli J, Ibrahim RA and Antonia
SJ: Clinical activity and biomarkers of MEDI4736, an anti-PD-L1
antibody, in patients with NSCLC. J Clin Oncol. 32 (15
Suppl)(S8021)2014.
|
|
9
|
Weber J, Mandala M, Del Vecchio M, Gogas
HJ, Arance AM, Cowey CL, Dalle S, Schenker M, Chiarion-Sileni V,
Marquez-Rodas I, et al: Adjuvant nivolumab versus ipilimumab in
resected stage III or IV melanoma. N Engl J Med. 377:1824–1835.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ansell SM, Lesokhin AM, Borrello I,
Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry
D, Freeman GJ, et al: PD-1 blockade with nivolumab in relapsed or
refractory Hodgkin's lymphoma. N Engl J Med. 372:311–319.
2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Antonia SJ, Villegas A, Daniel D, Vicente
D, Murakami S, Hui R, Yokoi T, Chiappori A, Lee KH, de Wit M, et
al: Durvalumab after chemoradiotherapy in stage III non-small-cell
lung cancer. N Engl J Med. 377:1919–1929. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Overman MJ, McDermott R, Leach JL, Lonardi
S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al:
Nivolumab in patients with metastatic DNA mismatch repair-deficient
or microsatellite instability-high colorectal cancer (CheckMate
142): An open-label, multicentre, phase 2 study. Lancet Oncol.
18:1182–1191. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y,
Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK,
et al: Pembrolizumab as second-line therapy for advanced urothelial
carcinoma. N Engl J Med. 376:1015–1026. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kaufman HL, Russell J, Hamid O, Bhatia S,
Terheyden P, D'Angelo SP, Shih KC, Lebbé C, Linette GP, Milella M,
et al: Avelumab in patients with chemotherapy-refractory metastatic
Merkel cell carcinoma: A multicentre, single-group, open-label,
phase 2 trial. Lancet Oncol. 17:1374–1385. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
El-Khoueiry AB, Sangro B, Yau T, Crocenzi
TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, et al:
Nivolumab in patients with advanced hepatocellular carcinoma
(CheckMate 040): An open-label, non-comparative, phase 1/2 dose
escalation and expansion trial. Lancet. 389:2492–2502.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fuchs CS, Doi T, Jang RW, Muro K, Satoh T,
Machado M, Sun W, Jalal SI, Shah MA, Metges JP, et al: Safety and
efficacy of pembrolizumab monotherapy in patients with previously
treated advanced gastric and gastroesophageal junction cancer:
Phase 2 clinical KEYNOTE-059 trial. JAMA Oncol.
4(e180013)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sunshine J and Taube JM: PD-1/PD-L1
inhibitors. Curr Opin Pharmacol. 23:32–38. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Diem S, Hasan Ali O, Ackermann CJ, Bomze
D, Koelzer VH, Jochum W, Speiser DE, Mertz KD and Flatz L: Tumor
infiltrating lymphocytes in lymph node metastases of stage III
melanoma correspond to response and survival in nine patients
treated with ipilimumab at the time of stage IV disease. Cancer
Immunol Immunother. 67:39–45. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kansy BA, Concha-Benavente F, Srivastava
RM, Jie HB, Shayan G, Lei Y, Moskovitz J, Moy J, Li J, Brandau S,
et al: PD-1 Status in CD8+ T cells associates with
survival and anti-PD-1 therapeutic outcomes in head and neck
cancer. Cancer Res. 77:6353–6364. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hellmann MD, Callahan MK, Awad MM, Calvo
E, Ascierto PA, Atmaca A, Rizvi NA, Hirsch FR, Selvaggi G,
Szustakowski JD, et al: Tumor mutational burden and efficacy of
nivolumab monotherapy and in combination with ipilimumab in
small-cell lung cancer. Cancer Cell. 33:853–861.e4. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Meng X, Huang Z, Teng F, Xing L and Yu J:
Predictive biomarkers in PD-1/PD-L1 checkpoint blockade
immunotherapy. Cancer Treat Rev. 41:868–876. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gandini S, Massi D and Mandalà M: PD-L1
expression in cancer patients receiving anti PD-1/PD-L1 antibodies:
A systematic review and meta-analysis. Crit Rev Oncol Hematol.
