Introduction
Research on genome-wide association analyses and the
association of single-nucleotide polymorphisms (SNPs) with various
diseases have seen great advances (1). The association between the
pathophysiology of chronic liver disease and SNPs has also been
reported. Patatin-like phospholipase domain-containing 3
(PNPLA3) rs738409 (2-4),
tolloid-like protein 1 (TLL1) rs17047200 (5-7)
and interleukin-28B (IL28b) rs809917 (8-14)
are associated with liver fibrosis, inflammation and steatosis in
hepatitis C virus (HCV)-infected patients.
Liver biopsy is considered the gold standard for
evaluating liver histology, including steatosis, inflammation and
fibrosis; however, it is an invasive procedure that can cause
various complications such as pain, fever, and bleeding (15). To address this limitation, FibroScan
and Virtual Touch tissue quantification (VTTQ) were developed as
modalities for the non-invasive assessment of liver fibrosis
(16,17). In addition, FibroScan includes
software for assessing hepatic steatosis using controlled
attenuation parameter (CAP) (16).
These modalities can be used to repeatedly evaluate liver
steatosis, inflammation and fibrosis.
Hepatitis C is a disease that causes cirrhosis and
hepatocellular carcinoma. It is estimated that 177.5 million
individuals worldwide are infected with HCV, which is 2.5% of the
world's population (17). HCV
infection poses a significant risk of cirrhosis and liver cancer.
Originally, interferon-based treatment was used as the treatment
for HCV, but the elimination rate of HCV was low, and adverse
events frequently appeared (18).
Direct-acting antivirals (DAAs) have been used as a treatment for
HCV, and a high treatment success rate has been reported (19). Ogasawara et al (20) reported the influence of DAA therapy
on liver stiffness (LS) and CAP, finding that the treatment of HCV
with DAA decreases LS and increases CAP after treatment. Therefore,
in this study, the impact of three SNPs (PNPLA3, TLL1
and IL28B) on changes in CAP and LS following DAA therapy
were examined.
Materials and methods
Patients
A total of 78 seven patients (39 females and 38
males; median age, 68 years; age range, 33-88 years) who received
DAA therapy for chronic HCV infection at Nagasaki University
Hospital (Nagasaki, Japan) and were subsequently confirmed as
HCV-negative 12 weeks after the end of treatment [sustained
virologic response (SVR12)] between September 2014 and December
2017 were enrolled in the study.
The patients' profiles and the results of laboratory
data at the start of DAA therapy are summarized in Table I. Written informed consent was
obtained from all patients, and the study protocol conformed to the
ethical guidelines of the 1975 Declaration of Helsinki (21). This study was approved by the Ethics
Committee of Nagasaki University (approval no. 15012688-3).
 | Table IBaseline characteristics of the
patients (n=77). |
Table I
Baseline characteristics of the
patients (n=77).
| Parameter | Value |
|---|
| Age, years
(range) | 68 (33-88) |
| Sex, n | |
|
Males | 38 |
|
Females | 39 |
| Chronic hepatitis,
n | 53 |
| Liver cirrhosis,
n | 24 |
| Total bilirubin,
mg/dla | 0.9 (0.3-2.5) |
| Albumin,
g/dla | 3.9 (2.5-4.9) |
| Low density
lipoprotein cholesterol, mg/dla | 88.8 (31-158) |
| Platelet count,
104/µla | 17.8
(3.9-47.0) |
| Aspartate
aminotransferase, IU/la | 49.2 (10-188) |
| Alanine
aminotransferase, IU/la | 40.0
(26.5-56.3) |
| Total cholesterol,
mg/dla | 166.1
(112-241) |
| Mac-2 binding
protein glycosylation isomera | 2.39
(0.37-14.57) |
| Body mass index,
kg/m2a | 22.5
(15.6-29.0) |
| Liver stiffness
measured using Virtual Touch Tissue Quantification,
m/sa | 1.57
(0.75-3.77) |
| Liver stiffness
measured using FibroScan, kPaa | 6.30
(4.40-11.63) |
| Controlled
attenuation parameter, dB/ma | 213.7
(100-373) |
| Direct acting
antiviral agent, n | 14/7/32/9/13/2 |
|
Daclatasvir/asunaprevir | 14 |
|
Sofosbuvir +
ribavirin | 7 |
|
Sofosbuvir/ledipasvir | 32 |
|
Ombitasvir/paritaprevir/ritonavir | 9 |
|
Elbasvir/grazoprevir | 13 |
|
Glecaprevir/pibrentasvir | 2 |
| PNPLA3
(rs738409) genotype, n | |
|
CC | 32 |
|
CG | 30 |
|
GG | 15 |
| TLL1
(rs17047200) genotype, n | |
|
AA | 57 |
|
AT | 20 |
|
TT | 0 |
| IL28B
(rs8099917) genotype, n | |
|
TT | 57 |
|
TG | 18 |
|
GG | 2 |
Measurement of LS and CAP
LS was measured twice, at the baseline (before DAA
therapy) and 12 weeks after the end of the treatment (sustained
virologic response, 12 weeks after the end of direct-acting
antiviral treatment; SVR12). LS was measured by VTTQ [LS (VTTQ)]
using an ACUSON S2000 (Siemens AG) and by transient elastography
with the M-probe of FibroScan 502 Touch (Echosens) [LS
(FibroScan)]. The CAP was also measured using FibroScan. The
patient was examined in a supine position with their right arm
raised. The tip of the probe was placed on the skin of the patient
between the ribs and the right lobe of the liver. For VTTQ
measurement, the region of interest was located 2-4 cm under the
capsule in the right lobe to avoid major blood vessels.
Measurements were taken five times and the median was used for
analysis. The LS results after VTTQ are presented as m/s (22). For FibroScan measurement, the probe
was placed on the skin between the ribs and aimed at a location
similar to that used for VTTQ. Measurements were taken 10 times,
and the median was used for analysis. The LS results after
FibroScan are presented as kPa, and CAP results are presented as
dB/m (23,24).
