Introduction
Cervical cancer is frequently diagnosed in women
and, with a 6.5% incidence rate, ranks as the fourth most common
type of cancer globally. Furthermore, it also ranks fourth in
mortality statistics, with a rate of 7.7% (1). Cervical cancer incidence and mortality
rates are higher in developing than in developed countries. The
vast majority of the highest cervical cancer rates are observed in
countries in sub-Saharan Africa, Melanesia, South America and
South-Eastern Asia (1). Previous
studies have demonstrated that cervical cancer is a disease caused
by multiple interacting factors, and thus, human papillomavirus
(HPV) is a necessary but not sufficient cause of cervical cancer
occurrence (2). Infections, such as
human immunodeficiency virus, and environmental factors, such as
smoking or long-term use of oral contraceptives, are important
co-factors (3). Cervical cancer is
considered highly preventable, and HPV vaccination and screening
programs are widely used for preventative purposes (4,5).
Countries that successfully implement these measures have lower
cervical cancer rates. For example, in the United States, cervical
cancer incidence rates declined 1.9% per year between 2007 and
2012(6). However, numerous
lower-income countries have not fulfilled mass screening programs,
and the cervical cancer incidence and mortality rates remain high
(7). Therefore, to reduce the gap
between developing and developed countries, and help diagnose
cervical cancer during the early stages, predict the course of the
disease and predict the outcome, more accessible screening options
are required.
Mitochondria are essential and complex organelles
involved in cellular processes, such as energy generation through
oxidative phosphorylation, apoptosis and metabolism. Therefore,
mitochondria are considered to serve an essential role in
carcinogenesis (8). Mitochondrial
transcription factor A (TFAM) is a nuclear-encoded protein,
which serves an important role in mitochondria, since it is
required for the transcription and replication of mitochondrial DNA
(9,10). TFAM expression has been
reported to be altered and associated with breast and lung cancer
(11-13),
and polymorphisms in the TFAM gene have been reported to be
associated with prostate (14),
colorectal (15), breast (16), uterine, ovary and cervical cancer
(17). Numerous studies have
investigated the associations between various single nucleotide
polymorphisms (SNPs) and the risk of cervical cancer; however,
there are not enough studies analyzing mitochondria-related SNPs
and the associations with the clinical/pathological characteristics
of the patients or the morphological characteristics of their
tumors (18). In the present study,
genotyping analysis was used to investigate TFAM SNP
distribution in a cervical cancer cohort, and to explore their
potential as biomarkers for the tumor phenotype or outcomes of
patients.
Materials and methods
Study subjects
The study population consisted of 172 female
patients who were diagnosed with cervical cancer. All patients were
Lithuanians with a median age of 56 years (age range, 22-83 years).
Patients were included in the present study if they matched all of
the following criteria: Cervical cancer was diagnosed as a primary
disease, the patient signed a consent form, a blood sample was
taken at the time of the diagnosis and clinical information was
available. In our previous study, the same patient group was
analyzed focusing on the polymorphisms of a different gene (DNA
polymerase γ, catalytic subunit) and their associations with
cervical cancer (18). The research
was conducted at the Institute of Oncology, Lithuanian University
of Health Sciences (Kaunas, Lithuania) between 2015 January and
2021 May. The present study was performed in accordance to the
guidelines of the Declaration of Helsinki (19) and was approved by the Kaunas
Regional Biomedical Research Ethics Committee (approval nos.
BE-2-10 and P1-BE-2-10/2014; Kaunas, Lithuania). Clinical data were
gathered retrospectively from medical records.
Tumor grading
At present no particular grading system has achieved
universal acceptance, and grading of cervical carcinomas remains of
uncertain clinical value. In the present study histopathological
grading (G) was based on the degree of cell and tissue atypia and
mitotic activity. The definitions of the G categories apply to all
carcinomas, these are: GX, grade of differentiation cannot be
assessed; G1, well differentiated; G2, moderately differentiated;
and G3, poorly differentiated or undifferentiated (20).