100:88–98. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wallis CJD, Lawson K, Butaney M,
Satkunasivam R, Parikh J, Freedland SJ, Patel SP, Hamid O, Pal SK
and Klaassen Z: Association between PD-L1 status and immune
checkpoint inhibitor response in advanced malignancies: A
systematic review and meta-analysis of overall survival data. Jpn J
Clin Oncol. 50:800–809. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Davis AA and Patel VG: The role of PD-L1
expression as a predictive biomarker: An analysis of all US Food
and Drug Administration (FDA) approvals of immune checkpoint
inhibitors. J Immunother Cancer. 7(278)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu P, Wu D, Li L, Chai Y and Huang J:
PD-L1 and survival in solid tumors: A meta-analysis. PLoS One.
10(e0131403)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pyo JS, Kang G and Kim JY: Prognostic role
of PD-L1 in malignant solid tumors: A meta-analysis. Int J Biol
Markers. 32:e68–e74. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen L and Han X: Anti-PD-1/PD-L1 therapy
of human cancer: Past, present, and future. J Clin Invest.
125:3384–3391. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Behrends C, Sowa ME, Gygi SP and Harper
JW: Network organization of the human autophagy system. Nature.
466:68–76. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sosman JA, Kim KB, Schuchter L, Gonzalez
R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ,
Flaherty KT, et al: Survival in BRAF V600-mutant advanced melanoma
treated with vemurafenib. N Engl J Med. 366:707–714.
2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kaushik S and Cuervo AM: The coming of age
of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 19:365–381.
2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dash S, Aydin Y and Moroz K:
Chaperone-mediated autophagy in the liver: Good or bad? Cells.
8(1308)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Feng Y, He D, Yao Z and Klionsky DJ: The
machinery of macroautophagy. Cell Res. 24:24–41. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shibutani ST and Yoshimori T: A current
perspective of autophagosome biogenesis. Cell Res. 24:58–68.
2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hailey DW, Rambold AS, Satpute-Krishnan P,
Mitra K, Sougrat R, Kim PK and Lippincott-Schwartz J: Mitochondria
supply membranes for autophagosome biogenesis during starvation.
Cell. 141:656–667. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huang J, Sun R, Qi X, Liu L, Yang Y and
Sun B: Effect of autophagy on expression of neutrophil programmed
death ligand-1 in mice with sepsis. Zhonghua Wei Zhong Bing Ji Jiu
Yi Xue. 31:1091–1096. 2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
40
|
Booth L, Roberts JL, Poklepovic A and Dent
P: (Pemetrexed + sildenafil), via autophagy-dependent HDAC
downregulation, enhances the immunotherapy response of NSCLC cells.
Cancer Biol Ther. 18:705–714. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Dent P, Booth L, Roberts JL, Poklepovic A
and Hancock JF: (Curcumin + sildenafil) enhances the efficacy of
5FU and anti-PD1 therapies in vivo. J Cell Physiol. 235:6862–6874.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen MC, Lin YC, Liao YH, Liou JP and Chen
CH: MPT0G612, a Novel HDAC6 inhibitor, induces apoptosis and
suppresses IFN-γ-induced programmed death-ligand 1 in human
colorectal carcinoma cells. Cancers (Basel).
11(1617)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Booth L, Roberts JL, West C, Von Hoff D
and Dent P: GZ17-6.02 initiates DNA damage causing
autophagosome-dependent HDAC degradation resulting in enhanced
anti-PD1 checkpoint inhibitory antibody efficacy. J Cell Physiol.
235:8098–8113. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Booth L, Roberts JL, Poklepovic A,
Avogadri-Connors F, Cutler RE, Lalani AS and Dent P: HDAC
inhibitors enhance neratinib activity and when combined enhance the
actions of an anti-PD-1 immunomodulatory antibody in vivo.
Oncotarget. 8:90262–90277. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang X, Wu WKK, Gao J, Li Z, Dong B, Lin
X, Li Y, Li Y, Gong J, Qi C, et al: Autophagy inhibition enhances
PD-L1 expression in gastric cancer. J Exp Clin Cancer Res.
