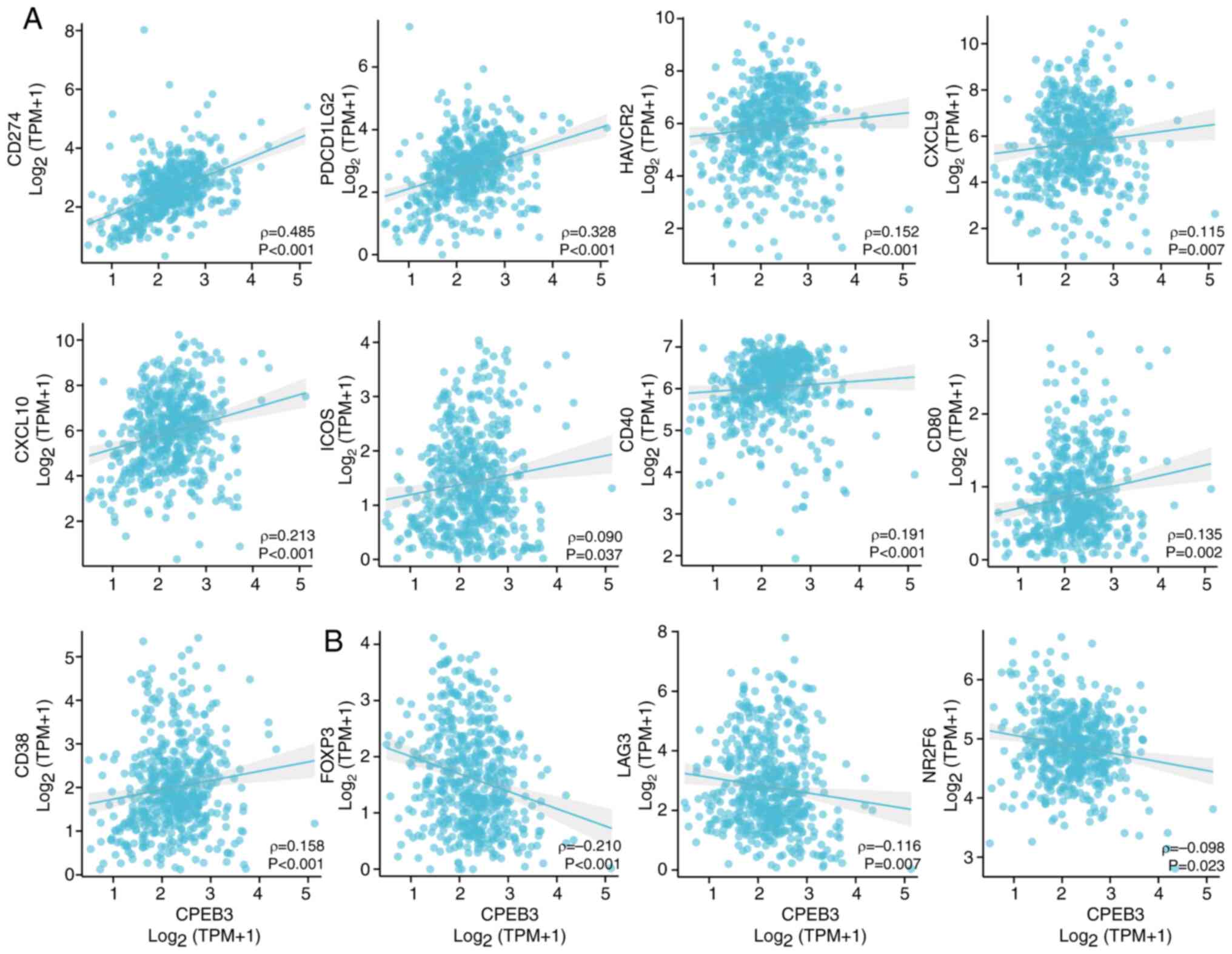
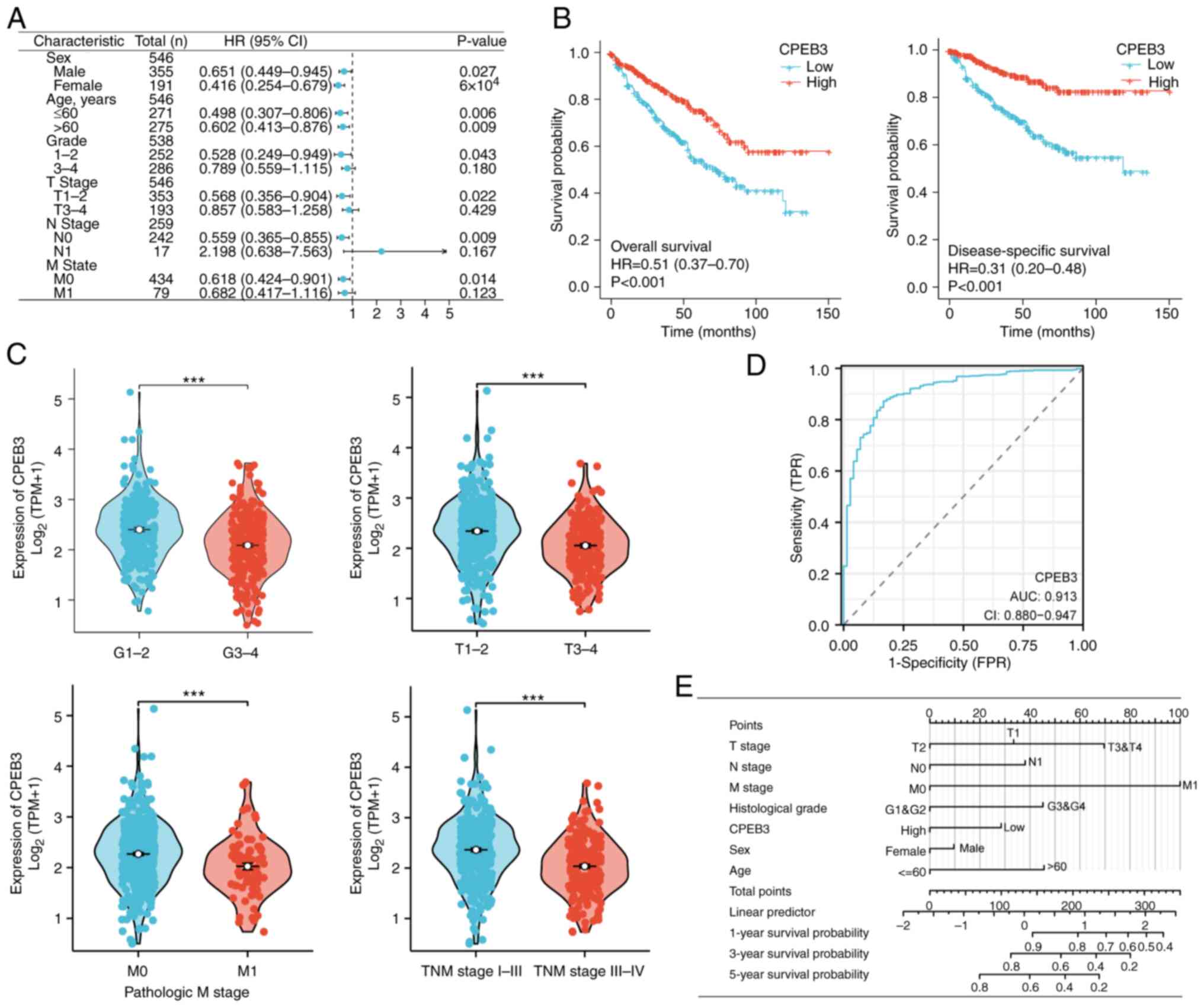
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Leibovich BC, Lohse CM, Crispen PL,
Boorjian SA, Thompson RH, Blute ML and Cheville JC: Histological
subtype is an independent predictor of outcome for patients with
renal cell carcinoma. J Urol. 183:1309–1315. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cella D, Motzer RJ, Suarez C, Blum SI,
Ejzykowicz F, Hamilton M, Wallace JF, Simsek B, Zhang J, Ivanescu
C, et al: Patient-reported outcomes with first-line nivolumab plus
cabozantinib versus sunitinib in patients with advanced renal cell
carcinoma treated in CheckMate 9ER: an open-label, randomised,
phase 3 trial. Lancet Oncol. 23:292–303. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Motzer R, Alekseev B, Rha SY, Porta C, Eto
M, Powles T, Grünwald V, Hutson TE, Kopyltsov E, Méndez-Vidal MJ,
et al: Lenvatinib plus pembrolizumab or everolimus for advanced
renal cell carcinoma. N Engl J Med. 384:1289–1300. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Choueiri TK, Powles T, Burotto M, Escudier
B, Bourlon MT, Zurawski B, Oyervides Juárez VM, Hsieh JJ, Basso U,
Shah AY, et al: Nivolumab plus cabozantinib versus sunitinib for
advanced renal-cell carcinoma. N Engl J Med. 384:829–841.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Powles T, Plimack ER, Soulières D, Waddell
T, Stus V, Gafanov R, Nosov D, Pouliot F, Melichar B, Vynnychenko
I, et al: Pembrolizumab plus axitinib versus sunitinib monotherapy
as first-line treatment of advanced renal cell carcinoma
(KEYNOTE-426): Extended follow-up from a randomised, open-label,