Genotyping
DNA was isolated from EDTA-preserved peripheral
blood using a GeneJet Genomic DNA purification kit (cat. no. K0721;
Thermo Fisher Scientific, Inc.). SNPs (rs11006132, rs11006129,
rs1937, rs16912174, rs16912202 and rs3900887) in the TFAM
gene were determined using TaqMan probe SNP Genotyping assays (cat.
no. 4351379; Thermo Fisher Scientific, Inc.) and a QuantStudio 3
Real-Time PCR system (cat. no. A28137; Thermo Fisher Scientific,
Inc.). The reaction mixture contained 15 ng purified DNA sample,
6.125 µl TaqMan Universal MasterMix (cat. no. 4304437; Thermo
Fisher Scientific, Inc.), 0.625 µl TaqMan SNP Genotyping assay and
nuclease-free water to reach a total volume of 12 µl. Nuclease-free
water was used as a no-template control for every plate. The
standard genotyping PCR program: Pre-read step at 60˚C for 30 sec,
95˚C for 10 min; followed by 40 cycles of 95˚C for 15 sec and 60˚C
for 1 min; with a final post-read step at 60˚C for 30 sec, was used
to determine the genotype, and this relied on VIC and FAM
fluorescence intensity.
Statistical analysis
Associations between all TFAM polymorphisms
included in the present study (rs11006132, rs11006129, rs1937,
rs16912174, rs16912202 and rs3900887) and clinical data,
pathological tumor size (T), tumor differentiation grade (G), lymph
node status (N), distant metastasis (M), histological tumor type,
progression and death, were analyzed. Additionally, patients were
divided into two groups according to median age at diagnosis
(<56 and ≥56 years old) and pathological tumor size [small
tumors (T1 + T2; n=110) and larger tumors (T3 + T4; n=62)].
Furthermore, patients were divided into groups based on tumor
differentiation grade; the first group included patients with
well-differentiated and moderately differentiated tumors (G1 + G2),
and the second group included patients with poorly differentiated
tumors (G3).
SPSS version 22 software (IBM Corp.) was used for
association analysis. Associations between genotype and clinical
data were evaluated using a Pearson's χ2 test
(rs11006132, rs11006129, rs1937 and rs3900887) or Fisher's Exact
(rs16912174 and rs16912202) tests and binary logistic regression.
Univariate logistic regression analysis adjusted for age at
diagnosis (Model A) and multivariate logistic regression analysis
with additional confounding factor N (Model B), were used. Survival
analysis was performed using Kaplan-Meier analysis and the
difference between survival curves was analyzed using a log-rank
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Distribution of clinical
characteristics
The present study included 172 patients with
cervical cancer with a median age of 56 years (age range, 22-83
years). The majority of the patients had squamous cell carcinoma
(n=157), whereas adenocarcinoma (n=9) or mucinous adenocarcinoma
(n=6) were less frequent. The tumor differentiation grade was
mostly G2 (n=113). The majority of the patients did not have
distant metastasis (n=162) or affected regional lymph nodes (n=95).
Additional information on the clinical characteristics are provided
in Table I.
 | Table IDemographic and clinicopathological
characteristics of the patients. |
Table I
Demographic and clinicopathological
characteristics of the patients.
| Characteristics | Frequency, n (%) |
|---|
| Stage | |
|
I | 16 (9.3) |
|
II | 60 (34.9) |
|
III | 83 (48.3) |
|
IV | 13 (7.6) |
| Tumor size | |
|
T1 | 26 (15.1) |
|
T2 | 84 (48.8) |
|
T3 | 55(32) |
|
T4 | 7 (4.1) |
| Lymph node
status | |
|
Negative | 95 (55.2) |
|
Positive | 77 (44.8) |
| Distant
metastasis | |
|
Absent | 162 (94.2) |
|
Present | 10 (5.8) |
| Differentiation
grade | |
|
G1 | 13 (7.6) |
|
G2 | 113 (65.7) |
|
G3 | 44 (25.6) |
|
Missinga | 2 (1.2) |
| Survival status | |
|
Alive | 132 (76.7) |
|
Deceased | 40 (23.3) |
| Fact of
progression | |
|
Absent | 123 (71.5) |
|
Present | 49 (28.5) |
Distribution of genotypes
The distributions of all genotypes in the present
cohort were estimated using the Hardy-Weinberg equilibrium (HWE),
and it was revealed that rs11006132, rs11006129, rs1937, rs16912174
and rs16912202 SNPs were in HWE, whereas rs3900887 was not. There
were 13 missing data points from rs3900887 genotyping due to
amplification failure. The distribution of TFAM genotypes is
shown in Fig. 1 and Table II.
