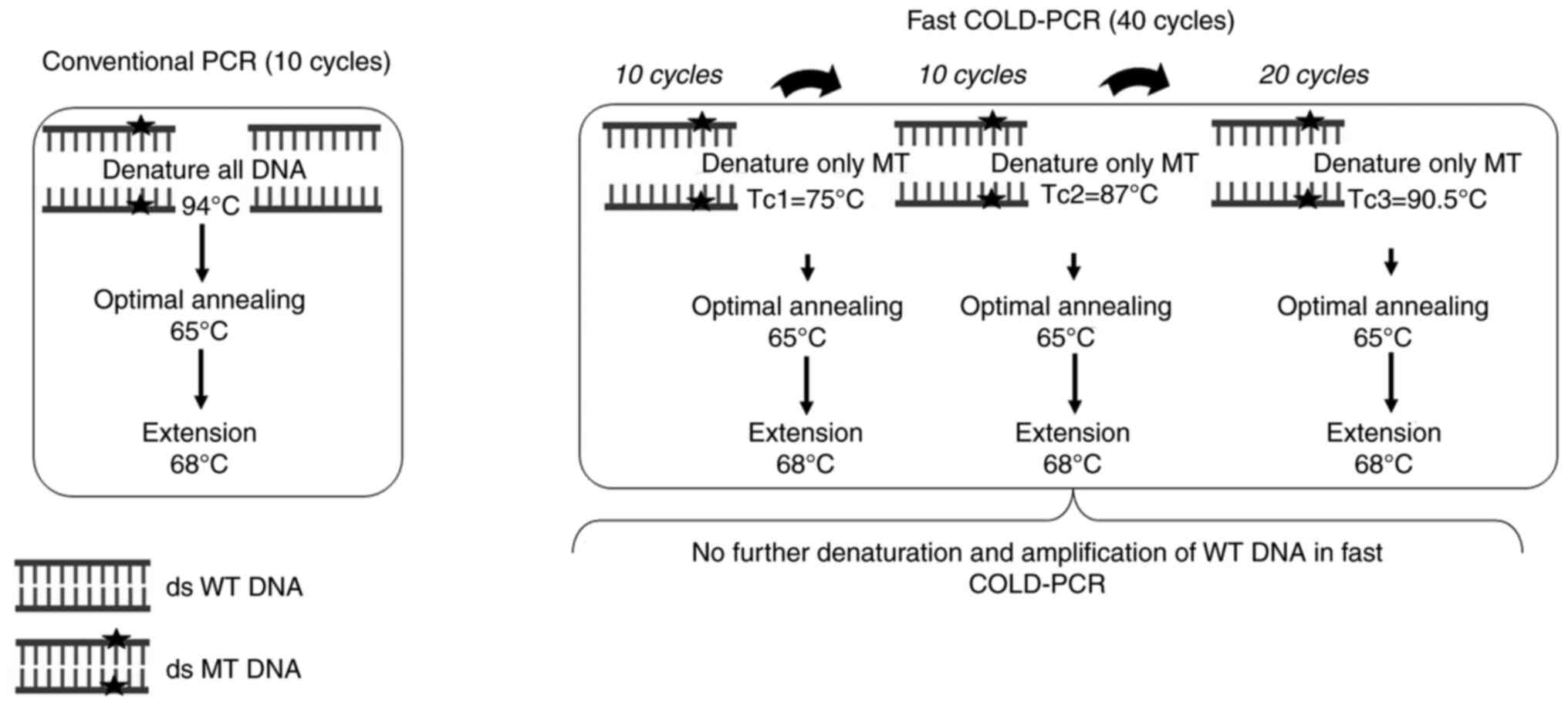
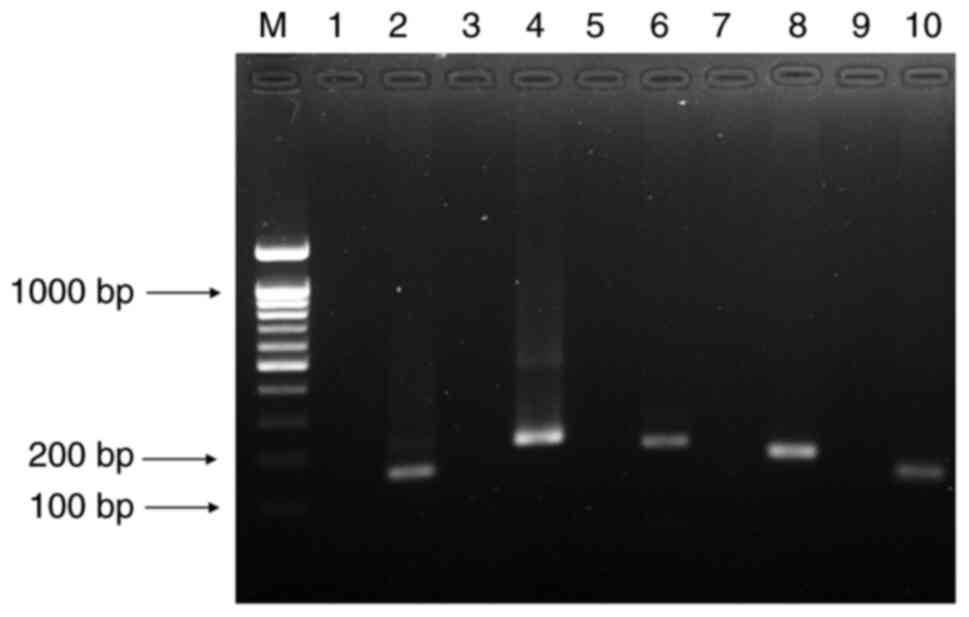
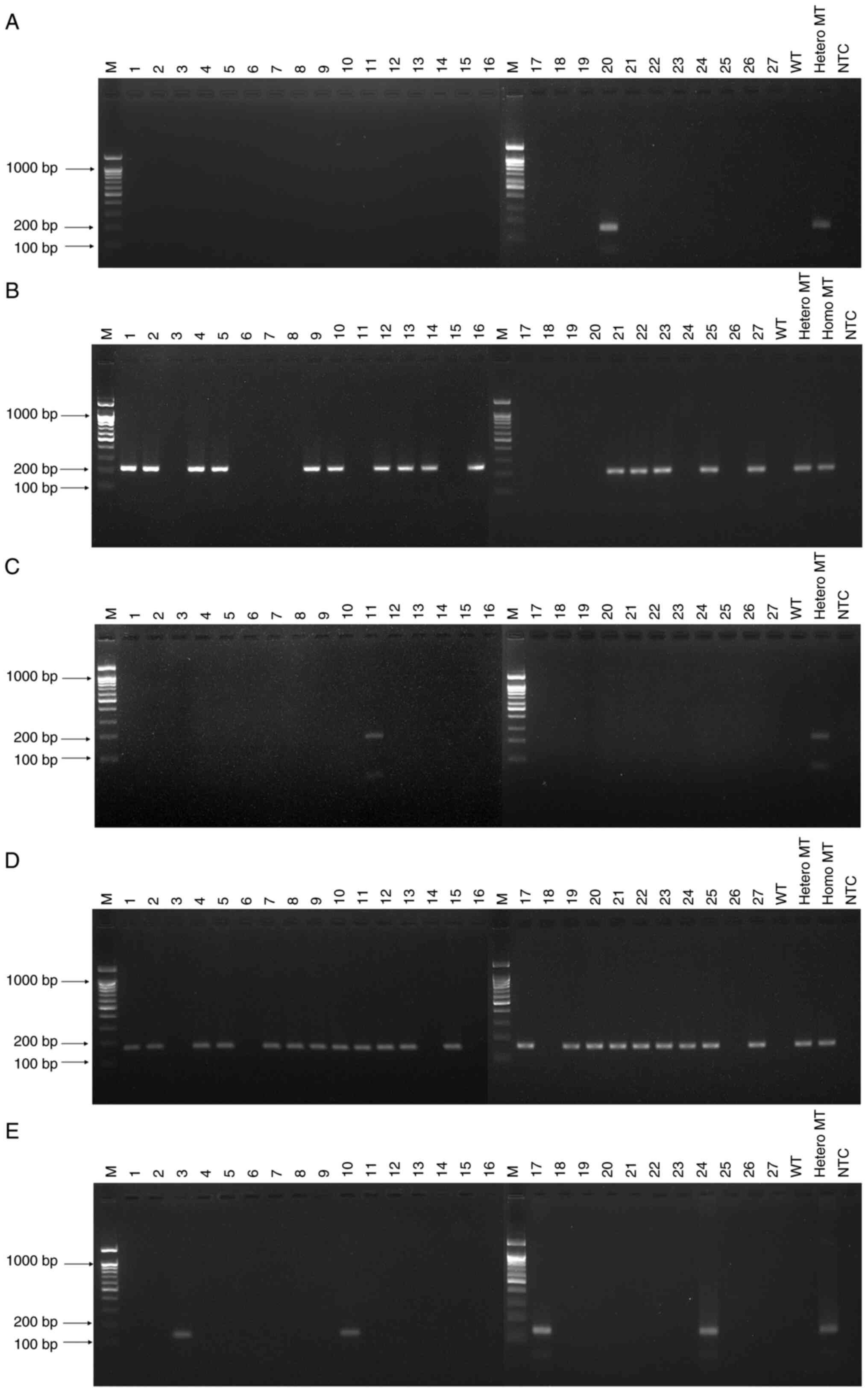
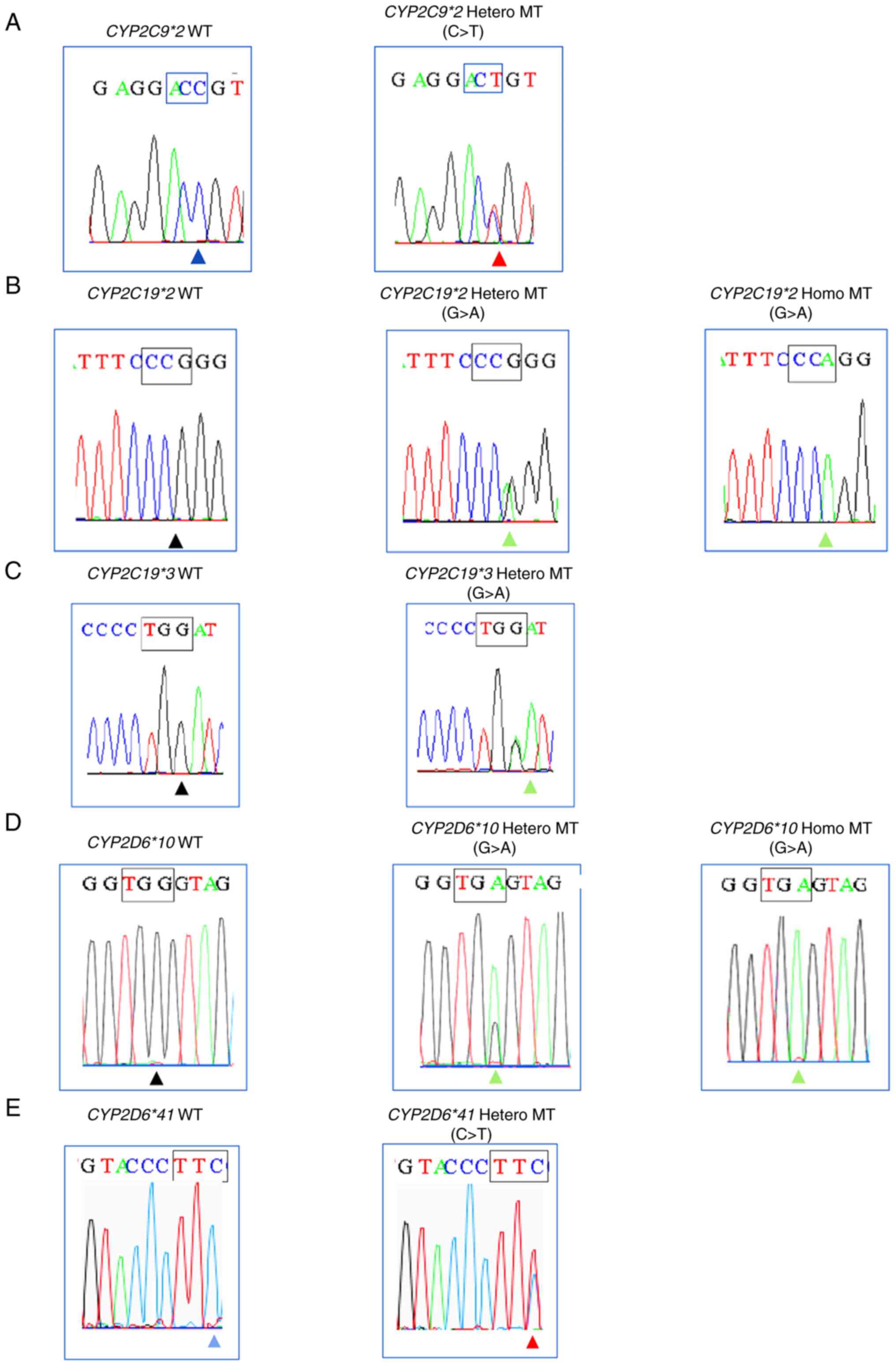
|
1
|
Arici M and Özhan G: CYP2C9, CYPC19 and
CYP2D6 gene profiles and gene susceptibility to drug response and
toxicity in Turkish population. Saudi Pharm J. 25:376–380.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brockmöller J, Kirchheiner J, Meisel C and
Roots I: Pharmacogenetic diagnostics of cytochrome P450
polymorphisms in clinical drug development and in drug treatment.
