Introduction
Drug-metabolizing cytochrome P450 (CYP) phase I
bioactivation system affects drug responses. Among >50 CYPs,
genetic variations of CYP2C9, CYP2C19 and CYP2D6 enzymes
potentially affect drug efficacy and toxicity. CYP2C9,
CYP2C19 and CYP2D6 polymorphisms comprise the most
frequent enzyme variations because nearly 80% of drugs used in
today are metabolized by these enzymes. Accordingly, these
CYP genetic mutations lead to different phenotypes of
metabolism status, such as ultra-rapid (UMs), normal, intermediate
(IMs) and poor metabolizers (PMs). In UMs, individuals metabolize
drugs very rapidly, resulting in lack of response and
subtherapeutic plasma concentrations at normal doses whereas in IMs
or PMs, these lead to altered risk for adverse drug reactions
(1-3).
For example, altered CYP2D6 activity affects antidepressant
treatment (4) and CYP2C19*17
leading to UM phenotype causes risk of therapeutic failure in drug
treatment (5). Therefore,
annotations of these CYP genes and pharmacogenomics
(PGx)-based drug-dosing guidelines are being constantly updated to
make dose adjustment to avoid toxicity and increase drug efficacy
(6,7). This may maximize drug efficacy and
minimize toxicity for individuals from drugs, thereby improving
patient compliance and safety. Thus, PGx testing has been
implemented in these three CYP polymorphisms (CYP2C9,
CYP2C19 and CYP2D6) to achieve optimal quality use of
medicines (8). Various approaches
have been conducted, such as allele-specific PCR, invader assay,
pyrosequencing and oligonucleotide microarray (9). A variety of testing kits for
CYP450 genotyping have also been approved by the U.S. FDA,
including Amplicon Chip CYP450 GeneChip®, TaqMan
real-time PCR and Luminex CYP2D6 and CY2C19 xTAG
detection kits (10).
Nevertheless, use of PGx testing as a routine
practice is still challenging due to an underestimation of clinical
importance, lack of health information and high cost in developing
countries (11). Even though
physicians are educated on healthcare, optimistic attitudes to PGx
are still demanding because of lack of participation in controlled
trials and clinical validity. One example of variations between
pharmacogenetic clinical guidelines and recommendations was found
in clopidogrel in which clinical guidelines and FDA demonstrated
different recommendations upon the interpretation of PGx testing
(12). The key role of PGx is to
divide drug responders from non-responders for physicians.
The aim of this study was to qualitatively evaluate
pharmacoeconomic characteristics. The objective of this study was
to introduce an alternative pipeline to detect mutation before
traditional genotyping for PGx. Co-amplification at lower
denaturation temperature (COLD)-PCR technology is considered to be
a better qualitative detection method in minority allele detection
than conventional PCR because of its feasibility, simplicity,
time-efficiency and cost-effectiveness with preferential
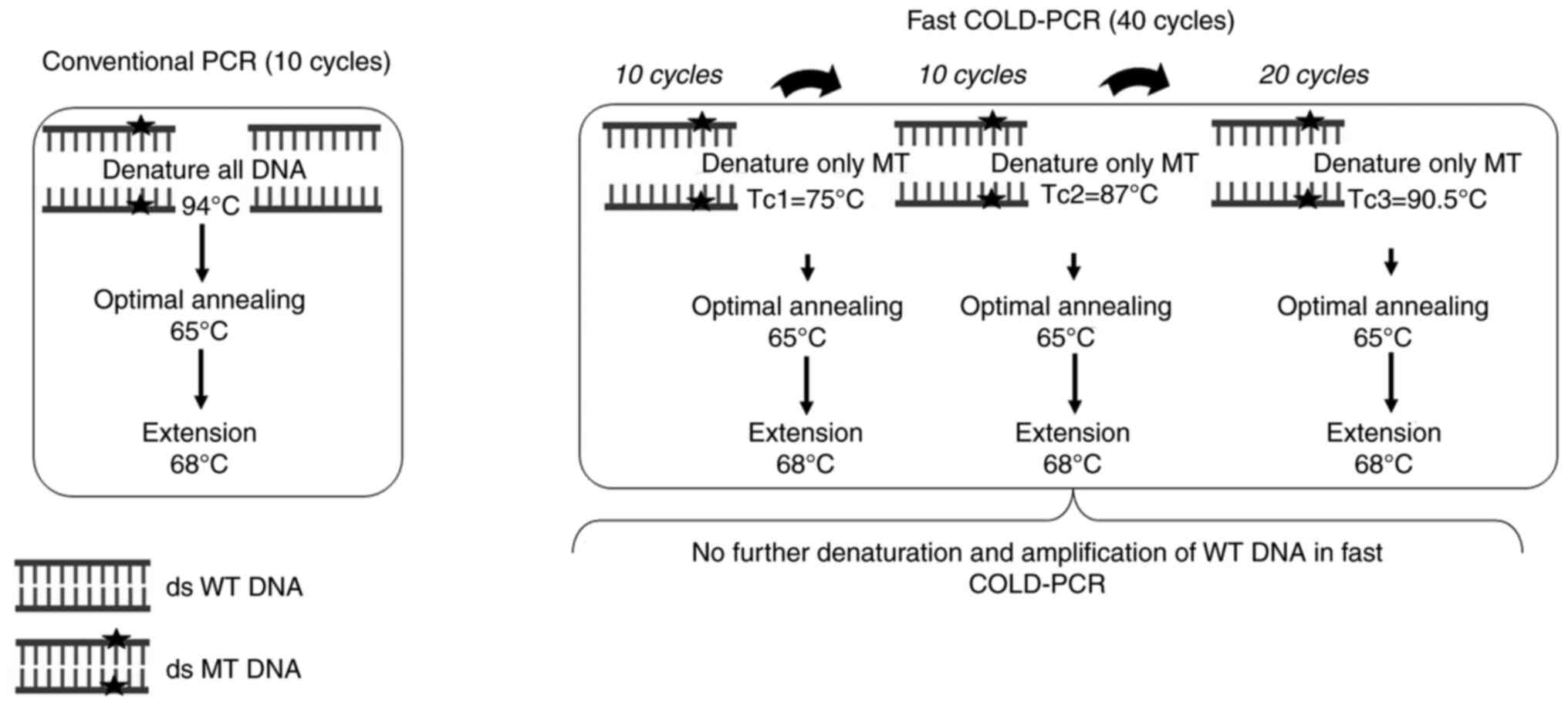
denaturation on mismatch-forming variants (13). There are several forms of COLD-PCR,
among them, fast COLD-PCR is cheapest and easiest. Fast COLD-PCR is
a modified form of conventional PCR involving an additional
parameter, the critical denaturation temperature (Tc), primarily
suitable for Tm-reducing mutations (for example, G:C>A:T or
G:C>T:A) (14). Following this
selective denaturation, only mutant (MT) A/T-containing alleles are
obtained and wild-type (WT) G/C alleles are left double-stranded
(ds). This can result in increased sensitivity in the detection of
low-abundance variants over conventional PCR (15). It not only enables robust enrichment
but is also easily accessible with high reproducibility. As a
consequence of this, COLD-PCR has been widely applied to detect
cancer mutations (16-23).