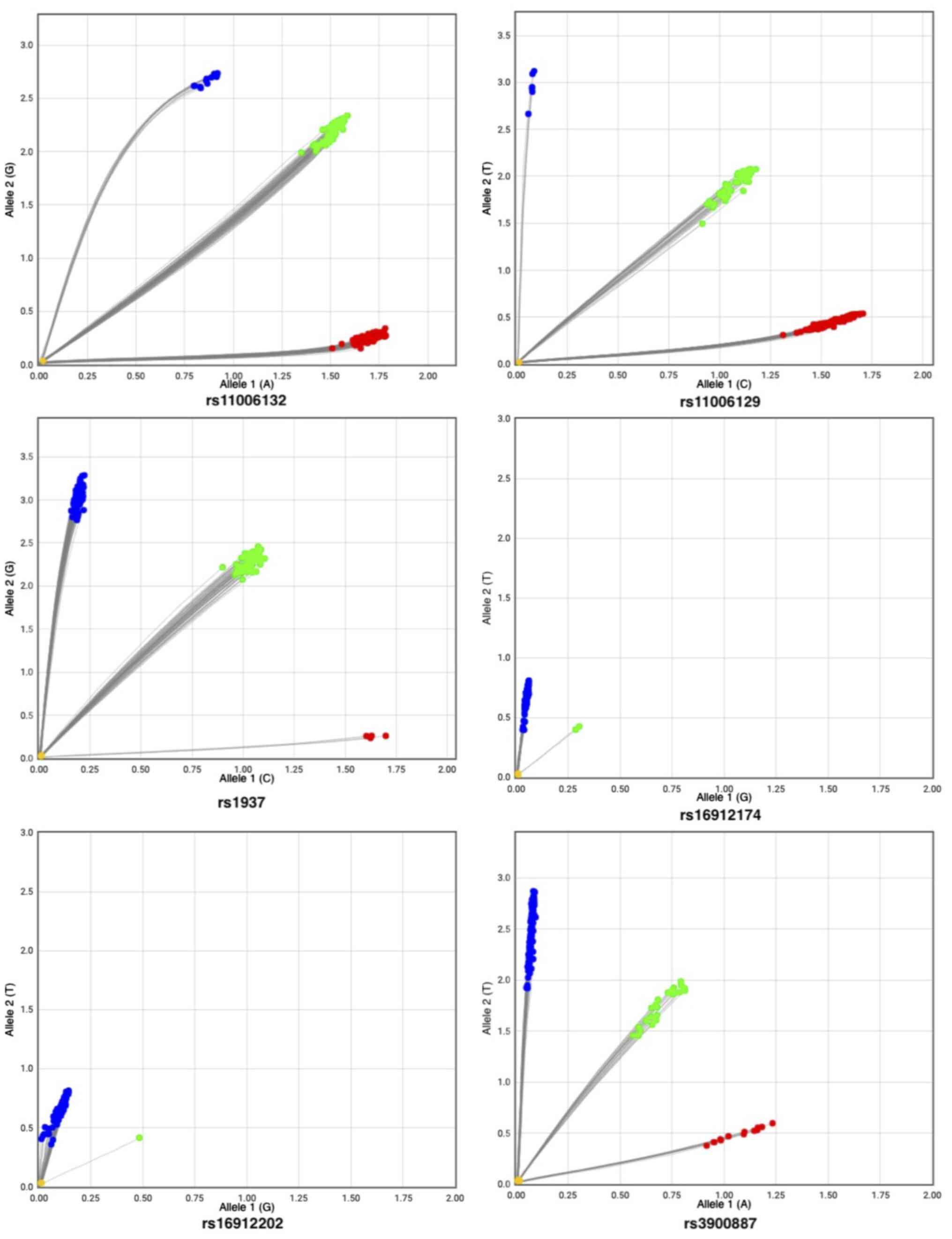 | Figure 1Distribution of TFAM genotypes.
Allelic discrimination plots representing the TFAM
rs11006132, rs11006129, rs1937, rs16912174, rs16912202, rs3900887
genotypes in the cervical cancer cohort. The x-axis represents
Allele 1 labeled with VIC dye, and the y-axis represents Allele 2
labeled with FAM dye. The colored dots represent different
genotypes called according to VIC and FAM fluorescence intensities.
Red dots represents the homozygous Allele 1 genotype, blue dots
represents the homozygous Allele 2 genotype, and the green dots
represents the heterozygous genotype. The no-template control is
marked as a yellow dot at the bottom left corner of the plot.