phase 3 trial. Lancet Oncol. 21:1563–1573. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rini BI, Plimack ER, Stus V, Gafanov R,
Hawkins R, Nosov D, Pouliot F, Alekseev B, Soulières D, Melichar B,
et al: Pembrolizumab plus axitinib versus sunitinib for advanced
renal-cell carcinoma. N Engl J Med. 380:1116–1127. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Motzer RJ, Penkov K, Haanen J, Rini B,
Albiges L, Campbell MT, Venugopal B, Kollmannsberger C, Negrier S,
Uemura M, et al: Avelumab plus axitinib versus sunitinib for
advanced renal-cell carcinoma. N Engl J Med. 380:1103–1115.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Motzer RJ, Tannir NM, McDermott DF, Arén
Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P,
Porta C, George S, et al: Nivolumab plus ipilimumab versus
sunitinib in advanced renal-cell carcinoma. N Engl J Med.
378:1277–1290. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Choueiri TK, Tomczak P, Park SH, Venugopal
B, Ferguson T, Chang YH, Hajek J, Symeonides SN, Lee JL, Sarwar N,
et al: Adjuvant pembrolizumab after nephrectomy in renal-cell
carcinoma. N Engl J Med. 385:683–694. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bi K, He MX, Bakouny Z, Kanodia A,
Napolitano S, Wu J, Grimaldi G, Braun DA, Cuoco MS, Mayorga A, et
al: Tumor and immune reprogramming during immunotherapy in advanced
renal cell carcinoma. Cancer Cell. 39:649–661.e5. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu F, Fan J, He Y, Xiong A, Yu J, Li Y,
Zhang Y, Zhao W, Zhou F, Li W, et al: Single-cell profiling of
tumor heterogeneity and the microenvironment in advanced non-small
cell lung cancer. Nat Commun. 12(2540)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ma L, Hernandez MO, Zhao Y, Mehta M, Tran