SNP genotyping
Genomic DNA was extracted from mononuclear cells in
peripheral blood samples of each patient using a FlexiGene DNA kit
(Qiagen GmbH). The SNPs in PNPLA3, TLL1 and
IL28B were genotyped in each sample using TaqMan SNP
genotyping assays kit (Thermo Fisher Scientific, Inc.) containing
two allele-specific TaqMan MGB probes labeled with different
fluorochromes and a PCR primer pair according to the manufacturer's
protocol. The following primers were used: PNPLA3 rs738409,
AGGCCTTGGTATGTTCCTGCTTCAT[C/G]CCCTTCTACAGTGGCCTTATCCCTC (cat. no.
4351379); TLL1 rs17047200,
TTTTGCCCACTTATGTCCATTTCAC[A/T]GTTCATTGACATCTATTTCTGAAGG (cat. no.
4351379); IL28B rs8099917,
TTTTGTTTTCCTTTCTGTGAGCAAT[G/T]TCACCCAAATTGGAACCATGCTGTA (cat. no.
4351379). As an example, the data for PNPLA3 are shown in Fig. S1. The protocol was the same as that
described in a previous study (25).
Statistical analysis
Data are presented as the median (inter-quartile
range). Pre- and post-treatment data were analyzed by a Wilcoxon
signed-rank test. P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using the StatFlex software (version 6.0; Artech).
Results
Whole group analysis
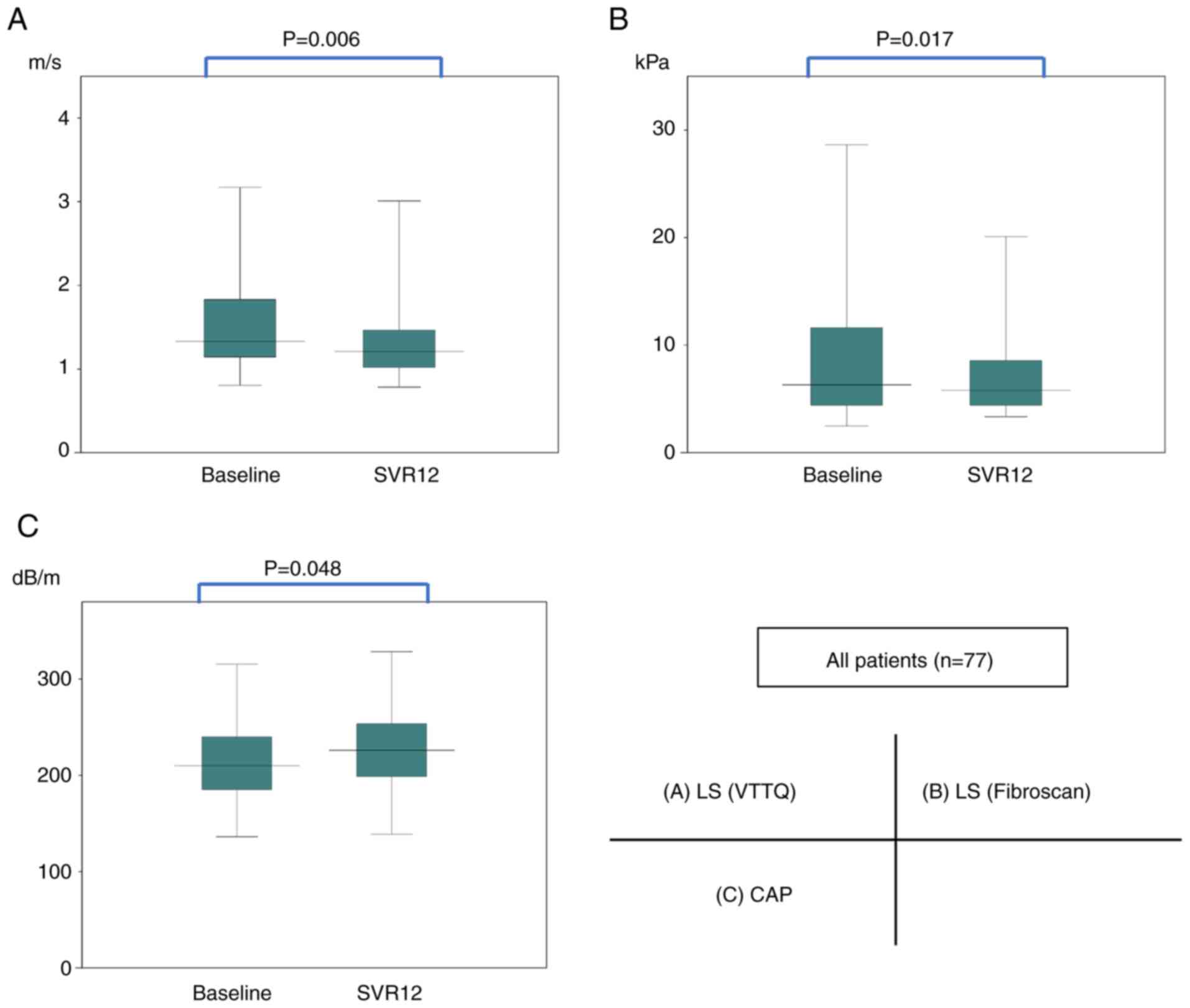
Fig. 1 shows the
changes in LS (VTTQ), LS (FibroScan) and CAP from before treatment
and at SVR12. In the whole group, the median LS (VTTQ) values
decreased significantly (P=0.006) from 1.33 to 1.21 m/s from the
start of the treatment to SVR12. The median LS (FibroScan) values
decreased significantly (P=0.017) from 6.10 to 5.80 kPa, and the
median CAP values increased significantly (P=0.048) from 210 to 226
dB/m.