TFAM, TFAM, mitochondrial transcription factor A. |
 | Table IIGenotype and allele frequencies of
the analyzed TFAM polymorphisms in the study population. |
Table II
Genotype and allele frequencies of
the analyzed TFAM polymorphisms in the study population.
| TFAM | Patient count | Genotype,
frequency | Allele,
frequency |
|---|
| rs11006132 | 90 | AA, 0.52 | A, 0.72 |
| | 68 | AG, 0.4 | G, 0.28 |
| | 14 | GG, 0.08 | |
| rs11006129 | 130 | CC, 0.76 | C, 0.86 |
| | 36 | CT, 0.21 | T, 0.14 |
| | 6 | TT, 0.03 | |
| rs1937 | 115 | GG, 0.67 | G, 0.82 |
| | 53 | CG, 0.31 | C, 0.18 |
| | 4 | CC, 0.02 | |
| rs16912174 | 170 | TT, 0.99 | T, 0.99 |
| | 2 | GT, 0.01 | G, 0.01 |
| rs16912202 | 171 | TT, 0.99 | T, 1.00 |
| | 1 | GT, 0.01 | G, 0.00 |
| rs3900887 | 118 | TT, 0.74 | T, 0.83 |
| | 27 | AT, 0.17 | A, 0.17 |
| | 14 | AA, 0.09 | |
Association analysis
Associations between TFAM polymorphisms and
clinical data were evaluated using Pearson's χ2 or
Fisher's exact tests and are presented in Table III.
 | Table IIIAssociations between mitochondrial
transcription factor A polymorphisms and clinical data. |
Table III
Associations between mitochondrial
transcription factor A polymorphisms and clinical data.
| Variable | rs11006132 | rs11006129 | rs1937 | rs16912174 | rs16912202 | rs3900887 |
|---|
| T (T1 + T2 vs. T3 +
T4 | 0.789 | 0.105 | 0.309 | 0.408 | 0.640 | 0.006 |
| N (negative vs.
positive) | 0.979 | 0.893 | 0.954 | 0.696 | 0.448 | 0.468 |
| M (negative vs.
positive) | 0.427 | 0.382 | 0.211 | 0.887 | 0.058 | 0.166 |
| Differentiation
grade (G1 + G2 vs. G3) | 0.417 | 0.910 | 0.439 | 0.452 | 0.741 | 0.898 |
| Stage (I + II vs.
III + IV) | 0.775 | 0.225 | 0.966 | 0.690 | 0.558 | 0.065 |
| Survival status
(alive vs. deceased) | 0.565 | 0.833 | 0.661 | 0.588 | 0.767 | 0.457 |
| Fact of
progression | 0.310 | 0.209 | 0.986 | 0.510 | 0.715 | 0.258 |
| Age group (<56
vs. ≥56) | 0.249 | 0.546 | 0.605 | 0.254 | 0.494 | 0.905 |
| Adenocarcinoma | 0.472 | 0.150 | 0.734 | 0.898 | 0.948 | 0.376 |
| Squamous cell
carcinoma | 0.328 | 0.615 | 0.812 | 0.833 | 0.913 | 0.480 |
The present study revealed that rs3900887 was
associated with grouped tumor size T1 + T2 vs. T3 + T4 (P=0.006).
However, no associations with age group (P=0.905), lymph node
status (P=0.468), distant metastasis (P=0.166), differentiation
grade (P=0.898), adenocarcinoma (P=0.376), squamous cell carcinoma
(P=0.480), stage (P=0.065), progression (P=0.258) or death
(P=0.457) were observed. To further evaluate the associations
amongst rs3900887 and clinical data, logistic regression analysis
was performed. It was revealed that rs3900887 remained
significantly associated with grouped tumor size in logistic
regression analysis of genotype and allele models. Patients with
the rs3900887 TT [odds ratio (OR)=0.328; 95% CI, 0.127-0.849;
P=0.022] or AT (OR=0.091; 95% CI, 0.023-0.367; P=0.001) genotypes
were less likely to have larger tumors than those with the AA
genotype. Univariate and multivariate logistic regression analyses
are presented in Table IV.
 | Table IVUnivariate and multivariate logistic
regression analysis of the associations of mitochondrial
transcription factor A rs3900887 genotypes, alleles and clinical
data with potential confounding factors. |
Table IV
Univariate and multivariate logistic
regression analysis of the associations of mitochondrial
transcription factor A rs3900887 genotypes, alleles and clinical
data with potential confounding factors.