B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, et al: Tumor
cell biodiversity drives microenvironmental reprogramming in liver
cancer. Cancer Cell. 36:418–430.e6. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Barkley D, Moncada R, Pour M, Liberman DA,
Dryg I, Werba G, Wang W, Baron M, Rao A, Xia B, et al: Cancer cell
states recur across tumor types and form specific interactions with
the tumor microenvironment. Nat Genet. 54:1192–1201.
2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nakamura K and Smyth MJ: Myeloid
immunosuppression and immune checkpoints in the tumor
microenvironment. Cell Mol Immunol. 17:1–12. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kumagai S, Koyama S, Itahashi K,
Tanegashima T, Lin YT, Togashi Y, Kamada T, Irie T, Okumura G, Kono
H, et al: Lactic acid promotes PD-1 expression in regulatory T
cells in highly glycolytic tumor microenvironments. Cancer Cell.
40:201–218.e9. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Naser R, Fakhoury I, El-Fouani A,
Abi-Habib R and El-Sibai M: Role of the tumor microenvironment in
cancer hallmarks and targeted therapy (review). Int J Oncol.
62(23)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Josefowicz SZ, Lu LF and Rudensky AY:
Regulatory T cells: Mechanisms of differentiation and function.
Annu Rev Immunol. 30:531–564. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Khattri R, Cox T, Yasayko SA and Ramsdell
F: Pillars article: An essential role for scurfin in CD4+CD25+ T
regulatory cells. Nat. Immunol. 2003.4:337-342. J Immunol.
198:993–998. 2017.PubMed/NCBI
|
|
21
|
Hori S, Nomura T and Sakaguchi S: Control
of regulatory T cell development by the transcription factor Foxp3.