To date, there are only a few reports of
pharmacogenomics (PGx) studies (2,7,12) in
which fast COLD-PCR has not yet been applied. Therefore, the
present study introduce an affordable methodology to determine
whether the patient carries a variant without requiring
heterozygous or homozygous variant typing. Accordingly, this can
decrease unnecessary expensive direct genotyping in uncharacterized
patients. Moreover, the present study aimed to demonstrate how to
multiplex fast COLD-PCR based on precise Tc values, which has
previously been difficult because the critical denaturation
temperature of COLD-PCR must be controlled precisely (within
±0.2˚C). Therefore, it is critical to set up a thermocycler with
precise temperature. The novel assay panel used selected
gene-associated single nucleotide polymorphisms (SNPs) specific to
Asian populations published in previous studies (24-28).
The present method may promote use of more
pharmacogenetic-associated SNPs. To assess the efficiency, the
testing results were compared with those from Sanger sequencing,
which is the reliable available gold-standard method (29) to determine heterozygous or
homozygous patients.
Materials and methods
Database for searching for sequences
of CYP genes
The reference sequences of CYP2C9 and
CYP2C19 on chromosome 10 (accession no. NC_000010.11) and
CYP2D6 on chromosome 22 (accession no NC_000022.11) from
Homo sapiens genome assembly, GHCh38.p13, were downloaded from the
National Center for Biotechnology Information (ncbi.nlm.nih.gov, accession date 22 June 2022) to
perform target gene analysis. Genetic polymorphisms of cytochrome
related pharmacogenomic studies focusing on Asian populations were
selected (24-28).
Respective reference SNP (rs) numbers were obtained from the Human
CYP Allele Nomenclature Database and shown in Table I (pharmvar.org/htdocs/archive/index_original.htm,
accession date 22 June 2022). SNPs on reference sequences of these
genes were mapped on the reference chromosome sequences and
identified using NCBI BLAST tool (blast.ncbi.nlm.nih.gov/Blast.cgi).
 | Table IGene panel selection of
CYP2C9, CYP2C19 and CYP2D6 variants. |
Table I
Gene panel selection of
CYP2C9, CYP2C19 and CYP2D6 variants.
| Allele | Mutation | Rs number | Position of
synthetic oligonucleotide sequence (length) | Primer sequence,
5'3' |
|---|
|
CYP2C9*2 | 430C>T | rs1799853 | 94942072
GAA---------CGT---------- | F:
GAAATGGAAGGAGATCCGGC |
| | | | TTC 94942561
(490 bp) | R:
GATATGGAGTAGGGTCACCC |
|
CYP2C19*2 | 681G>A | rs4244285 | 94781615
CAT---------GGG--------- | F:
CGCCAACCAGAGCTTGGCAT |
| | | | GAC 94782055
(441 bp) | R:
CGGGCCATCGATTCTTGGTG |
|
CYP2C19*3 | 636G>A | rs4986893 | 94780502
CAC---------GAT---------- | F:
GGCCGCCAGAAACGTTTCGA |
| | | | TGC 94780955
(454 bp) | R:
CGGTACTTCAGGGCTTGGTC |
|
CYP2D6*10 | 100C>T | rs1065852 | 42130444
CAG--------GGT--------- | F:
GGAAGTCCACATGCAGCAGG |
| | | | CGC 42130890
(447 bp) | R:
GCAGGTATGGGGCTAGAAGC |
|
CYP2D6*41 | 2988G>A | rs28371725 | 42127601
CCT---------CCT---------- | F:
GGTCAAGCCTGTGCTTGGAG |
| | | | GTC 42128090
(490 bp) | R:
CCTACATCCGGATGTGCAGC |
Selection of SNPs, new primers and
synthetic oligonucleotide designs
The novel primers were designed to develop a gene
panel focusing on CYP2C9, CYP2C19, and CYP2D6
variants. Homology to function and evolution with other gene
families were assessed using Basic Local Alignment Search Tool
(BLAST) against the GHCh38.p13 assembly (ncbi.nlm.nih.gov/tools/primer-blast/, 22 June
2022). DNA melting temperature (Tm) of WT and MT variants was
predicted using the web-based tool uMelt version 3.6.2, developed
by the Wittwer lab (dna-utah.org/,
22 June 2022). Synthetic ds DNA fragments (gBlocks®,
Integrated DNA Technologies) of 250-500 bp for each variant were
used for quality control, as previously described (30). Position of synthetic ds DNA and
primer sequences are shown in Table
I.