| | Model A | Model B |
|---|
| Single nucleotide
polymorphism | Parameter | Covariates | Odds ratio | 95% confidence
interval | P-value | Odds ratio | 95% confidence
interval | P-value |
|---|
| rs3900887 | Tumor size | TT vs. AA | 0.328 | 0.127-0849 | 0.022a | 0.350 | 0.097-1.261 | 0.108 |
| | (T1 + T2 vs. | AT vs. AA | 0.091 | 0.023-0.367 | 0.001b | 0.093 | 0.018-0.490 | 0.005b |
| | T3 + T4) | Age | 1.011 | 0.996-1.028 | 0.156 | 1.038 | 1.008-1.070 | 0.013a |
| | | Lymph node status
(negative vs. positive) | - | - | - | 7.242 | 3.199-16.395 |
<0.0001c |
| rs3900887 T
allele | Tumor size | Carrier vs.
non-carrier | 0.277 | 0.109-0.709 | 0.007b | 0.286 | 0.080-1.020 | 0.054 |
| | (T1 + T2 vs. T3 +
T4) | | | | | | | |
| | | Age | 1.011 | 0.995-1.027 | 0.168 | 1.037 | 1.007-1.068 | 0.014a |
| | | Lymph node status
(negative vs. positive) | - | - | - | 7.105 | 3.195-15.799 |
<0.0001c |
No statistically significant associations of
rs11006132, rs11006129, rs1937, rs16912174 and rs16912202 with
clinical data were observed.
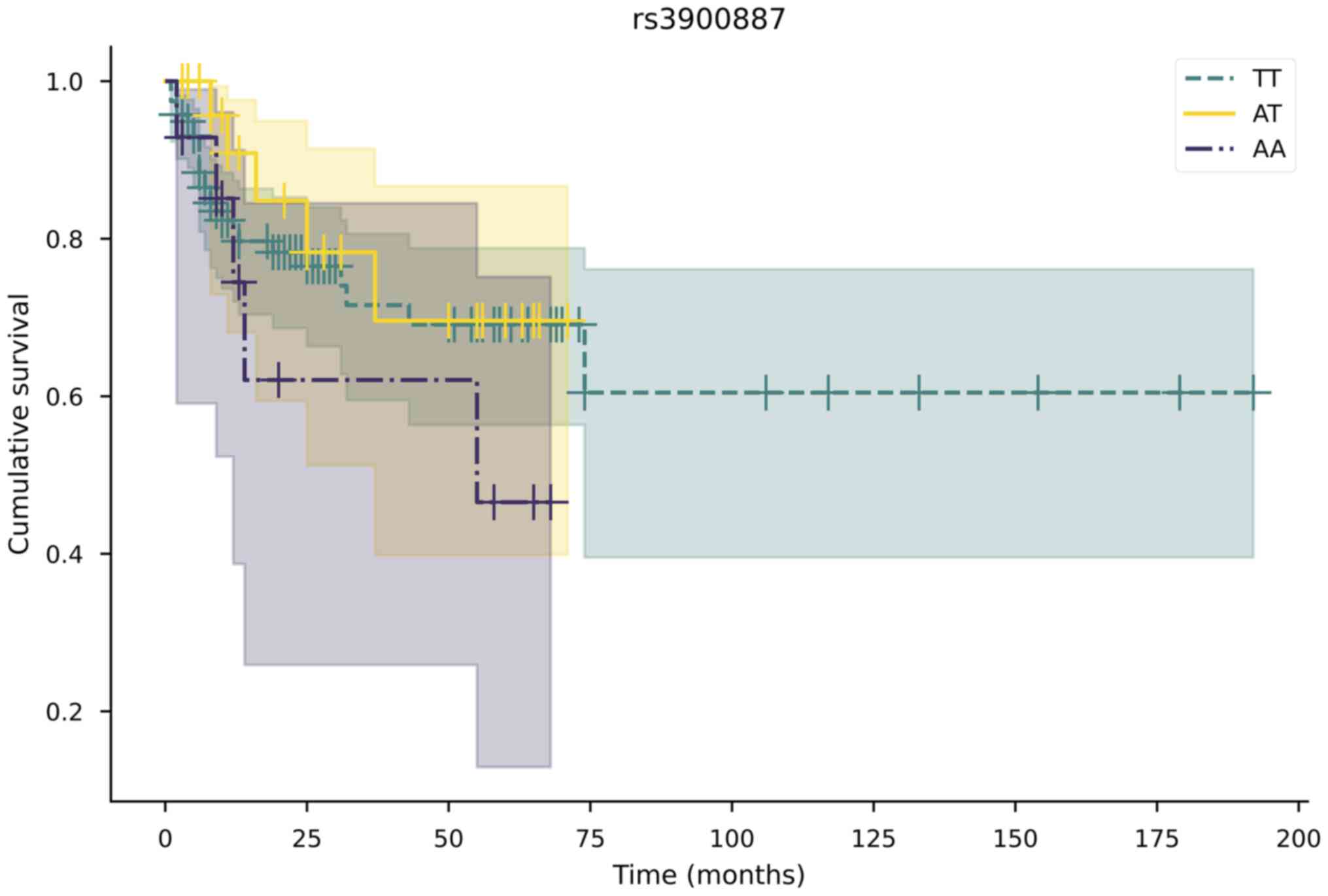
Survival analysis
Kaplan-Meier survival analysis was used to determine
the association between rs3900887 and overall survival; however, it
was not statistically significant: TT vs. AT (P=0.548), TT vs. AA
(P=0.357), AT vs. AA (P=0.195). The Kaplan-Meier survival plot is
shown in Fig. 2.