Pharmacogenomics. 1:125–151. 2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Malki MA and Pearson ER: Drug-drug-gene
interactions and adverse drug reactions. Pharmacogenomics J.
20:355–366. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
D'Empaire I, Guico-Pabia CJ and Preskorn
SH: Antidepressant treatment and altered CYP2D6 activity: Are
pharmacokinetic variations clinically relevant? J Psychiatr Pract.
17:330–339. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sim SC, Risinger C, Dahl ML, Aklillu E,
Christensen M, Bertilsson L and Ingelman-Sundberg M: A common novel
CYP2C19 gene variant causes ultrarapid drug metabolism relevant for
the drug response to proton pump inhibitors and antidepressants.
Clin Pharmacol Ther. 79:103–113. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Westergaard N, Søgaard Nielsen R,
Jørgensen S and Vermehren C: Drug use in denmark for drugs having
pharmacogenomics (PGx) based dosing guidelines from CPIC or DPWG
for CYP2D6 and CYP2C19 Drug-gene pairs: Perspectives for
introducing PGx test to polypharmacy patients. J Pers Med.
10(3)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Abdullah-Koolmees H, van Keulen AM,
Nijenhuis M and Deneer VHM: Pharmacogenetics guidelines: Overview
and comparison of the DPWG, CPIC, CPNDS, and RNPGx guidelines.
Front Pharmacol. 11(595219)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Verbeurgt P, Mamiya T and Oesterheld J:
How common are drug and gene interactions? Prevalence in a sample
of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping.
Pharmacogenomics. 15:655–665. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Dong Y, Xiao H, Wang Q, Zhang C, Liu X,
Yao N, Sheng H and Li H: Analysis of genetic variations in CYP2C9,
CYP2C19, CYP2D6 and CYP3A5 genes using oligonucleotide microarray.