Ethical considerations
All participants were recruited from unrelated Thai
volunteers with the following inclusion criteria: i) Age 18-60
years old, ii) no history of drug ADRs and SCARs and iii) have
history of drug ADRs or SCARs (but at the time of recruiting
participants, have no symptom of ADRs). A total of 27 volunteers
including 13 males and 14 females was recruited between January to
March 2022 at Faculty of Associated Medical Sciences, Chiang Mai
University, Chiang Mai, Thailand. All participants provided written
consent to participate in the study before collecting blood samples
from vein. The study was approved by Research Ethics Committee,
Faculty of Associated Medical Sciences, Chiang Mai University,
Thailand (approval no. AMSEC-64EX-130; date of approval: 28
December 2021).
DNA extraction
A total of 6 ml of blood samples were collected from
vein and stored in EDTA tube and DNA extraction was performed using
the PureLink™ Genomic DNA mini kit (Invitrogen; Thermo
Fisher Scientific, Inc.), following the manufacturer's
instructions. At least 200 µl of buffy coat from EDTA blood was
used to a final elution volume of 100 µl extracted DNA. The quality
of extracted DNA was determined using an Epoch Microplate
Spectrophotometer (BioTek Instruments, Inc.) with a 260/280
absorbance ratio of 1.65-1.80. The DNA concentration was normalized
to 50 ng/µl.
Optimization of annealing temperature
(Ta) and critical denaturation temperature (Tc) of fast
COLD-PCR
Optimization of Ta was performed by conventional PCR
for primer annealing to a target sequence. A total of 50 ng genomic
DNA template was used in a total reaction volume of 12.5 µl. PCR
reaction was performed using 1X Quick Taq™ HS Dye Mix
(Toyobo Life Science) with 0.2 µM all primers (Table I) according to the manufacturer's
instructions. The conditions for PCR cycling were 94˚C for 2 min
followed by 30 cycles at 94˚C for 30 sec, 50-65˚C with gradient PCR
for 30 sec, 68˚C for 1 min/kb and 68˚C for 7 min.
Selective denaturation stage (Tc) is vital for fast
COLD-PCR to precisely denature the mutated sequence (14,23).
First, 10 rounds of conventional PCR were performed to amplify and
generate a sufficient template for COLD-PCR. Afterwards, the
precise Tc was determined by gradual reduction of the denaturation
temperature (Tm). The amplified PCR products were analyzed by 2%
agarose gel electrophoresis in 10x Tris-Borate-EDTA (TBE) buffer
for 40 min. As a result, only MT PCR products were observed in
comparison with WT and MT synthetic DNA templates. The reaction
mixture and total volume of fast COLD-PCR were the same as those of
conventional PCR. A total of 10 cycles of conventional PCR and fast
COLD-PCR conditions were optimized as follows: 30 cycles of precise
Tc (gradually decreasing Tm until only MT DNA was enriched) for 30
sec, 65˚C for 30 sec, 68˚C for 12 sec and final extension of 68˚C
for 7 min.
Tc combination of fast COLD-PCR assay
evaluation
To multiplex CYP2C9, CYP2C19 and
CYP2D6, combined-Tc fast COLD-PCR was performed, starting
from the lowest to highest Tc by sequentially adding 10+10+20
cycles. The optimization condition was the as the precise Tc
determination. An initial denaturation of 94˚C for 2 min was
followed by 10 cycles of conventional PCR, as aforementioned. Next,
10 cycles of Tc1 (75.0˚C) for 30 sec with annealing and extension
steps were performed as aforementioned. Another 10 cycles at Tc2
(87.0˚C) for 30 sec, followed by 20 cycles of Tc3 (90.5˚C) for 30
sec with annealing and extension were performed, as shown in
Fig. 1. Afterwards, the resulting
assay was tested and evaluated on 27 samples in comparison with
Sanger sequencing.
Sanger sequencing
After screening 27 samples for all variants with
fast COLD-PCR, the resulting positive MT samples were determined by
traditional Sanger sequencing to determine homozygous or
heterozygous status. To evaluate the efficiency of fast COLD-PCR
screening, all 27 samples were subjected to Sanger sequencing. To
perform Sanger sequencing, DNA samples were amplified using the
aforementioned conventional PCR. The obtained PCR amplicons were
sequenced by the Sanger reference method at Macrogen, Inc.
(31). Sanger sequence assemblies
were analyzed using SeqMan Ultra DNASTAR Bioinformatics Software
version 17.2 (dnastar.com/software/lasergene/seqman-ultra/, 22
June 2022).
Statistical analysis
As data of COLD-PCR and Sanger sequencing were
examined each clinical sample for variants to determine whether
they will be found to match. Data will be converted from category
data into quantitative data (data 0-1 means negative-positive). The
Cohen's Kappa (κ) agreement between two assays was calculated using
SPSS software v. 22.0. Moreover, we have also calculated the
allelic frequency of five variants found in this study.
Results
Determination of Ta and Tc for initial
fast COLD-PCR screening test
Total five variants were selected from
CYP2C9, CYP2C19, and CYP2D6 genes and these
five variants were optimized at the Ta, 50-65˚C. Following this,
65˚C was selected as the optimal Ta as all five genetic variants
were shown the same annealing reaction at 65˚C. No amplification in
WT and a positive band in MT variants were obtained with three
different Tc values for the five targets: Tc=75.0˚C for
CYP2C9*2 (150 bp), 19*2 (206 bp) and 19*3 (192
bp); Tc=87.0˚C for CYP2D6*41 (124 bp) and Tc=90.5˚C for
CYP2D6*10 (160 bp). These Tc results were validated using
synthetic WT and MT oligonucleotide templates at a concentration of
50 ng/µl with 10 cycles of conventional PCR at Ta=65˚C (Fig. 2). Accordingly, optimal Ta at 65˚C
and three Tcs (75.0˚C, 87.0˚C and 90.5˚C) for fast COLD-PCR
screening test were obtained.