The PNPLA3 genotype frequencies were as
follows: CC, 32 patients (42%); CG, 30 patients (39%); and GG, 15
patients (19%). The TLL1 genotype frequencies were as
follows: AA, 57 patients (74%); AT, 20 patients (26%); and TT, no
patients (0%). The IL28B genotype frequencies were as
follows: TT, 57 patients (74%); TG, 18 patients (23%); and GG, two
patients (3%). We divided PNPLA3 into CC and CG/GG groups,
TLL1 into AA and AT(/TT) groups, and IL28B into TT
and TG/GG groups.
Changes in LS (VTTQ)
Fig. 2 shows the
changes in the LS (VTTQ) for each SNP. In PNPLA3, the median
values of CC decreased significantly (P=0.035) from 1.39 to 1.19
m/s. The median values of CG/GG did not change significantly from
1.31 to 1.25 m/s. Even after dividing CG/GG genotypes into CG and
GG, no significant difference was found between CG and GG. In
TLL1, the median values of AA decreased significantly
(P=0.011) from 1.31 to 1.19 m/s. The median values of AT did not
change significantly from 1.43 to 1.26 m/s. In IL28B, the
median values of TT decreased significantly (P=0.005) from 1.36 to
1.19 m/s. The median values of TG/GG did not change significantly
from 1.31 to 1.36 m/s.
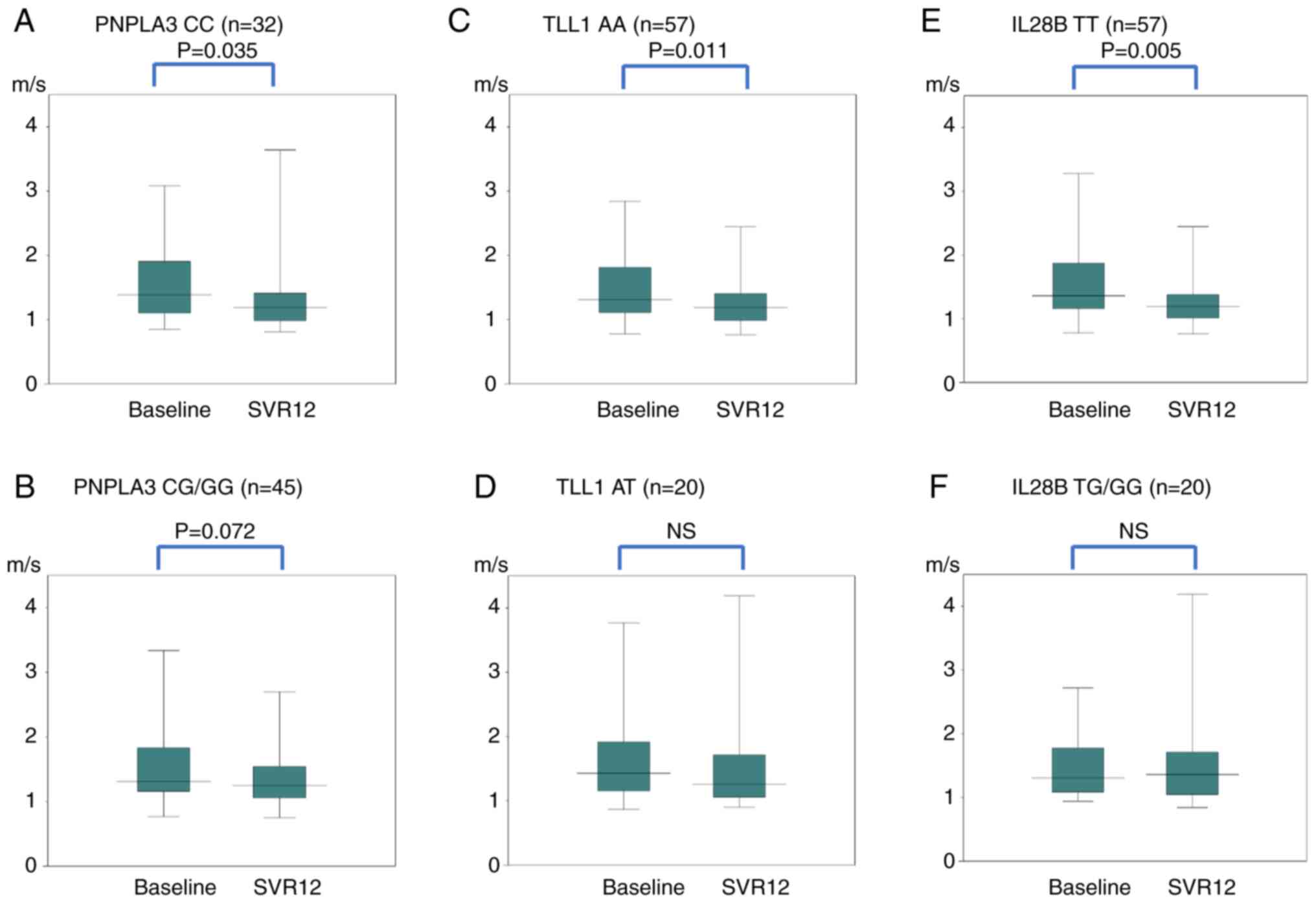 | Figure 2Changes in LS (VTTQ) for each SNP in
PNPLA3: (A) CC, (B) CG/GG, TLL1; (C) AA, (D) AT,
IL28B; (E) TT and (F) TG/GG genes. In the PNPLA3
gene, the median values of CC decreased significantly (P=0.035)
from 1.39 m/s before treatment to 1.19 m/s at SVR12. The median
values of CG/GG did not change significantly; 1.31 to 1.25 m/s. In
the TLL1 gene, the median values of AA decreased
significantly (P=0.011) from 1.31 to 1.19 m/s. The median values of
AT did change significantly; 1.43 to 1.26 m/s. In the IL28B
gene, the median values of TT decreased significantly (P=0.005)
from 1.36 to 1.19 m/s. The median values of TG/GG did not change
significantly; 1.31 to 1.36 m/s. LS, liver stiffness; VTTQ, virtual
touch tissue quantification; SNP, single-nucleotide polymorphism;
SVR12, sustained virologic response, 12 weeks after the end of
direct-acting antiviral treatment. |
Changes in LS (FibroScan)
Fig. 3 shows changes
in LS (FibroScan) for each SNP. In PNPLA3, the median values
of CC decreased from 6.15 kPa before treatment to 5.75 kPa at
SVR12, but the decrease was not significant. The median values of
CG/GG did not change significantly from 6.10 to 5.80 kPa. Even
after dividing CG/GG into CG and GG, no significant difference was
found between CG and GG. In TLL1, the median values of AA
decreased significantly (P=0.028) from 6.10 to 5.75 kPa. The median
values of AT did not change significantly from 6.60 to 5.10 kPa. In
IL28B, the median values of TT decreased significantly
(P=0.032) from 6.10 to 5.50 kPa. The median values of TG/GG did not
change significantly from 6.70 to 7.40 kPa.