Int J Clin Exp Med. 8:18917–18926. 2015.PubMed/NCBI
|
|
10
|
Samer CF, Lorenzini KI, Rollason V, Daali
Y and Desmeules JA: Applications of CYP450 testing in the clinical
setting. Mol Diagn Ther. 17:165–184. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sukri A, Salleh MZ, Masimirembwa C and Teh
LK: A systematic review on the cost effectiveness of
pharmacogenomics in developing countries: Implementation
challenges. Pharmacogenomics J. 22:147–159. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Luzum JA and Luzum MJ: Physicians'
attitudes toward pharmacogenetic testing before and after
pharmacogenetic education. Per Med. 13:119–127. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Galbiati S, Monguzzi A, Damin F, Soriani
N, Passiu M, Castellani C, Natacci F, Curcio C, Seia M, Lalatta F,
et al: COLD-PCR and microarray: Two independent highly sensitive
approaches allowing the identification of fetal paternally
inherited mutations in maternal plasma. J Med Genet. 53:481–487.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Byrou S, Makrigiorgos GM, Christofides A,
Kallikas I, Papasavva T and Kleanthous M: Fast Temperature-gradient
COLD PCR for the enrichment of the paternally inherited SNPs in
cell free fetal DNA; an application to non-invasive prenatal
diagnosis of β-thalassaemia. PLoS One. 13(e0200348)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li J, Wang L, Mamon H, Kulke MH, Berbeco R
and Makrigiorgos GM: Replacing PCR with COLD-PCR enriches variant
DNA sequences and redefines the sensitivity of genetic testing. Nat
Med. 14:579–584. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Boisselier B, Marie Y, Labussière M,
Ciccarino P, Desestret V, Wang X, Capelle L, Delattre JY and Sanson
M: COLD PCR HRM: A highly sensitive detection method for IDH1
mutations. Hum Mutat. 31:1360–1365. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li J, Milbury CA, Li C and Makrigiorgos
GM: Two-round coamplification at lower denaturation temperature-PCR
(COLD-PCR)-based sanger sequencing identifies a novel spectrum of
low-level mutations in lung adenocarcinoma. Hum Mutat.
30:1583–1590. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zuo Z, Chen SS, Chandra PK, Galbincea JM,
Soape M, Doan S, Barkoh BA, Koeppen H, Medeiros LJ and Luthra R:
Application of COLD-PCR for improved detection of KRAS mutations in
clinical samples. Mod Pathol. 22:1023–1031. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kristensen LS, Daugaard IL, Christensen M,
Hamilton-Dutoit S, Hager H and Hansen LL: Increased sensitivity of
KRAS mutation detection by high-resolution melting analysis of
COLD-PCR products. Hum Mutat. 31:1366–1373. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Milbury CA, Chen CC, Mamon H, Liu P,
Santagata S and Makrigiorgos GM: Multiplex amplification coupled
with COLD-PCR and high resolution melting enables identification of
low-abundance mutations in cancer samples with low DNA content. J
Mol Diagn. 13:220–232. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Castellanos-Rizaldos E, Liu P, Milbury CA,
Guha M, Brisci A, Cremonesi L, Ferrari M, Mamon H and Makrigiorgos
GM: Temperature-tolerant COLD-PCR reduces temperature stringency
and enables robust mutation enrichment. Clin Chem. 58:1130–1138.
2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Castellanos-Rizaldos E, Milbury CA and
Makrigiorgos GM: Enrichment of mutations in multiple DNA sequences
using COLD-PCR in emulsion. PLoS One. 7(e51362)2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Carotenuto P, Roma C, Cozzolino S, Fenizia
F, Rachiglio AM, Tatangelo F, Iannaccone A, Baron L, Botti G and
Normanno N: Detection of KRAS mutations in colorectal cancer with
Fast COLD-PCR. Int J Oncol. 40:378–384. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sistonen J, Fuselli S, Palo JU, Chauhan N,
Padh H and Sajantila A: Pharmacogenetic variation at CYP2C9,
CYP2C19, and CYP2D6 at global and microgeographic scales.
Pharmacogenet Genomics. 19:170–179. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bradford LD: CYP2D6 allele frequency in
European Caucasians, Asians, Africans and their descendants.
Pharmacogenomics. 3:229–243. 2002.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dorji PW, Tshering G and Na-Bangchang K:
CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in south-east and
east asian populations: A systematic review. J Clin Pharm Ther.