Science. 299:1057–1061. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fontenot JD, Gavin MA and Rudensky AY:
Foxp3 programs the development and function of CD4+CD25+ regulatory
T cells. Nat Immunol. 4:330–336. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Sakaguchi S, Sakaguchi N, Asano M, Itoh M
and Toda M: Immunologic self-tolerance maintained by activated T
cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a
single mechanism of self-tolerance causes various autoimmune
diseases. J Immunol. 155:1151–1164. 1995.PubMed/NCBI
|
|
24
|
Tada Y, Togashi Y, Kotani D, Kuwata T,
Sato E, Kawazoe A, Doi T, Wada H, Nishikawa H and Shitara K:
Targeting VEGFR2 with Ramucirumab strongly impacts
effector/activated regulatory T cells and CD8+ T cells
in the tumor microenvironment. J Immunother Cancer.
6(106)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Togashi Y and Nishikawa H: Regulatory T
cells: Molecular and cellular basis for immunoregulation. Curr Top
Microbiol Immunol. 410:3–27. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Saito T, Nishikawa H, Wada H, Nagano Y,
Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et
al: Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the
prognosis of colorectal cancers. Nat Med. 22:679–684.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Mendez R, Barnard D and Richter JD:
Differential mRNA translation and meiotic progression require
Cdc2-mediated CPEB destruction. EMBO J. 21:1833–1844.
2002.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen J, Li L, Liu TY, Fu HF, Lai YH, Lei
X, Xu JF, Yu JS, Xia YJ, Zhang TH, et al: CPEB3 suppresses gastric
cancer progression by inhibiting ADAR1-mediated RNA editing via
localizing ADAR1 mRNA to P bodies. Oncogene. 41:4591–4605.
2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng J, Ma H, Yan M, Zhang Z and Xing W:
Circ_0007624 suppresses the development of esophageal squamous cell
carcinoma via targeting miR-224-5p/CPEB3 to inactivate the
EGFR/PI3K/AKT signaling. Cell Signal. 99(110448)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhong Q, Fang Y, Lai Q, Wang S, He C, Li
A, Liu S and Yan Q: CPEB3 inhibits epithelial-mesenchymal
transition by disrupting the crosstalk between colorectal cancer
cells and tumor-associated macrophages via IL-6R/STAT3 signaling. J
Exp Clin Cancer Res. 39(132)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cancer Genome Atlas Research Network.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120.
2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
GTEx Consortium: Human genomics. The
genotype-tissue expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
R Core Team. R: A Language and Environment
for Statistical Computing. R Foundation for Statistical Computing,
Vienna, Austria, 2019.
|
|
36
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Villanueva RAM and Chen ZJ: ggplot2:
Elegant graphics for data analysis (2nd ed.). Meas: Inter Res
Perspect. 17:160–167. 2019.
|
|
40
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gene Ontology Consortium. Aleksander SA,
Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M,
Gaudet P, Harris NL, et al: The gene ontology knowledgebase in
2023. Genetics. 224(iyad031)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Charoentong P, Finotello F, Angelova M,
Mayer C, Efremova M, Rieder D, Hackl H and Trajanoski Z: Pan-cancer
immunogenomic analyses reveal genotype-immunophenotype
relationships and predictors of response to checkpoint blockade.
Cell Rep. 18:248–262. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48 (W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Therneau TM: A package for survival
analysis in R. https://CRAN.R-project.org/package=survival.
|
|
51
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC (eds), et al: AJCC cancer staging manual. 8th edition.
New York: Springer, 2017.
|
|
53
|
Pablo C, Marcela G, Lía EA and María IA:
Correlation between MVD and two prognostic factors: Fuhrman grade
and tumoral size, in clear cell renal cell carcinoma. J Cancer Sci
Ther. 4:313–316. 2012.
|
|
54
|
Kim SP, Alt AL, Weight CJ, Costello BA,
Cheville JC, Lohse C and Leibovich BC: Independent validation of
the 2010 American joint committee on cancer TNM classification for
renal cell carcinoma: Results from a large, single institution
cohort. J Urol. 185:2035–2039. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Klatte T, Patard JJ, Goel RH, Kleid MD,
Guille F, Lobel B, Abbou CC, De La Taille A, Tostain J, Cindolo L,
et al: Prognostic impact of tumor size on pT2 renal cell carcinoma:
An international multicenter experience. J Urol. 178:35–40.
2007.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ged Y, Markowski MC, Singla N and Rowe SP:
The shifting treatment paradigm of metastatic renal cell carcinoma.