Discussion
Despite extensive prevention and vaccination
programs, cervical cancer remains the fourth most common cancer in
women globally and has the fourth highest mortality rate amongst
all types of cancer (1). In more
developed countries with better established screening and
vaccination programs, the incidence rates of cervical cancer have
been declining each year (6);
however, it is considerably more common in developing countries
(1).
Mitochondria are organelles responsible for energy
generation, apoptosis and metabolism, and serve an essential role
in cancer development (8). The
TFAM gene encodes mitochondrial transcription factor, a
protein encoded in nuclear DNA that is responsible for the
transcription and translation of mitochondrial DNA (9,10).
Research has revealed that SNPs in the TFAM gene are
associated with various types of cancer (14-16),
including cervical cancer (17).
Numerous studies have assessed the association between the risk of
developing cervical cancer and SNPs; however, few studies have
investigated the possible associations between SNPs and clinical
and morphological cervical tumor characteristics. In the present
study, the associations of the rs11006132, rs11006129, rs1937,
rs16912174, rs16912202 and rs3900887 polymorphisms in the
TFAM gene with cervical tumor clinical and morphological
characteristics, including tumor histological type, stage, size,
differentiation grade, regional lymph node involvement, distant
metastases, progression and survival status, were analyzed.
It was revealed that the TFAM rs3900887
polymorphism was associated with tumor characteristics. Logistic
regression analysis indicated that patients with TT and TA
genotypes had a lower risk of possessing larger tumors than
patients with the AA genotype. rs3900887 is located in an intron of
the TFAM gene and it may affect splicing. However, it is
also reported to be benign by VarSome (21) and Franklin (22) databases. The imum allele frequency
(MAF) of this gene in the 1,000 Genome project was 0.11, and a
similar MAF (0.17) was calculated in the present study. Our
previous study identified several associations between this
polymorphism and morphological characteristics of breast cancer. It
was revealed that TT and TA genotypes were associated with an
increased risk of positive lymph nodes, and TT genotype carriers
also had an increased risk of positive estrogen receptors (ER) and
lymphatic invasion compared with patients with the AA genotype
(16). Additionally, there has been
only one study investigating associations between this polymorphism
and late-onset Alzheimer's disease, in which no significant
associations were observed (23).
Our recent study revealed significant associations between this SNP
and tumor size, which remained significant after multivariate
analysis. The results may indicate the role of the rs3900887 SNP in
cervical tumor development, and thus, more in-depth studies are
required to confirm its role in cervical tumor development.
Other SNPs in the present study (rs11006132,
rs11006129, rs1937, rs16912174 and rs16912202) exhibited no
significant associations with cervical tumor morphological
parameters. The rs11006132 SNP is located in the 3' untranslated
region of the gene. Our previous study indicated no associations
between this SNP and morphological characteristics of breast cancer
(16). There was also no observed
association with aggressive prostate cancer (14). However, researchers have found a
link between this SNP and the age at onset of Huntington's disease
(24). rs11006129 is located in an
intron of the gene. Our previous study demonstrated an association
between this SNP and breast cancer. It was revealed that T allele
carriers were less likely to be predisposed to ER-positive tumors
and carriers of the C allele were less likely to develop tumors
with vascular invasion than non-carriers of the respective allele
(16). To the best of our
knowledge, no other studies have investigated this SNP to date.
rs1937 is a missense variant of the TFAM gene. Our previous
study found no significant associations between this SNP and breast
tumor parameters (16). Other
studies have investigated the association between the rs1937 and
early-onset myocardial infarction (25), late-onset Alzheimer's disease
(26) and aggressive prostate
cancer (14); however, no
significant results were determined. One study has reported a link
between the rs1937 polymorphism and cervical cancer development in
women (27). The rs16912174 SNP is
located in the 5' untranslated region of the gene. Our previous
study revealed no significant association between this polymorphism
and breast cancer (16). The
rs16912202 polymorphism is located in the 3' untranslated region of
the gene (c.*3236T>G) and, therefore, it may
interfere with mRNA stability or translation. Our previous study
did not find any significant associations between this SNP and
morphological characteristics of breast cancer (16). To the best of our knowledge, no
other studies have been performed for this SNP in cervical cancer
to date.