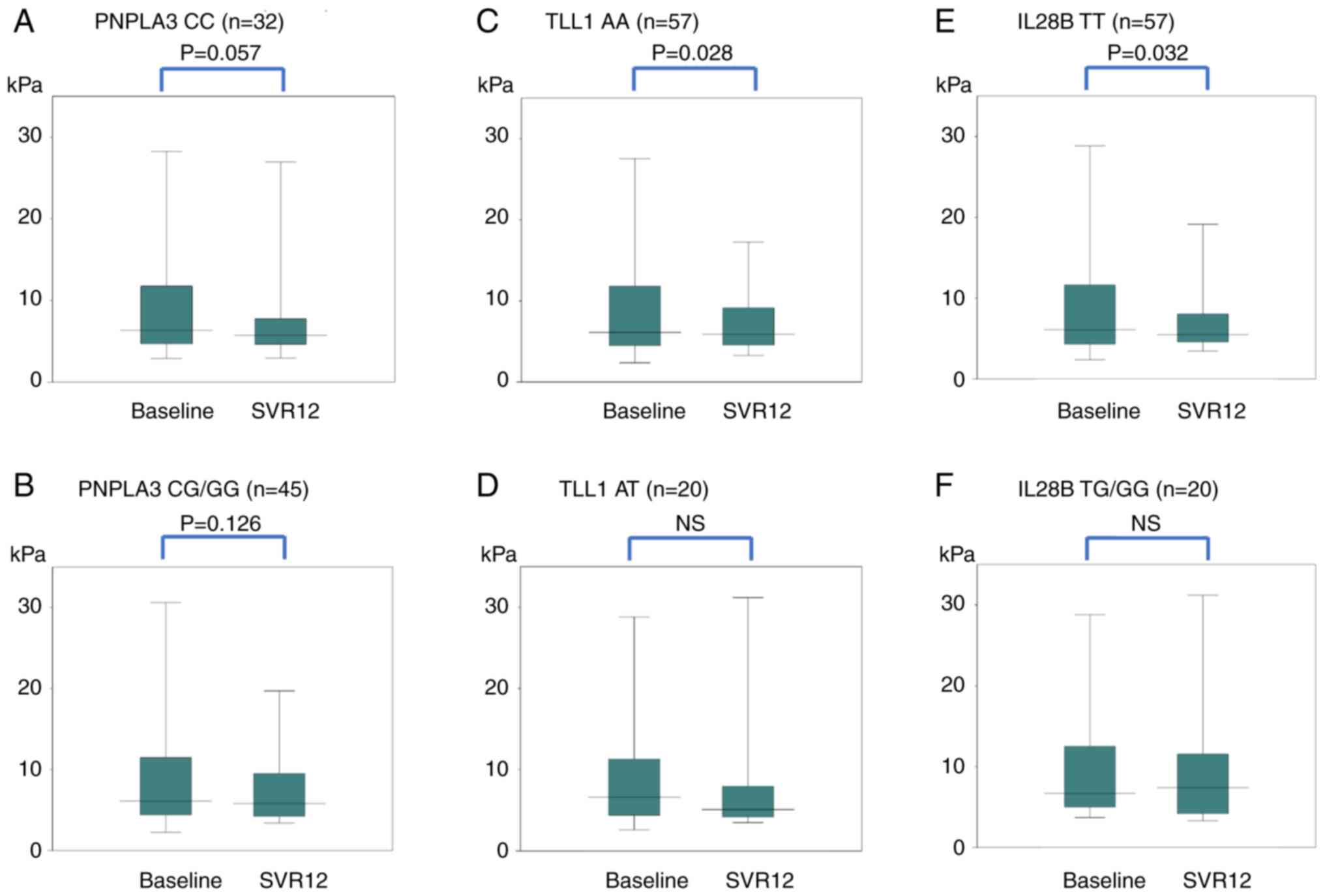 | Figure 3Changes in LS (FibroScan) for each
SNP in PNPLA3: (A) CC, (B) CG/GG, TLL1; (C) AA, (D)
AT, IL28B; (E) TT, and (F) TG/GG genes. In the PNPLA3
gene, the median values of CC were 6.15 kPa before DAA treatment
and 5.75 kPa at SVR12. There was no significant difference, but
there was a decreasing trend (P=0.057). The median values of CG/GG
did not change significantly; 6.10 to 5.80 kPa. In the TLL1
gene, the median values of AA decreased significantly (P=0.028)
from 6.10 to 5.75 kPa. The median values of AT did change not
significantly; 6.60 to 5.10 kPa. In the IL28B gene, the
median values of TT decreased significantly (P=0.032) from 6.10 to
5.50 kPa. The median values of TG/GG did not change significantly;
6.70 to 7.40 kPa. LS, liver stiffness; VTTQ, virtual touch tissue
quantification; SNP, single-nucleotide polymorphism; SVR12,
sustained virologic response, 12 weeks after the end of
direct-acting antiviral treatment. |
Changes in CAP
Fig. 4 shows the
changes in CAP for each SNP. In PNPLA3, the median values of
CC did not change significantly from 217 to 209 dB/m. The median
values of CG/GG increased significantly (P=0.004) from 204 to 233
dB/m. After dividing CG/GG into CG and GG, both CG (P=0.039) and GG
(P<0.05) increased significantly. In TLL1, the median
values of AA did not change significantly from 207 to 226 dB/m. The
median values of AT did not change significantly from 215 to 225
dB/m. In IL28B, the median values of TT increased
significantly (P=0.014) from 213 to 226 dB/m. The median values of
TG/GG did not change significantly from 206 to 218 dB/m.