38(140)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Buchser WJ, Laskow TC, Pavlik PJ, Lin HM
and Lotze MT: Cell-mediated autophagy promotes cancer cell
survival. Cancer Res. 72:2970–2979. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Tang D, Zhao D, Wu Y, Yao R, Zhou L, Lu L,
Gao W and Sun Y: The miR-3127-5p/p-STAT3 axis up-regulates PD-L1
inducing chemoresistance in non-small-cell lung cancer. J Cell Mol
Med. 22:3847–3856. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhu J, Li Y, Luo Y, Xu J, Liufu H, Tian Z
and Huang C, Li J and Huang C: A feedback loop formed by
ATG7/autophagy, FOXO3a/miR-145 and PD-L1 regulates stem-like
properties and invasion in human bladder cancer. Cancers (Basel).
11(349)2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Maher CM, Thomas JD, Haas DA, Longen CG,
Oyer HM, Tong JY and Kim FJ: Small-molecule sigma1 modulator
induces autophagic degradation of PD-L1. Mol Cancer Res.
16:243–255. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Liang J, Wang L, Wang C, Shen J, Su B,
Marisetty AL, Fang D, Kassab C, Jeong KJ, Zhao W, et al:
Verteporfin Inhibits PD-L1 through autophagy and the
STAT1-IRF1-TRIM28 signaling axis, exerting antitumor efficacy.
Cancer Immunol Res. 8:952–965. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang N and Bevan MJ: CD8(+) T cells: Foot
soldiers of the immune system. Immunity. 35:161–168.
2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Patel SP, Osada T, Osada K, Hurwitz H,
Lyerly HK and Morse MA: Modulation of immune system inhibitory
checkpoints in colorectal cancer. Curr Colorectal Cancer Rep.
9:391–397. 2013.
|
|
53
|
Dunn GP, Old LJ and Schreiber RD: The
three Es of cancer immunoediting. Annu Rev Immunol. 22:329–360.
2004.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Green MR, Monti S, Rodig SJ, Juszczynski
P, Currie T, O'Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub
TR, et al: Integrative analysis reveals selective 9p24.1
amplification, increased PD-1 ligand expression, and further
induction via JAK2 in nodular sclerosing Hodgkin lymphoma and
primary mediastinal large B-cell lymphoma. Blood. 116:3268–3277.
2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ikeda S, Okamoto T, Okano S, Umemoto Y,
Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y,
et al: PD-L1 is upregulated by simultaneous amplification of the
PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol.
11:62–71. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Roemer MG, Advani RH, Ligon AH, Natkunam
Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, et
al: PD-L1 and PD-L2 genetic alterations define classical Hodgkin
lymphoma and predict outcome. J Clin Oncol. 34:2690–2697.
2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Twa DD, Chan FC, Ben-Neriah S, Woolcock
BW, Mottok A, Tan KL, Slack GW, Gunawardana J, Lim RS, McPherson
AW, et al: Genomic rearrangements involving programmed death
ligands are recurrent in primary mediastinal large B-cell lymphoma.
Blood. 123:2062–2065. 2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Prestipino A, Emhardt AJ, Aumann K,
O'Sullivan D, Gorantla SP, Duquesne S, Melchinger W, Braun L,
Vuckovic S, Boerries M, et al: Oncogenic JAK2V617F
causes PD-L1 expression, mediating immune escape in
myeloproliferative neoplasms. Sci Transl Med.
10(eaam7729)2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Sato H, Niimi A, Yasuhara T, Permata TBM,
Hagiwara Y, Isono M, Nuryadi E, Sekine R, Oike T, Kakoti S, et al:
DNA double-strand break repair pathway regulates PD-L1 expression
in cancer cells. Nat Commun. 8(1751)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Sun LL, Yang RY, Li CW, Chen MK, Shao B,
Hsu JM, Chan LC, Yang Y, Hsu JL, Lai YJ and Hung MC: Inhibition of
ATR downregulates PD-L1 and sensitizes tumor cells to T
cell-mediated killing. Am J Cancer Res. 8:1307–1316.