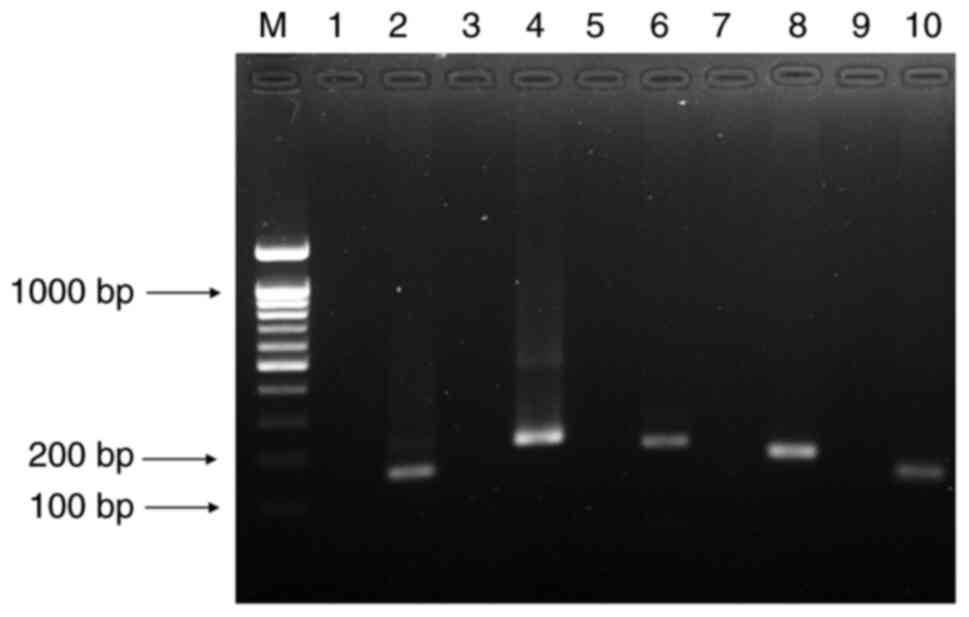 | Figure 2Precise Tc validation of five
variants obtained by fast COLD-PCR. Lane M, DNA ladder 100 bp;
lanes 1 and 2, synthetic WT and MT of CYP2C9*2 (150 bp);
lanes 3 and 4, synthetic WT and MT of CYP2C19*2 (206 bp);
lanes 5 and 6, synthetic WT and MT of CYP2C19*3 (192 bp);
lanes 7 and 8, synthetic WT and MT of CYP2D6*10 (160 bp) and
lanes 9 and 10, synthetic WT and MT of CYP2D6*41 (124 bp).
COLD, co-amplification at lower denaturation temperature; WT,
wild-type; MT, mutant; CYP, cytochrome P450. |
Establishment and evaluation of
combined fast COLD-PCR
The proposed assay was modified to discriminate all
variants within a single reaction through multiplex fast COLD-PCR
by combining three Tc values (75.0, 87.0 and 90.5˚C) with 10+10+20
cycles. A total of 27 randomly collected samples were tested by
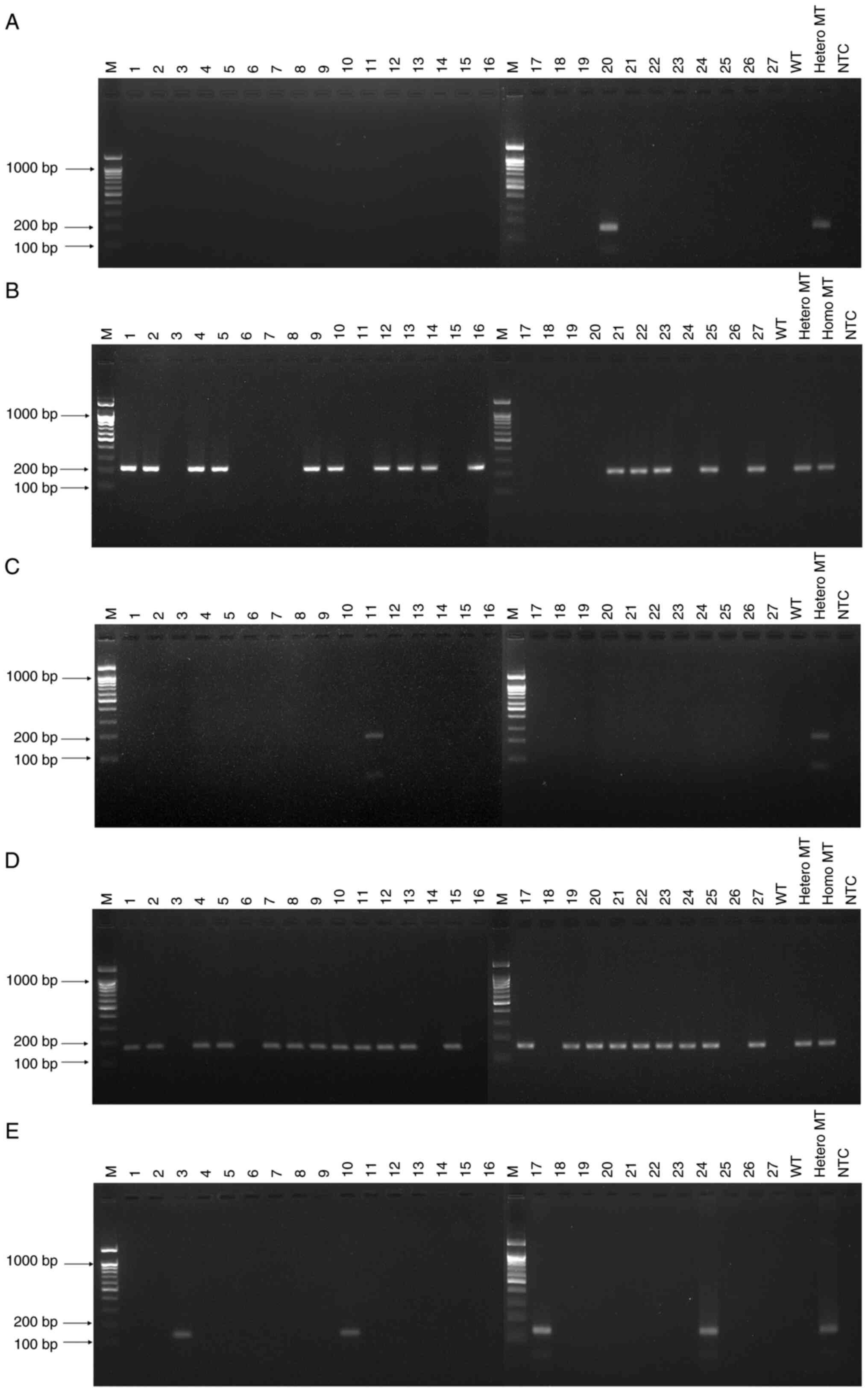
multiplex performance with control samples (Fig. 3). For the detection of
CYP2C9*2, only one sample (sample 20) showed a positive band
(Fig. 3A). For CYP2C19*2,
positive bands are observed for 15 samples (samples 1, 2, 4, 5, 9,
10, 12-14, 16, 21-23, 25 and 27; Fig.