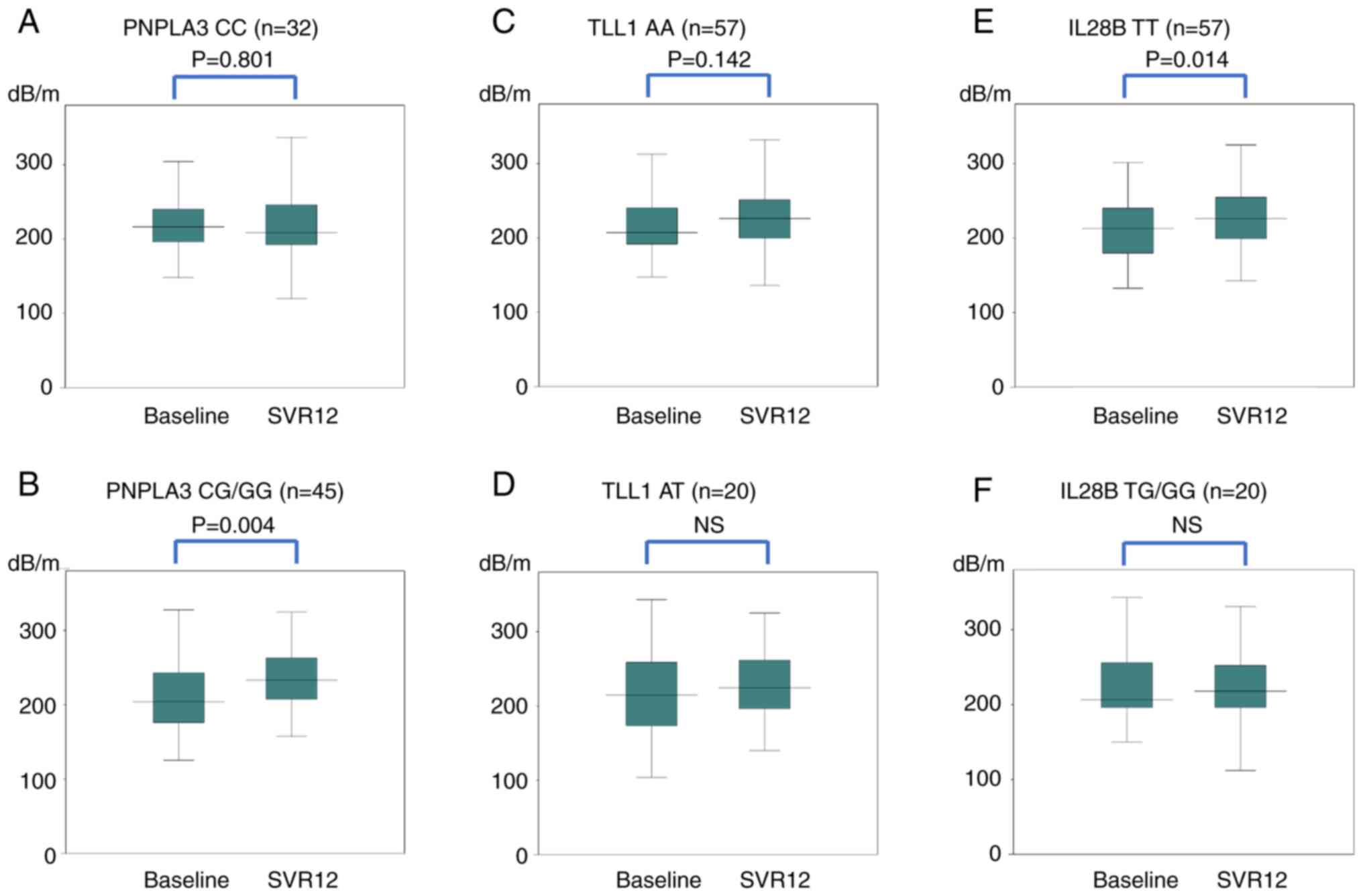 | Figure 4Changes in CAP for each SNP in
PNPLA3: (A) CC, (B) CG/GG, TLL1; (C) AA, (D) AT,
IL28B; (E) TT and (F) TG/GG genes. In the PNPLA3
gene, the median values of CC did not change significantly; 217
dB/m before treatment and 209 dB/m at SVR12. The median values of
CG/GG increased significantly (P=0.004) from 204 to 233 dB/m. In
the TLL1 gene, the median values of AA did not change
significantly; 207/m to 226 dB/m. The median values of AT did not
change significantly; 215 to 225 dB/m. In the IL28B, the
median values of TT increased significantly (P=0.014) from 213 to
226 dB/m. The median values of TG/GG did not change significantly;
206 to 218 dB/m. CAP, controlled attenuation parameter; SNP,
single-nucleotide polymorphism; SVR12, sustained virologic
response, 12 weeks after the end of direct-acting antiviral
treatment. |
Discussion
In this study, the impact of SNPs on changes in LS
and CAP after DAA therapy in patients with HCV infection were
determined. LS was evaluated using VTTQ and FibroScan, and CAP was
evaluated using FibroScan. LS (VTTQ) and LS (FibroScan)
significantly decreased, and CAP significantly increased at SVR12
in all cases. These results are consistent with those of a previous
study (20). LS reflects liver
fibrosis staging and liver inflammation; therefore, a decrease in
LS indicates an improvement in fibrosis and inflammation.