2018.PubMed/NCBI
|
|
62
|
Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y
and Kwok HF: The roles of microRNAs in regulating the expression of
PD-1/PD-L1 immune checkpoint. Int J Mol Sci.
18(2540)2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Xie G, Li W, Li R, Wu K, Zhao E, Zhang Y,
Zhang P, Shi L, Wang D, Yin Y, et al: Helicobacter pylori promote
B7-H1 expression by suppressing miR-152 and miR-200b in gastric
cancer cells. PLoS One. 12(e0168822)2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang
T, Song W, Chen Y, OuYang J, Chen J, et al: MiR-424(322) reverses
chemoresistance via T-cell immune response activation by blocking
the PD-L1 immune checkpoint. Nat Commun. 7(11406)2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kataoka K, Shiraishi Y, Takeda Y, Sakata
S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S,
et al: Aberrant PD-L1 expression through 3'-UTR disruption in
multiple cancers. Nature. 534:402–406. 2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Deng S, Hu Q, Zhang H, Yang F, Peng C and
Huang C: HDAC3 inhibition upregulates PD-L1 expression in B-cell
lymphomas and augments the efficacy of anti-PD-L1 therapy. Mol
Cancer Ther. 18:900–908. 2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Woods DM, Sodré AL, Villagra A, Sarnaik A,
Sotomayor EM and Weber J: HDAC inhibition upregulates PD-1 ligands
in melanoma and augments immunotherapy with PD-1 blockade. Cancer
Immunol Res. 3:1375–1385. 2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Lu C, Paschall AV, Shi H, Savage N, Waller
JL, Sabbatini ME, Oberlies NH, Pearce C and Liu K: The MLL1-H3K4me3
axis-mediated PD-L1 expression and pancreatic cancer immune
evasion. J Natl Cancer Inst. 109(djw283)2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Casey SC, Tong L, Li Y, Do R, Walz S,
Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M and Felsher
DW: MYC regulates the antitumor immune response through CD47 and
PD-L1. Science. 352:227–231. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Marzec M, Zhang Q, Goradia A, Raghunath
PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA and
Wasik MA: Oncogenic kinase NPM/ALK induces through STAT3 expression
of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad
Sci USA. 105:20852–20857. 2008.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Akbay EA, Koyama S, Carretero J, Altabef
A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp
EM, Pugh TJ, et al: Activation of the PD-1 pathway contributes to
immune escape in EGFR-driven lung tumors. Cancer Discov.
3:1355–1363. 2013.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Atefi M, Avramis E, Lassen A, Wong DJ,
Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et
al: Effects of MAPK and PI3K pathways on PD-L1 expression in
melanoma. Clin Cancer Res. 20:3446–3457. 2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Barsoum IB, Smallwood CA, Siemens DR and
Graham CH: A mechanism of hypoxia-mediated escape from adaptive
immunity in cancer cells. Cancer Res. 74:665–674. 2014.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Jiang X, Zhou J, Giobbie-Hurder A, Wargo J
and Hodi FS: The activation of MAPK in melanoma cells resistant to
BRAF inhibition promotes PD-L1 expression that is reversible by MEK
and PI3K inhibition. Clin Cancer Res. 19:598–609. 2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Peng J, Hamanishi J, Matsumura N, Abiko K,
Murat K, Baba T, Yamaguchi K, Horikawa N, Hosoe Y, Murphy SK, et
al: Chemotherapy induces programmed cell death-ligand 1
overexpression via the nuclear factor-κB to foster an
immunosuppressive tumor microenvironment in ovarian cancer. Cancer
Res. 75:5034–5045. 2015.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y,
Chen Z, Herter-Sprie GS, Akbay EA, Tchaicha JH, Altabef A, et al:
Loss of Lkb1 and pten leads to lung squamous cell carcinoma with
elevated PD-L1 expression. Cancer Cell. 25:590–604. 2014.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Garcia-Diaz A, Shin DS, Moreno BH, Saco J,
Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X,
et al: Interferon receptor signaling pathways regulating PD-L1 and
PD-L2 Expression. Cell Rep. 19:1189–1201. 2017.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Carbotti G, Barisione G, Airoldi I,
Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M and
Ferrini S: IL-27 induces the expression of IDO and PD-L1 in human
cancer cells. Oncotarget. 6:43267–43280. 2015.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Lienlaf M, Perez-Villarroel P, Knox T,
Pabon M, Sahakian E, Powers J, Woan K V, Lee C, Cheng F, Deng S, et
al: Essential role of HDAC6 in the regulation of PD-L1 in melanoma.