44:508–524. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tassaneeyakul W, Mahatthanatrakul W,
Niwatananun K, Na-Bangchang K, Tawalee A, Krikreangsak N, Cykleng U
and Tassaneeyakul W: CYP2C19 genetic polymorphism in thai, burmese
and karen populations. Drug Metab Pharmacokinet. 21:286–290.
2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lo C, Nguyen S, Yang C, Witt L, Wen A,
Liao TV, Nguyen J, Lin B, Altman RB and Palaniappan L:
Pharmacogenomics in asian subpopulations and impacts on commonly
prescribed medications. Clin Transl Sci. 13:861–870.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Crossley BM, Bai J, Glaser A, Maes R,
Porter E, Killian ML, Clement T and Toohey-Kurth K: . Guidelines
for Sanger sequencing and molecular assay monitoring. J Vet Diagn
Invest. 32:767–775. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Blazejewski T, Ho HI and Wang HH:
Synthetic sequence entanglement augments stability and containment
of genetic information in cells. Science. 365:595–598.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Macrogen. Capillary Electrophoresis
Sequencing (CES). Available online: https://dna.macrogen.com/pageLinkDnaSys.do?layout=page_sub&link=/support/retrieveGuideCes
(Accession date 16 October 2022).
|
|
32
|
Sim SC and Ingelman-Sundberg M: The human
cytochrome P450 (CYP) allele nomenclature website: A peer-reviewed
database of CYP variants and their associated effects. Hum
Genomics. 4(278)2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kubota A, Stegeman JJ, Goldstone JV,
Nelson DR, Kim EY, Tanabe S and Iwata H: Cytochrome P450 CYP2 genes
in the common cormorant: Evolutionary relationships with 130
diapsid CYP2 clan sequences and chemical effects on their
expression. Comp Biochem Physiol C Toxicol Pharmacol. 153:280–289.
2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gaedigk A: Complexities of CYP2D6 gene
analysis and interpretation. Int Rev Psychiatry. 25:534–553.
2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gotoh O: Substrate recognition sites in
cytochrome P450 family 2 (CYP2) proteins inferred from comparative
analyses of amino acid and coding nucleotide sequences. J Biol
Chem. 267:83–90. 1992.PubMed/NCBI
|
|
36
|
Söderbäck E, Zackrisson AL, Lindblom B and
Alderborn A: Determination of CYP2D6 gene copy number by
pyrosequencing. Clin Chem. 51:522–531. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nakamura N, Fukuda T, Nonen S, Hashimoto
K, Azuma J and Gemma N: Simple and accurate determination of CYP2D6
gene copy number by a loop-mediated isothermal amplification method
and an electrochemical DNA chip. Clin Chim Acta. 411:568–573.
2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Puaprasert K, Chu C, Saralamba N, Day NPJ,
Nosten F, White NJ, Dondorp AM and Imwong M: Real time PCR
detection of common CYP2D6 genetic variants and its application in
a Karen population study. Malar J. 17(427)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carvalho Henriques B, Buchner A, Hu X,
Wang Y, Yavorskyy V, Wallace K, Dong R, Martens K, Carr MS, Asl B,
et al: Methodology for clinical genotyping of CYP2D6 and CYP2C19.
Transl Psychiatry. 11(596)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
de Lecea MGM and Rossbach M: Translational
genomics in personalized medicine-scientific challenges en route to
clinical practice. Hugo J. 6(2)2012.
|
|
41
|
Sukprasong R, Chuwongwattana S, Koomdee N,
Jantararoungtong T, Prommas S, Jinda P, Rachanakul J,
Nuntharadthanaphong N, Jongjitsook N, Puangpetch A and Sukasem C:
Allele frequencies of single nucleotide polymorphisms of clinically
important drug-metabolizing enzymes CYP2C9, CYP2C19, and CYP3A4 in
a Thai population. Sci Rep. 11(12343)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hongkaew Y, Gaedigk A, Wilffert B,
Ngamsamut N, Kittitharaphan W, Limsila P and Sukasem C:
Relationship between CYP2D6 genotype, activity score and phenotype
in a pediatric Thai population treated with risperidone. Sci Rep.
11(4158)2021.PubMed/NCBI View Article : Google Scholar
|