PNPLA3 rs738409 is considered a risk factor
for liver steatosis, fibrosis progression, and the development of
hepatocellular carcinoma in patients with non-alcoholic fatty liver
disease (26,27). Moreover, a previous study reported
that PNPLA3 is a significant risk factor for steatosis,
inflammation and fibrosis in patients with hepatitis C (2-4).
Furthermore, PNPLA3 is reportedly involved in lipase activity, and
a C to G substitution causes an amino acid substitution from
isoleucine to methionine at position 148 of the coding sequence
(I148M), with the resulting loss of function possibly involved in
hepatic steatosis. It is also possible that I148M affects the
activation of hepatic stellate cells related to fibrosis; however,
the associated mechanism has not yet been elucidated (2-4).
Previous studies suggested that TLL1 rs17047200 may
contribute to hepatocarcinogenesis through the progression of liver
fibrosis and that the AT/TT genotype is a risk factor for the
development of HCC after SVR (5-7).
Furthermore, TLL1 may be involved in the development of
hepatic fibrosis through extracellular matrix production and TGF-β
signal activation (5-7).
Another report indicated that SNPs located close to IL28B
significantly influenced the therapeutic outcome of combination
therapy involving pegylated IFN and ribavirin in HCV-infected
patients (28). Additionally, liver
fibrosis is reportedly exacerbated by the minor (14) and major alleles (8,13) of
IL28B rs80991, with a previous report suggesting that
changes in the IL28B allele alter the expression levels of
IFN-stimulating genes, including some inflammatory cytokines, and
may be involved in fibrosis. Moreover, increased IFN expression
reportedly suppresses lipoprotein lipase activity and reduces
very-low-density lipoprotein levels through low-density lipoprotein
conversion, which may be involved in hepatic steatosis (8,13).
These findings suggest the involvement of these SNPs in steatosis,
inflammation and fibrosis in patients with chronic hepatitis, and
the observed changes in LS and CAP in this study are consistent
with previous findings.
Previous studies showed that HCV eradication by IFN
and DAA therapy led to downstaging of fibrosis and reduced HCC
incidence, particularly in patients who achieved SVR (29-31).
However, HCC development can occur even in patients with SVR, and
the major risk factors include older age, alcohol intake,
pathological factors and diabetes (32-35).
Motoyama et al (36)
reported that stagnation of fibrosis regression is associated with
a high risk of HCC development after SVR. Additionally, liver
steatosis is reportedly a risk factor for HCC development after SVR
(37). Given these findings,
elevations in LS and CAP following DAA may represent a risk factor
for HCC development. Therefore, patients found to possess these
risk alleles need careful follow-ups with their doctors considering
the risk of HCC.
In the present study, median LS (VTTQ) and LS
(FibroScan) increased after treatment in IL28B TG/GG
patients, although no significant difference was observed. After
treatment, LS (VTTQ) increased in 26 out of 77 cases, and LS
(FibroScan) increased in 29 out of 77 cases. Of the 20 cases of
IL28B TG/GG, 7 cases exhibited an increase in LS (VTTQ) and
8 cases exhibited an increase in LS (FibroScan). Further analysis
is difficult due to the small number of cases, and it is necessary
to investigate whether IL28B TG/GG exacerbates fibrosis
after DAA treatment using a larger cohort. The median decrease in
CAP for PNPLA3 CC showed a similar result; therefore, this
should be investigated using a larger cohort further.
In PNPLA3 CC, the value of LS (VTTQ)
decreased significantly after treatment, but there was no
significant difference in LS (FibroScan). However, the
post-treatment value of LS (FibroScan) apparently showed a downward
trend, and therefore, it is necessary to investigate whether this
trend may be significant with a larger cohort.
In addition, as LS levels are affected by
inflammation, it may be useful to compare the parameters
immediately after treatment and 3 months after treatment. However,
it was difficult to compare these because the protocol of this
survey did not include the measurement of the LS value immediately
after the treatment. In future studies, the parameters should be
measured immediately after treatment as well.
The present study has certain limitations. The study
was conducted in a single center, and the sample size was small. To
validate the results observed in this study, a larger number of
samples should be investigated in the future. Additionally, this
study had a short observation period; therefore, it will be
necessary to consider longer observation periods in future studies.
Moreover, transient elastography was performed using only the
M-probe, which is intended for patients whose distance from the
measurement position on the skin to the liver is <25 mm,
suggesting a possible disadvantage in that measurement errors are
more likely at distances of ≥25 mm. An XL probe can be used for
such patients (38).
In conclusion, the results suggested that certain
SNPs were associated with the changes in liver fibrosis and
steatosis after DAA treatment for HCV infection.
Supplementary Material
TaqMan SNP genotyping of the
PNPLA3 gene. Vertical axis is the FAM C allele; horizontal
axis is VIC G allele. SNP, single nucleotide polymorphism;
PNPLA3, patatin-like phospholipase domain-containing 3; VIC,
Victoria dye; FAM, fluorescein amidites.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
KM was involved in data curation, formal analysis,
investigation and of writing the original draft. HM was involved in
conceptualization, methodology and project administration. MF, RS,
MH and SM were involved in data curation and formal analysis. KN
was involved in data curation and supervision. All authors have
read and approved the final manuscript. KM and HM confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. This study was approved by the Ethics Committee of
Nagasaki University (Nagasaki, Japan; approval no. 15012688-3).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li R, Li Y, Fang X, Yang H and Wang J,
Kristiansen K and Wang J: SNP detection for massively parallel
whole-genome resequencing. Genome Res. 19:1124–1132.