Mol Oncol. 10:735–750. 2016.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN and
Gao FG: TGF-β of lung cancer microenvironment upregulates B7H1 and
GITRL expression in dendritic cells and is associated with
regulatory T cell generation. Oncol Rep. 28:615–621.
2012.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Pulko V, Liu X, Krco CJ, Harris KJ,
Frigola X, Kwon ED and Dong H: TLR3-stimulated dendritic cells
up-regulate B7-H1 expression and influence the magnitude of CD8 T
cell responses to tumor vaccination. J Immunol. 183:3634–3641.
2009.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Quandt D, Jasinski-Bergner S, Müller U,
Schulze B and Seliger B: Synergistic effects of IL-4 and TNFα on
the induction of B7-H1 in renal cell carcinoma cells inhibiting
allogeneic T cell proliferation. J Transl Med.
12(151)2014.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Wang X, Yang L, Huang F, Zhang Q, Liu S,
Ma L and You Z: Inflammatory cytokines IL-17 and TNF-α up-regulate
PD-L1 expression in human prostate and colon cancer cells. Immunol
Lett. 184:7–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu
Z and Huang JA: The EGFR pathway is involved in the regulation of
PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in
EGFR-mutated non-small cell lung cancer. Int J Oncol. 49:1360–1368.
2016.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo
CW, Khoo KH, Chang SS, Cha JH, Kim T, et al: Glycosylation and
stabilization of programmed death ligand-1 suppresses T-cell
activity. Nat Commun. 7(12632)2016.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha
JH, Wang HL, Yang WH, Yen EY, Chang WC, et al: IL-6/JAK1 pathway
drives PD-L1 Y112 phosphorylation to promote cancer immune evasion.
J Clin Invest. 129:3324–3338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Mezzadra R, Sun C, Jae LT, Gomez-Eerland
R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland
I, et al: Identification of CMTM6 and CMTM4 as PD-L1 protein
regulators. Nature. 549:106–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu
JL, Wei Y, Xia W, Hou J, Qiu Y and Hung MC: Palmitoylation
stabilizes PD-L1 to promote breast tumor growth. Cell Res.
29:83–86. 2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Schmelzle T and Hall MN: TOR, a central
controller of cell growth. Cell. 103:253–262. 2000.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Lastwika KJ, Wilson W III, Li QK, Norris
J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et
al: Control of PD-L1 expression by oncogenic activation of the
AKT-mTOR pathway in non-small cell lung cancer. Cancer Res.
76:227–238. 2016.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Hay N: The Akt-mTOR tango and its
relevance to cancer. Cancer Cell. 8:179–183. 2005.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Aoki M and Fujishita T: Oncogenic roles of
the PI3K/AKT/mTOR Axis. Curr Top Microbiol Immunol. 407:153–189.
2017.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Song M, Chen D, Lu B, Wang C, Zhang J,
Huang L, Wang X, Timmons CL, Hu J, Liu B, et al: PTEN Loss
Increases PD-L1 protein expression and affects the correlation
between PD-L1 expression and clinical parameters in colorectal
cancer. PLoS One. 8(e65821)2013.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning
W, Zeng H, Zhang N, Du W, Chen C and Huang JA: PD-L1 induced by
IFN-γ from tumor-associated macrophages via the JAK/STAT3 and
PI3K/AKT signaling pathways promoted progression of lung cancer.