3B). For CYP2C19*3, a positive band is observed for one
sample (sample 11; Fig. 3C). A
total of 21 samples (samples 1, 2, 4, 5, 7-13, 15, 17, 19-25 and
27) was CYP2D6*10-positive (Fig.
3D). A total of four samples (samples 3, 10, 17 and 24) was
CYP2D6*41-positive (Fig.
3E). In comparison of fast COLD-PCR with Sanger sequencing, the
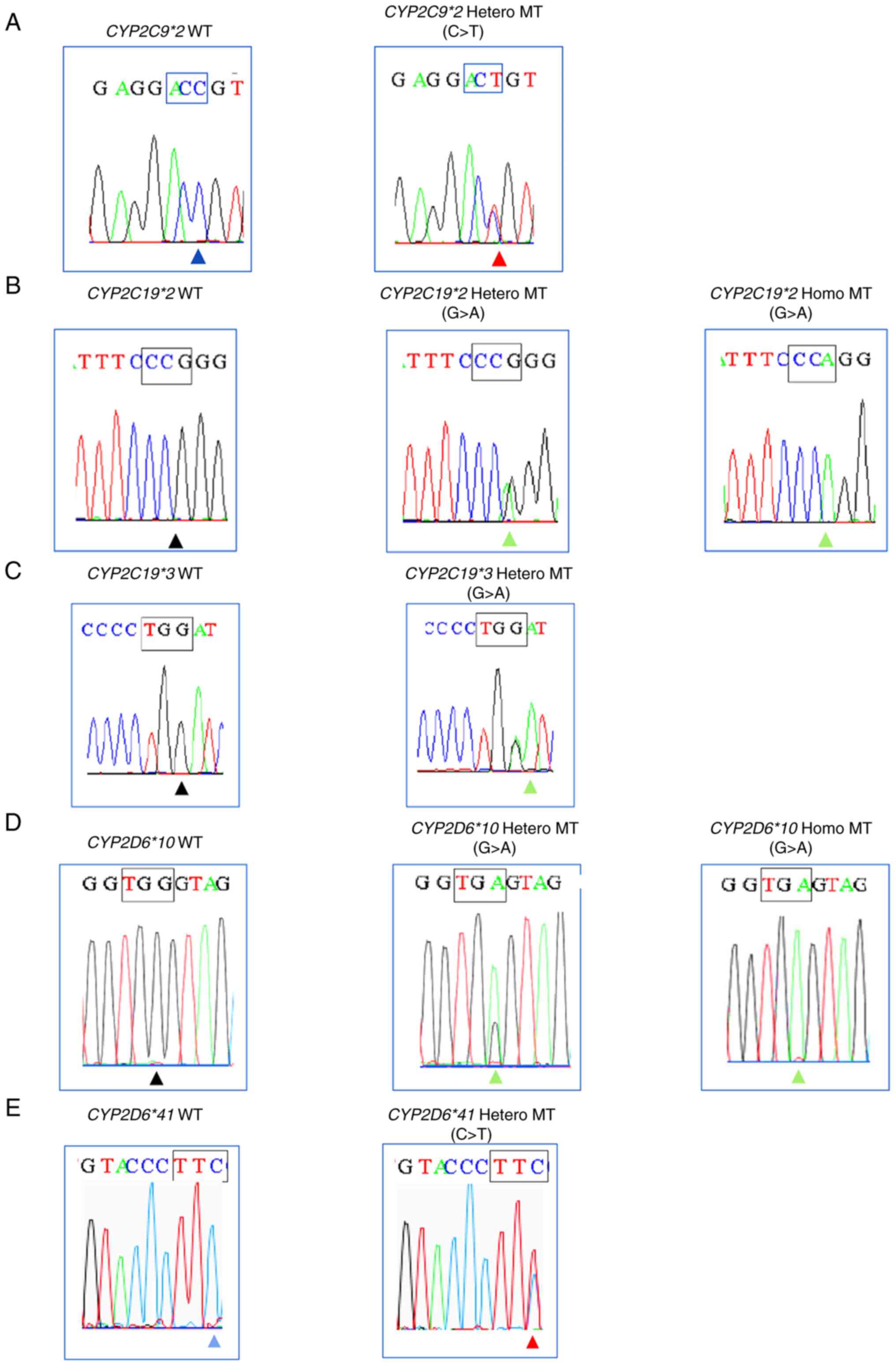
results show 100% consistency (κ=1.0) for all variants (Table II). The results of Sanger
sequencing in five variants are shown in Fig. 4. Fast COLD-PCR correctly identified
heterozygous or homozygous SNPs variant in Sanger results as
‘Positive’ and WT sample of Sanger sequencing results as
‘Negative’. The percentage of five variants present in 27 samples
as follows: 3.7% of CYP2C9*2 and CYP2C19*3, 55.5% of
CYP2C19*2, 77.7% of CYP2D6*10, and 14.8% of
CYP2D6*41.