2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sato M, Kato N, Tateishi R, Muroyama R,
Kowatari N, Li W, Goto K, Otsuka M, Shiina S, Yoshida H, et al:
Impact of PNPLA3 polymorphisms on the development of hepatocellular
carcinoma in patients with chronic hepatitis C virus infection.
Hepatol Res. 44:E137–E144. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yasui K, Kawaguchi T, Shima T, Mitsuyoshi
H, Seki K, Sendo R, Mizuno M, Itoh Y, Matsuda F and Okanoue T:
Effect of PNPLA3 rs738409 variant (I148 M) on hepatic steatosis,
necroinflammation, and fibrosis in Japanese patients with chronic
hepatitis C. J Gastroenterol. 50:887–893. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Crisan D, Grigorescu M, Crisan N, Craciun
R, Lupsor M, Radu C, Grigorescu MD, Suciu A, Epure F, Avram L and
Leach N: Association between PNPLA3[G]/I148M variant, steatosis and
fibrosis stage in hepatitis C virus-genetic matters. J Physiol
Pharmacol. 70:2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Matsuura K, Sawai H, Ikeo K, Ogawa S, Iio
E, Isogawa M, Shimada N, Komori A, Toyoda H, Kumada T, et al:
Genome-wide association study identifies TLL1 variant associated
with development of hepatocellular carcinoma after eradication of
hepatitis C virus infection. Gastroenterology. 152:1383–1394.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Huang CF, Yeh ML, Huang CI, Lin ZY, Chen
SC, Huang JF, Dai CY, Chuang WL, Chen JJ and Yu ML: Tolloid-like 1
genetic variants determine fibrosis regression in chronic hepatitis
C patients with curative antivirals. Sci Rep.
8(15058)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Iio E, Matsuura K, Shimada N, Atsukawa M,
Itokawa N, Abe H, Kato K, Takaguchi K, Senoh T, Eguchi Y, et al:
TLL1 variant associated with development of hepatocellular
carcinoma after eradication of hepatitis C virus by interferon-free
therapy. J Gastroenterol. 54:339–346. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Abe H, Ochi H, Maekawa T, Hayes CN, Tsuge
M, Miki D, Mitsui F, Hiraga N, Imamura M, Takahashi S, et al:
Common variation of IL28 affects gamma-GTP levels and inflammation
of the liver in chronically infected hepatitis C virus patients. J
Hepatol. 53:439–443. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bochud PY, Bibert S, Negro F, Haagmans B,
Soulier A, Ferrari C, Missale G, Zeuzem S, Pawlotsky JM, Schalm S,
et al: IL28B polymorphisms predict reduction of HCV RNA from the
first day of therapy in chronic hepatitis C. J Hepatol. 55:980–988.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kurosaki M, Tanaka Y, Nishida N, Sakamoto
N, Enomoto N, Honda M, Sugiyama M, Matsuura K, Sugauchi F, Asahina
Y, et al: Pre-treatment prediction of response to
pegylated-interferon plus ribavirin for chronic hepatitis C using
genetic polymorphism in IL28B and viral factors. J Hepatol.
54:439–448. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ohnishi M, Tsuge M, Kohno T, Zhang Y, Abe
H, Hyogo H, Kimura Y, Miki D, Hiraga N, Imamura M, et al: IL28B
polymorphism is associated with fatty change in the liver of
chronic hepatitis C patients. J Gastroenterol. 47:834–844.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Asahina Y, Tsuchiya K, Nishimura T,
Muraoka M, Suzuki Y, Tamaki N, Yasui Y, Hosokawa T, Ueda K,
Nakanishi H, et al: Genetic variation near interleukin 28B and the
risk of hepatocellular carcinoma in patients with chronic hepatitis
C. J Gastroenterol. 49:1152–1162. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sato M, Kondo M, Tateishi R, Fujiwara N,
Kato N, Yoshida H, Taguri M and Koike K: Impact of IL28B genetic
variation on HCV-induced liver fibrosis, inflammation, and
steatosis: A meta-analysis. PLoS One. 9(e91822)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tamaki N, Kurosaki M, Higuchi M, Takada H,
Nakakuki N, Yasui Y, Suzuki S, Tsuchiya K, Nakanishi H, Itakura J,
et al: Genetic polymorphisms of IL28B and PNPLA3 are predictive for
HCV related rapid fibrosis progression and identify patients who
require urgent antiviral treatment with new regimens. PLoS One.
10(e0137351)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Myers RP, Fong A and Shaheen AA:
Utilization rates, complications and costs of percutaneous liver
biopsy: A population-based study including 4275 biopsies. Liver
Int. 28:705–712. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sasso M, Beaugrand M, de Ledinghen V,
Douvin C, Marcellin P, Poupon R, Sandrin L and Miette V: Controlled
attenuation parameter (CAP): A novel VCTE™ guided
ultrasonic attenuation measurement for the evaluation of hepatic
steatosis: Preliminary study and validation in a cohort of patients
with chronic liver disease from various causes. Ultrasound Med
Biol. 36:1825–1835. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Petruzziello A, Marigliano S, Loquercio G,
Cozzolino A and Cacciapuoti C: Global epidemiology of hepatitis C
virus infection: An up-date of the distribution and circulation of
hepatitis C virus genotypes. World J Gastroenterol. 22:7824–7840.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Attar BM and Van Thiel DH: Hepatitis C
virus: A time for decisions. Who should be treated and when? World
J Gastrointest Pharmacol Ther. 7:33–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Leroy V, Angus P, Bronowicki JP, Dore GJ,
Hezode C, Pianko S, Pol S, Stuart K, Tse E, McPhee F, et al:
Daclatasvir, sofosbuvir, and ribavirin for hepatitis C virus
genotype 3 and advanced liver disease: A randomized phase III study
(ALLY-3+). Hepatology. 63:1430–1441. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ogasawara N, Kobayashi M, Akuta N,
Kominami Y, Fujiyama S, Kawamura Y, Sezaki H, Hosaka T, Suzuki F,
Saitoh S, et al: Serial changes in liver stiffness and controlled
attenuation parameter following direct-acting antiviral therapy
against hepatitis C virus genotype 1b. J Med Virol. 90:313–319.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
World Medical Association: World medical
association declaration of Helsinki. Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bamber J, Cosgrove D, Dietrich CF,
Fromageau J, Bojunga J, Calliada F, Cantisani V, Correas JM,
D'Onofrio M, Drakonaki EE, et al: EFSUMB guidelines and
recommendations on the clinical use of ultrasound elastography.