Despite the limitations of the present study,
including the small sample size and small number of SNPs, it was
possible to detect significant association, suggesting that the
TFAM gene may serve a role in cervical cancer development.
In this study the individual SNPs were analyzed as a pilot study to
understand the impact of TFAM SNPs on cervical cancer. In
future studies, larger cohorts will be employed to investigate the
mitochondria-related SNPs in other genes, examining grouped SNPs
and haplotypes, and their impact on cervical cancer. Additional
studies should be performed on broader and more ethnically diverse
cohorts of patients with cervical cancer to verify these
results.
In conclusion, the results of the present study
suggest that mitochondrial transcription factor A, encoded by the
nuclear TFAM gene, is important in cervical cancer, and the
rs3900887 SNP may serve as a potential biomarker of tumor size.
Acknowledgements
Not applicable.
Funding
This research was funded by the Lithuanian University of Health
Sciences (Kaunas, Lithuania).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
IG, RU and EJ conceived the study. IG and JC
developed the methodology. IG and JC performed the formal analysis.
JC performed the experiments. RU, EZ, AI, LP and EJ curated the
data. IG, RU and JC prepared the original draft of the manuscript.
EZ, AI, LP and EJ reviewed and edited the manuscript. IG prepared
the tables. RU and EJ supervised the study. All authors have read
and approved the final manuscript. JC and IG confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was performed in accordance with
the guidelines of the Declaration of Helsinki and approved by the
Kaunas Regional Biomedical Research Ethics Committee (approval nos.
BE-2-10 and P1-BE-2-10/2014). Informed written consent was obtained
from all subjects involved in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
de Freitas AC, Gurgel APAD, Chagas BS,
Coimbra EC and do Amaral CMM: Susceptibility to cervical cancer: An
overview. Gynecol Oncol. 126:304–311. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arbyn M, Weiderpass E, Bruni L, de Sanjosé
S, Saraiya M, Ferlay J and Bray F: Estimates of incidence and
mortality of cervical cancer in 2018: A worldwide analysis. Lancet
Glob Health. 8:e191–e203. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lei J, Ploner A, Elfström KM, Wang J, Roth
A, Fang F, Sundström K, Dillner J and Sparén P: HPV Vaccination and
the Risk of Invasive Cervical Cancer. N Engl J Med. 383:1340–1348.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kessler TA: Cervical Cancer: Prevention
and Early Detection. Semin Oncol Nurs. 33:172–183. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Benard VB, Thomas CC, King J, Massetti GM,
Doria-Rose VP and Saraiya M: Centers for Disease Control and
Prevention (CDC). Vital signs: Cervical cancer incidence,
mortality, and screening - United States, 2007-2012. MMWR Morb
Mortal Wkly Rep. 63:1004–1009. 2014.PubMed/NCBI
|
|
7
|
Vaccarella S, Laversanne M, Ferlay J and
Bray F: Cervical cancer in Africa, Latin America and the Caribbean
and Asia: Regional inequalities and changing trends. Int J Cancer.
141:1997–2001. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
van Gisbergen MW, Voets AM, Starmans MHW,
de Coo IFM, Yadak R, Hoffmann RF, Boutros PC, Smeets HJM, Dubois L
and Lambin P: How do changes in the mtDNA and mitochondrial
dysfunction influence cancer and cancer therapy? Challenges,
opportunities and models. Mutat Res Rev Mutat Res. 764:16–30.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kang D, Kim SH and Hamasaki N:
Mitochondrial transcription factor A (TFAM): Roles in maintenance
of mtDNA and cellular functions. Mitochondrion. 7:39–44.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hallberg BM and Larsson N-G: TFAM forces
mtDNA to make a U-turn. Nat Struct Mol Biol. 18:1179–1181.
2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gao W, Wu M-H, Wang N, Ying M-Z, Zhang
Y-Y, Hua J, Chuan L and Wang Y-J: Mitochondrial transcription
factor A contributes to cisplatin resistance in patients with
estrogen receptor positive breast cancer. Mol Med Rep.