 | Table IIEvaluation of fast COLD-PCR showing
100% consistency with Sanger sequencing on 27 collected
samples. |
Table II
Evaluation of fast COLD-PCR showing
100% consistency with Sanger sequencing on 27 collected
samples.
| |
CYP2C9*2 |
CYP2C19*2 |
CYP2C19*3 |
CYP2D6*10 |
CYP2D6*41 |
|---|
| Result | Fast COLD-PCR | Sanger
sequencing | Fast COLD-PCR | Sanger
sequencing | Fast COLD-PCR | Sanger
sequencing | Fast COLD-PCR | Sanger
sequencing | Fast COLD-PCR | Sanger
sequencing |
|---|
| Positive | 1 | 1 | 15 | 15 | 1 | 1 | 21 | 21 | 4 | 4 |
| Negative | 26 | 26 | 12 | 12 | 26 | 26 | 6 | 6 | 23 | 23 |
| κ value | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
Discussion
Genetic DNA variations of CYP genes alter
pharmacokinetics and responses to certain drugs. In The Human CYP
Allele Nomenclature Database (32),
CYP2 family is primarily involved in drug ‘physiology’,
‘toxicology’ and ‘diverse regulatory mechanisms’ (33). Of CYP2 family members, CYP2D6
presents highly polymorphic and complex structural variations
(34) and sequence similarities
>90% are seen in CYP2C9 and CYP2C19 (35). In previous studies, CYP gene
copy numbers (CYP2D6) have been determined by pyrosequencing
(36), loop-mediated isothermal
amplification, electrochemical DNA chip (37) and real-time PCR detection (38). Commercial kits and advanced
genotyping techniques have also been developed for clinical
implementation, including AmpliChip CYP450, TaqMan assays, Luminex
xTAG, next-generation sequencing platforms and MassARRAY as
systematic algorithms (39).
CYP genetic testing is used to monitor
patients effectively for therapeutic indication. However, the
greater challenge of PGx testing is economic (40). Therefore, the present aimed to
introduce a new basic platform to be able to use before traditional
genotyping methods in unknown patient samples. Although the present
data cannot demonstrate the extent of genotype predictable
phenotype, it may serve as the fundamental consideration whether
patient has genetic variants. Using fast COLD-PCR as initial
screening test and combination with other genotyping testing for
positive results may provide a more affordable approach in
precision medicine. Therefore, the present study aimed to evaluate
fast COLD-PCR as prescreening strategy to monitor patient safety.
In addition, fast COLD-PCR is also simple, easy, and cheap enough
to be widely used in routine lab work.
The present study observed a positive MT band with
no WT band on 2% agarose gel on the precise Tc of each SNP.
Previously, different PCR protocols for each reaction were required
for Tc per amplicon and multiplexing in fast COLD-PCR was
problematic due to precise Tc which only denatures mutant sequence
at its specific temperature. However, the present study combined
multiplexed fast COLD-PCR using 10+10+20 cycles with three
different Tc values of five variants (from low to high Tc). The
performance evaluation showed notable results in this single
system. The following estimated frequencies were obtained in our
Thai-population-focused study: 3.7% in CYP2C9*2 and
CYP2C19*3, 55.5% in CYP2C19*2, 77.7% in
CYP2D6*10, and 14.8% in CYP2D6*41. In previous
studies, frequencies of only 0.08% for CYP2C9*2, 25.6% for
CYP2C19*2 and 2.5% for CYP2C19*3 alleles were found
in a Thai population (26,41). For CYP2D6 in Thai population,
the decreased-function allele CYP2D6*10 is the most common
allele found in patients treated with risperidone, at 51.8%,
followed by CYP2D6*41 at 6.8% (42). Although the present allele
representation frequency was higher than previous studies (26,41,42),
it may be due to small sample size. Nevertheless, the present
COLD-PCR results showed 100% agreement with Sanger sequencing
results. Therefore, the present method may be applicable as an
initial test for unknown samples before haplotyping. Furthermore,
the cost (not including DNA extraction) of fast COLD-PCR in our
routine pharmacogenetic laboratory service is Thai baht (THB)
125/test (USD $3.6), as opposed to THB 1,200/test (USD $34.59) for
Sanger sequencing. Our study introduces an easily applicable
prescreening methodology in PGx settings.
However, the present methodology had limitations.
One constraint is in enriching only Tm-reducing variations.
CYP2C9*3 (1075A>C), a Tm-increasing mutant, was not
included and further COLD-PCR technique, such as full COLD-PCR, is
required to detect all types of mutations (Tm-increase,
Tm-equivalent, Tm-decrease). The present study used only 27
samples; thus, larger sample size is required to identify further
SNPs. The present method is a qualitative screening; for patients
with positive results, additional methods should be used to
distinguish homozygous or heterozygous genotypes. Nevertheless, the
aim of this study is to consider PGx testing in a cost-effective
way. Currently, the plurality of commercial assay is available,
however, our main objective is to view PGx testing by applying only
conventional PCR machine before sequencing or genetic testing.
The present study approached the first prescreening
pipeline in mutation detection before traditional genotyping
methods. Here, fast COLD-PCR methodology correctly identified all
WT samples. Only MT samples only need traditional genotyping to
distinguish homozygous or heterozygous type. For example, for
CYP2C9*2 and CYP2C19*3, 26 out of 27 samples were WT.
Therefore, sequencing or genetic testing for genotype (heterozygous
or homozygous) would not be needed in 26 samples. Therefore, cost
for the whole pipeline would be decreased and WT samples reported
faster with the present screening PCR test. This method may
decrease unnecessary costs of expensive genotyping in patients.