Int J Clin Oncol. 22:1026–1033. 2017.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Parsa AT, Waldron JS, Panner A, Crane CA,
Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et
al: Loss of tumor suppressor PTEN function increases B7-H1
expression and immunoresistance in glioma. Nat Med. 13:84–88.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
97
|
Mittendorf EA, Philips AV, Meric-Bernstam
F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM,
Akcakanat A, et al: PD-L1 expression in triple-negative breast
cancer. Cancer Immunol Res. 2:361–370. 2014.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Akula SM, Abrams SL, Steelman LS, Emma MR,
Augello G, Cusimano A, Azzolina A, Montalto G, Cervello M and
McCubrey JA: RAS/RAF/MEK/ERK, PI3K/PTEN/AKT/mTORC1 and TP53
pathways and regulatory miRs as therapeutic targets in
hepatocellular carcinoma. Expert Opin Ther Targets. 23:915–929.
2019.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Wang Z, Ma L, Su M, Zhou Y, Mao K, Li C,
Peng G, Zhou C, Shen B and Dou J: Baicalin induces cellular
senescence in human colon cancer cells via upregulation of DEPP and
the activation of Ras/Raf/MEK/ERK signaling. Cell Death Dis.
9(217)2018.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Wang A, Zhang H, Liang Z, Xu K, Qiu W,
Tian Y, Guo H, Jia J, Xing E, Chen R, et al: U0126 attenuates
ischemia/reperfusion-induced apoptosis and autophagy in myocardium
through MEK/ERK/EGR-1 pathway. Eur J Pharmacol. 788:280–285.
2016.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Wang J, Whiteman MW, Lian H, Wang G, Singh
A, Huang D and Denmark T: A non-canonical MEK/ERK signaling pathway
regulates autophagy via regulating Beclin 1. J Biol Chem.
284:21412–21424. 2009.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Corcelle E, Nebout M, Bekri S, Gauthier N,
Hofman P, Poujeol P, Fénichel P and Mograbi B: Disruption of
autophagy at the maturation step by the carcinogen lindane is
associated with the sustained mitogen-activated protein
kinase/extracellular signal-regulated kinase activity. Cancer Res.
66:6861–6870. 2006.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Liu J, Hamrouni A, Wolowiec D, Coiteux V,
Kuliczkowski K, Hetuin D, Saudemont A and Quesnel B: Plasma cells
from multiple myeloma patients express B7-H1 (PD-L1) and increase
expression after stimulation with IFN-{gamma} and TLR ligands via a
MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 110:296–304.
2007.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Loi S, Dushyanthen S, Beavis PA, Salgado
R, Denkert C, Savas P, Combs S, Rimm DL, Giltnane JM, Estrada MV,
et al: RAS/MAPK activation is associated with reduced
tumor-infiltrating lymphocytes in triple-negative breast cancer:
Therapeutic cooperation between MEK and PD-1/PD-L1 immune
checkpoint inhibitors. Clin Cancer Res. 22:1499–1509.
2016.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Karakhanova S, Meisel S, Ring S, Mahnke K
and Enk AH: ERK/p38 MAP-kinases and PI3K are involved in the
differential regulation of B7-H1 expression in DC subsets. Eur J
Immunol. 40:254–266. 2010.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Qian Y, Deng J, Geng L, Xie H, Jiang G,
Zhou L, Wang Y, Yin S, Feng X, Liu J, et al: TLR4 signaling induces
B7-H1 expression through MAPK pathways in bladder cancer cells.
Cancer Invest. 26:816–821. 2008.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Yamamoto R, Nishikori M, Tashima M, Sakai
T, Ichinohe T, Takaori-Kondo A, Ohmori K and Uchiyama T: B7-H1
expression is regulated by MEK/ERK signaling pathway in anaplastic
large cell lymphoma and Hodgkin lymphoma. Cancer Sci.
100:2093–2100. 2009.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Coelho MA, de Carné Trécesson S, Rana S,
Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E,
Barnouin K, et al: Oncogenic RAS signaling promotes tumor
immunoresistance by stabilizing PD-L1 mRNA. Immunity.
47:1083–1099.e6. 2017.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Gao H, Zhang J and Ren X: PD-L1 regulates
tumorigenesis and autophagy of ovarian cancer by activating mTORC
signaling. Biosci Rep. 39(BSR20191041)2019.PubMed/NCBI View Article : Google Scholar
|