14:5304–5310. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Peng H, Yang M, Chen ZY, Chen P, Guan CX,
Xiang XD, Cai S, Chen Y and Fang X: Expression and methylation of
mitochondrial transcription factor a in chronic obstructive
pulmonary disease patients with lung cancer. PLoS One.
8(e82739)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xie D, Wu X, Lan L, Shangguan F, Lin X,
Chen F, Xu S, Zhang Y, Chen Z, Huang K, et al: Downregulation of
TFAM inhibits the tumorigenesis of non-small cell lung cancer by
activating ROS-mediated JNK/p38MAPK signaling and reducing cellular
bioenergetics. Oncotarget. 7:11609–11624. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Granados JB, Méndez JP, Feria-Bernal G,
García-García E, Tejeda ME, Rojano-Mejía D, Tapia A and Canto P:
Association of a TFAM haplotype with aggressive prostate cancer in
overweight or obese Mexican Mestizo men. Urol Oncol.
35:111.e9–111.e14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Guo J, Zheng L, Liu W, Wang X, Wang Z,
Wang Z, French AJ, Kang D, Chen L, Thibodeau SN, et al: Frequent
truncating mutation of TFAM induces mitochondrial DNA depletion and
apoptotic resistance in microsatellite-unstable colorectal cancer.
Cancer Res. 71:2978–2987. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Golubickaite I, Ugenskiene R,
Korobeinikova E, Gudaitiene J, Vaitiekus D, Poskiene L and
Juozaityte E: The impact of mitochondria-related POLG and TFAM
variants on breast cancer pathomorphological characteristics and
patient outcomes. Biomarkers. 26:343–353. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hu W, Ma S-L, Liu L-L, Zhu Y-H, Zeng T-T,
Li Y and Guan X-Y: Impact of mitochondrial transcription factor A
expression on the outcomes of ovarian, endometrial and cervical
cancers. Am J Transl Res. 12:5343–5361. 2020.PubMed/NCBI
|
|
18
|
Golubickaite I, Ugenskiene R, Ziliene E,
Beniusyte J, Inciura A, Poskiene L and Juozaityte E: POLG Gene
Variants in Cervical Cancer Patients and Their Associations with
Clinical and Pathomorphological Tumor Characteristics. J Clin Med.
10(1838)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
World Medical Association. World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kurman RJ, Carcangiu ML, Herrington S and
Young RH (eds): WHO Classification of Tumours of Female
Reproductive Organs. IARC, Lyon, 2014.
|
|
21
|
Kopanos C, Tsiolkas V, Kouris A, Chapple
CE, Albarca Aguilera M, Meyer R and Massouras A: VarSome: The human
genomic variant search engine. Bioinformatics. 35:1978–1980.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Franklin by Genoox: The Future of Genomic
Medicine. https://franklin.genoox.com/clinical-db/home. Accessed
September 25, 2021.
|
|
23
|
Grupe A, Li Y, Rowland C, Nowotny P,
Hinrichs AL, Smemo S, Kauwe JSK, Maxwell TJ, Cherny S, Doil L, et
al: A scan of chromosome 10 identifies a novel locus showing strong
association with late-onset Alzheimer disease. Am J Hum Genet.
78:78–88. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Taherzadeh-Fard E, Saft C, Akkad DA,
Wieczorek S, Haghikia A, Chan A, Epplen JT and Arning L: PGC-1alpha
downstream transcription factors NRF-1 and TFAM are genetic
modifiers of Huntington disease. Mol Neurodegener.
6(32)2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Palacín M, Alvarez V, Martín M, Díaz M,
Corao AI, Alonso B, Díaz-Molina B, Lozano I, Avanzas P, Morís C, et
al: Mitochondrial DNA and TFAM gene variation in early-onset
myocardial infarction: Evidence for an association to haplogroup H.
Mitochondrion. 11:176–181. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Q, Yu J-T, Wang P, Chen W, Wu Z-C,
Jiang H and Tan L: Mitochondrial transcription factor A (TFAM)
polymorphisms and risk of late-onset Alzheimer's disease in Han
Chinese. Brain Res. 1368:355–360. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Günther C, von Hadeln K, Müller-Thomsen T,
Alberici A, Binetti G, Hock C, Nitsch RM, Stoppe G, Reiss J, Gal A,
et al: Possible association of mitochondrial transcription factor A
(TFAM) genotype with sporadic Alzheimer disease. Neurosci Lett.
369:219–223. 2004.PubMed/NCBI View Article : Google Scholar
|