To the best of our knowledge, the present study is
the first to propose fast COLD-PCR as a solution to the economic
barrier of PGx implementation. This method only needs conventional
PCR machine to perform and it shows the results as
positive/negative. Therefore, it can screen out WT samples and only
positive MT bands need traditional genetic testing. This method can
be used to assess genetic variants as an easy cost-effective
strategy.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Faculty of
Associated Medical Sciences, Chiang Mai University, Thailand (grant
no. R000025740).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YMN, SD, CS, AL, PP and NA conceptualized the study.
YMN, SD and NA designed the experiments. YMN and NA analyzed data.
YMN performed the experiments and visualized data. SD and CS
confirm the authenticity of all the raw data. Resources from CS.
YMN, AL, PP, SD, CS and NA drafted and edited the manuscript. NA
supervised the study. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee, Faculty of Associated Medical Sciences, Chiang
Mai University, Thailand (approval no. AMSEC-64EX-130; date of
approval: 28 December 2021). Written informed consent was obtained
from all subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arici M and Özhan G: CYP2C9, CYPC19 and
CYP2D6 gene profiles and gene susceptibility to drug response and
toxicity in Turkish population. Saudi Pharm J. 25:376–380.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brockmöller J, Kirchheiner J, Meisel C and
Roots I: Pharmacogenetic diagnostics of cytochrome P450
polymorphisms in clinical drug development and in drug treatment.
Pharmacogenomics. 1:125–151. 2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Malki MA and Pearson ER: Drug-drug-gene
interactions and adverse drug reactions. Pharmacogenomics J.
20:355–366. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
D'Empaire I, Guico-Pabia CJ and Preskorn
SH: Antidepressant treatment and altered CYP2D6 activity: Are
pharmacokinetic variations clinically relevant? J Psychiatr Pract.
17:330–339. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sim SC, Risinger C, Dahl ML, Aklillu E,
Christensen M, Bertilsson L and Ingelman-Sundberg M: A common novel
CYP2C19 gene variant causes ultrarapid drug metabolism relevant for
the drug response to proton pump inhibitors and antidepressants.
Clin Pharmacol Ther. 79:103–113. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Westergaard N, Søgaard Nielsen R,
Jørgensen S and Vermehren C: Drug use in denmark for drugs having
pharmacogenomics (PGx) based dosing guidelines from CPIC or DPWG
for CYP2D6 and CYP2C19 Drug-gene pairs: Perspectives for
introducing PGx test to polypharmacy patients. J Pers Med.
10(3)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Abdullah-Koolmees H, van Keulen AM,
Nijenhuis M and Deneer VHM: Pharmacogenetics guidelines: Overview
and comparison of the DPWG, CPIC, CPNDS, and RNPGx guidelines.
Front Pharmacol. 11(595219)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Verbeurgt P, Mamiya T and Oesterheld J:
How common are drug and gene interactions? Prevalence in a sample
of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping.
Pharmacogenomics. 15:655–665. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Dong Y, Xiao H, Wang Q, Zhang C, Liu X,
Yao N, Sheng H and Li H: Analysis of genetic variations in CYP2C9,
CYP2C19, CYP2D6 and CYP3A5 genes using oligonucleotide microarray.
Int J Clin Exp Med. 8:18917–18926. 2015.PubMed/NCBI
|
|
10
|
Samer CF, Lorenzini KI, Rollason V, Daali
Y and Desmeules JA: Applications of CYP450 testing in the clinical
setting. Mol Diagn Ther. 17:165–184. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sukri A, Salleh MZ, Masimirembwa C and Teh
LK: A systematic review on the cost effectiveness of
pharmacogenomics in developing countries: Implementation
challenges. Pharmacogenomics J. 22:147–159. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Luzum JA and Luzum MJ: Physicians'
attitudes toward pharmacogenetic testing before and after
pharmacogenetic education. Per Med. 13:119–127. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Galbiati S, Monguzzi A, Damin F, Soriani
N, Passiu M, Castellani C, Natacci F, Curcio C, Seia M, Lalatta F,
et al: COLD-PCR and microarray: Two independent highly sensitive
approaches allowing the identification of fetal paternally
inherited mutations in maternal plasma. J Med Genet. 53:481–487.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Byrou S, Makrigiorgos GM, Christofides A,
Kallikas I, Papasavva T and Kleanthous M: Fast Temperature-gradient
COLD PCR for the enrichment of the paternally inherited SNPs in
cell free fetal DNA; an application to non-invasive prenatal
diagnosis of β-thalassaemia. PLoS One. 13(e0200348)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li J, Wang L, Mamon H, Kulke MH, Berbeco R
and Makrigiorgos GM: Replacing PCR with COLD-PCR enriches variant
DNA sequences and redefines the sensitivity of genetic testing. Nat
Med. 14:579–584. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Boisselier B, Marie Y, Labussière M,
Ciccarino P, Desestret V, Wang X, Capelle L, Delattre JY and Sanson
M: COLD PCR HRM: A highly sensitive detection method for IDH1
mutations. Hum Mutat. 31:1360–1365. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li J, Milbury CA, Li C and Makrigiorgos
GM: Two-round coamplification at lower denaturation temperature-PCR
(COLD-PCR)-based sanger sequencing identifies a novel spectrum of
low-level mutations in lung adenocarcinoma. Hum Mutat.