Part 1: Basic principles and technology. Ultraschall Med.
34:169–184. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Boursier J, Zarski JP, de Ledinghen V,
Rousselet MC, Sturm N, Lebail B, Fouchard-Hubert I, Gallois Y,
Oberti F, Bertrais S, et al: Determination of reliability criteria
for liver stiffness evaluation by transient elastography.
Hepatology. 57:1182–1191. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kumagai E, Korenaga K, Korenaga M, Imamura
M, Ueyama M, Aoki Y, Sugiyama M, Murata K, Masaki N, Kanto T, et
al: Appropriate use of virtual touch quantification and FibroScan M
and XL probes according to the skin capsular distance. J
Gastroenterol. 51:496–505. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
De la Vega FM, Lazaruk KD, Rhodes MD and
Wenz MH: Assessment of two flexible and compatible SNP genotyping
platforms: TaqMan SNP genotyping assays and the SNPlex genotyping
system. Mutat Res. 573:111–135. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Romeo S, Kozlitina J, Xing C, Pertsemlidis
A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC and Hobbs HH:
Genetic variation in PNPLA3 confers susceptibility to nonalcoholic
fatty liver disease. Nat Genet. 40:1461–1465. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Seko Y, Sumida Y, Tanaka S, Mori K,
Taketani H, Ishiba H, Hara T, Okajima A, Umemura A, Nishikawa T, et
al: Development of hepatocellular carcinoma in Japanese patients
with biopsy-proven non-alcoholic fatty liver disease: Association
between PNPLA3 genotype and hepatocarcinogenesis/fibrosis
progression. Hepatol Res. 47:1083–1092. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tanaka Y, Nishida N, Sugiyama M, Kurosaki
M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S,
et al: Genome-wide association of IL28B with response to pegylated
interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat
Genet. 41:1105–1109. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Morgan TR, Ghany MG, Kim HY, Snow KK,
Shiffman ML, De Santo JL, Lee WM, Di Bisceglie AM, Bonkovsky HL,
Dienstag JL, et al: Outcome of sustained virological responders
with histologically advanced chronic hepatitis C. Hepatology.
52:833–844. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kanwal F, Kramer J, Asch SM, Chayanupatkul
M, Cao Y and El-Serag HB: Risk of hepatocellular cancer in HCV
patients treated with direct-acting antiviral agents.
Gastroenterology. 153:996–1005.e1. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ogata F, Kobayashi M, Akuta N, Osawa M,
Fujiyama S, Kawamura Y, Sezaki H, Hosaka T, Kobayashi M, Saitoh S,
et al: Outcome of all-oral direct-acting antiviral regimens on the
rate of development of hepatocellular carcinoma in patients with
hepatitis C virus genotype 1-related chronic liver disease.
Oncology. 93:92–98. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Makiyama A, Itoh Y, Kasahara A, Imai Y,
Kawata S, Yoshioka K, Tsubouchi H, Kiyosawa K, Kakumu S, Okita K,
et al: Characteristics of patients with chronic hepatitis C who
develop hepatocellular carcinoma after a sustained response to
interferon therapy. Cancer. 101:1616–1622. 2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Asahina Y, Tsuchiya K, Nishimura T,
Muraoka M, Suzuki Y, Tamaki N, Yasui Y, Hosokawa T, Ueda K,
Nakanishi H, et al: α-Fetoprotein levels after interferon therapy
and risk of hepatocarcinogenesis in chronic hepatitis C.
Hepatology. 58:1253–1262. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
El-Serag HB, Kanwal F, Richardson P and
Kramer J: Risk of hepatocellular carcinoma after sustained
virological response in Veterans with hepatitis C virus infection.
Hepatology. 64:130–137. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ioannou GN, Green PK, Beste LA, Mun EJ,
Kerr KF and Berry K: Development of models estimating the risk of
hepatocellular carcinoma after antiviral treatment for hepatitis C.
J Hepatol. 69:1088–1098. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Motoyama H, Tamori A, Kubo S,
Uchida-Kobayashi S, Takemura S, Tanaka S, Ohfuji S, Teranishi Y,
Kozuka R, Kawamura E, et al: Stagnation of histopathological
improvement is a predictor of hepatocellular carcinoma development
after hepatitis C virus eradication. PLoS One.
13(e0194163)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tanaka A, Uegaki S, Kurihara H, Aida K,
Mikami M, Nagashima I, Shiga J and Takikawa H: Hepatic steatosis as
a possible risk factor for the development of hepatocellular
carcinoma after eradication of hepatitis C virus with antiviral
therapy in patients with chronic hepatitis C. World J
Gastroenterol. 13:5180–5187. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Myers RP, Pomier-Layrargues G, Kirsch R,
Pollett A, Duarte-Rojo A, Wong D, Beaton M, Levstik M, Crotty P and
Elkashab M: Feasibility and diagnostic performance of the FibroScan
XL probe for liver stiffness measurement in overweight and obese
patients. Hepatology. 55:199–208. 2012.PubMed/NCBI View Article : Google Scholar
|