30:1583–1590. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zuo Z, Chen SS, Chandra PK, Galbincea JM,
Soape M, Doan S, Barkoh BA, Koeppen H, Medeiros LJ and Luthra R:
Application of COLD-PCR for improved detection of KRAS mutations in
clinical samples. Mod Pathol. 22:1023–1031. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kristensen LS, Daugaard IL, Christensen M,
Hamilton-Dutoit S, Hager H and Hansen LL: Increased sensitivity of
KRAS mutation detection by high-resolution melting analysis of
COLD-PCR products. Hum Mutat. 31:1366–1373. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Milbury CA, Chen CC, Mamon H, Liu P,
Santagata S and Makrigiorgos GM: Multiplex amplification coupled
with COLD-PCR and high resolution melting enables identification of
low-abundance mutations in cancer samples with low DNA content. J
Mol Diagn. 13:220–232. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Castellanos-Rizaldos E, Liu P, Milbury CA,
Guha M, Brisci A, Cremonesi L, Ferrari M, Mamon H and Makrigiorgos
GM: Temperature-tolerant COLD-PCR reduces temperature stringency
and enables robust mutation enrichment. Clin Chem. 58:1130–1138.
2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Castellanos-Rizaldos E, Milbury CA and
Makrigiorgos GM: Enrichment of mutations in multiple DNA sequences
using COLD-PCR in emulsion. PLoS One. 7(e51362)2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Carotenuto P, Roma C, Cozzolino S, Fenizia
F, Rachiglio AM, Tatangelo F, Iannaccone A, Baron L, Botti G and
Normanno N: Detection of KRAS mutations in colorectal cancer with
Fast COLD-PCR. Int J Oncol. 40:378–384. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sistonen J, Fuselli S, Palo JU, Chauhan N,
Padh H and Sajantila A: Pharmacogenetic variation at CYP2C9,
CYP2C19, and CYP2D6 at global and microgeographic scales.
Pharmacogenet Genomics. 19:170–179. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bradford LD: CYP2D6 allele frequency in
European Caucasians, Asians, Africans and their descendants.
Pharmacogenomics. 3:229–243. 2002.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dorji PW, Tshering G and Na-Bangchang K:
CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in south-east and
east asian populations: A systematic review. J Clin Pharm Ther.
44:508–524. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tassaneeyakul W, Mahatthanatrakul W,
Niwatananun K, Na-Bangchang K, Tawalee A, Krikreangsak N, Cykleng U
and Tassaneeyakul W: CYP2C19 genetic polymorphism in thai, burmese
and karen populations. Drug Metab Pharmacokinet. 21:286–290.
2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lo C, Nguyen S, Yang C, Witt L, Wen A,
Liao TV, Nguyen J, Lin B, Altman RB and Palaniappan L:
Pharmacogenomics in asian subpopulations and impacts on commonly
prescribed medications. Clin Transl Sci. 13:861–870.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Crossley BM, Bai J, Glaser A, Maes R,
Porter E, Killian ML, Clement T and Toohey-Kurth K: . Guidelines
for Sanger sequencing and molecular assay monitoring. J Vet Diagn
Invest. 32:767–775. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Blazejewski T, Ho HI and Wang HH:
Synthetic sequence entanglement augments stability and containment
of genetic information in cells. Science. 365:595–598.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Macrogen. Capillary Electrophoresis
Sequencing (CES). Available online: https://dna.macrogen.com/pageLinkDnaSys.do?layout=page_sub&link=/support/retrieveGuideCes
(Accession date 16 October 2022).
|
|
32
|
Sim SC and Ingelman-Sundberg M: The human
cytochrome P450 (CYP) allele nomenclature website: A peer-reviewed
database of CYP variants and their associated effects. Hum
Genomics. 4(278)2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kubota A, Stegeman JJ, Goldstone JV,
Nelson DR, Kim EY, Tanabe S and Iwata H: Cytochrome P450 CYP2 genes
in the common cormorant: Evolutionary relationships with 130
diapsid CYP2 clan sequences and chemical effects on their
expression. Comp Biochem Physiol C Toxicol Pharmacol. 153:280–289.
2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gaedigk A: Complexities of CYP2D6 gene
analysis and interpretation. Int Rev Psychiatry. 25:534–553.
2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gotoh O: Substrate recognition sites in
cytochrome P450 family 2 (CYP2) proteins inferred from comparative
analyses of amino acid and coding nucleotide sequences. J Biol
Chem. 267:83–90. 1992.PubMed/NCBI
|
|
36
|
Söderbäck E, Zackrisson AL, Lindblom B and
Alderborn A: Determination of CYP2D6 gene copy number by
pyrosequencing. Clin Chem. 51:522–531. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nakamura N, Fukuda T, Nonen S, Hashimoto
K, Azuma J and Gemma N: Simple and accurate determination of CYP2D6
gene copy number by a loop-mediated isothermal amplification method
and an electrochemical DNA chip. Clin Chim Acta. 411:568–573.
2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Puaprasert K, Chu C, Saralamba N, Day NPJ,
Nosten F, White NJ, Dondorp AM and Imwong M: Real time PCR
detection of common CYP2D6 genetic variants and its application in
a Karen population study. Malar J. 17(427)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carvalho Henriques B, Buchner A, Hu X,
Wang Y, Yavorskyy V, Wallace K, Dong R, Martens K, Carr MS, Asl B,
et al: Methodology for clinical genotyping of CYP2D6 and CYP2C19.
Transl Psychiatry. 11(596)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
de Lecea MGM and Rossbach M: Translational
genomics in personalized medicine-scientific challenges en route to
clinical practice. Hugo J. 6(2)2012.
|
|
41
|
Sukprasong R, Chuwongwattana S, Koomdee N,
Jantararoungtong T, Prommas S, Jinda P, Rachanakul J,
Nuntharadthanaphong N, Jongjitsook N, Puangpetch A and Sukasem C:
Allele frequencies of single nucleotide polymorphisms of clinically
important drug-metabolizing enzymes CYP2C9, CYP2C19, and CYP3A4 in
a Thai population. Sci Rep. 11(12343)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hongkaew Y, Gaedigk A, Wilffert B,
Ngamsamut N, Kittitharaphan W, Limsila P and Sukasem C:
Relationship between CYP2D6 genotype, activity score and phenotype
in a pediatric Thai population treated with risperidone. Sci Rep.
11(4158)2021.PubMed/NCBI View Article : Google Scholar
|