Nat Rev Urol. 19:631–632. 2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
No authors listed. New treatments emerge
for RCC. Cancer Discov. 11(OF10)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Zilionis R, Engblom C, Pfirschke C, Savova
V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV,
Kerwin CM, et al: Single-cell transcriptomics of human and mouse
lung cancers reveals conserved myeloid populations across
individuals and species. Immunity. 50:1317–1334.e10.
2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Rosenthal R, Cadieux EL, Salgado R, Bakir
MA, Moore DA, Hiley CT, Lund T, Tanić M, Reading JL, Joshi K, et
al: Neoantigen-directed immune escape in lung cancer evolution.
Nature. 567:479–485. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
McGranahan N and Swanton C: Cancer
evolution constrained by the immune microenvironment. Cell.
170:825–827. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Vesely MD, Kershaw MH, Schreiber RD and
Smyth MJ: Natural innate and adaptive immunity to cancer. Annu Rev
Immunol. 29:235–271. 2011.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: Integrating immunity's roles in cancer suppression
and promotion. Science. 331:1565–1570. 2011.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Koebel CM, Vermi W, Swann JB, Zerafa N,
Rodig SJ, Old LJ, Smyth MJ and Schreiber RD: Adaptive immunity
maintains occult cancer in an equilibrium state. Nature.
450:903–907. 2007.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wolchok JD, Chiarion-Sileni V, Gonzalez R,
Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J,
Dummer R, et al: Long-Term outcomes with nivolumab plus ipilimumab
or nivolumab alone versus ipilimumab in patients with advanced
melanoma. J Clin Oncol. 40:127–137. 2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Taylor MH, Betts CB, Maloney L, Nadler E,
Algazi A, Guarino MJ, Nemunaitis J, Jimeno A, Patel P,
Munugalavadla V, et al: Safety and efficacy of pembrolizumab in
combination with acalabrutinib in advanced head and neck squamous
cell carcinoma: Phase 2 proof-of-concept study. Clin Cancer Res.
28:903–914. 2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Slomski A: Pembrolizumab boosts breast and
cervical cancer survival. JAMA. 326(2001)2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Brahmer J, Reckamp KL, Baas P, Crinò L,
Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE,
Holgado E, et al: Nivolumab versus docetaxel in advanced
squamous-cell non-small-cell lung cancer. N Engl J Med.
373:123–135. 2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Ugel S, Canè S, De Sanctis F and Bronte V:
Monocytes in the tumor microenvironment. Annu Rev Pathol.
16:93–122. 2021.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Li C, Teixeira AF, Zhu HJ and Ten Dijke P:
Cancer associated-fibroblast-derived exosomes in cancer
progression. Mol Cancer. 20(154)2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Cheng HS, Lee JXT, Wahli W and Tan NS:
Exploiting vulnerabilities of cancer by targeting nuclear receptors
of stromal cells in tumor microenvironment. Mol Cancer.
18(51)2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Quail DF, Bowman RL, Akkari L, Quick ML,
Schuhmacher AJ, Huse JT, Holland EC, Sutton JC and Joyce JA: The
tumor microenvironment underlies acquired resistance to CSF-1R
inhibition in gliomas. Science. 352(aad3018)2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
De Henau O, Rausch M, Winkler D, Campesato
LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, et
al: Overcoming resistance to checkpoint blockade therapy by
targeting PI3Kγ in myeloid cells. Nature. 539:443–447.
2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Coussens LM, Zitvogel L and Palucka AK:
Neutralizing tumor-promoting chronic inflammation: A magic bullet?
Science. 339:286–291. 2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Mao X, Xu J, Wang W, Liang C, Hua J, Liu
J, Zhang B, Meng Q, Yu X and Shi S: Crosstalk between
cancer-associated fibroblasts and immune cells in the tumor
microenvironment: New findings and future perspectives. Mol Cancer.
20(131)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Erez N, Truitt M, Olson P, Arron ST and
Hanahan D: Cancer-associated fibroblasts are activated in incipient
neoplasia to orchestrate tumor-promoting inflammation in an
NF-kappaB-dependent manner. Cancer Cell. 17:135–147.
2010.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wade RJ and Burdick JA: Engineering ECM
signals into biomaterials. Mater Today. 15:454–459. 2012.
|
|
80
|
Baumeister SH, Freeman GJ, Dranoff G and
Sharpe AH: Coinhibitory pathways in immunotherapy for cancer. Annu
Rev Immunol. 34:539–573. 2016.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Masugi Y, Nishihara R, Hamada T, Song M,
da Silva A, Kosumi K, Gu M, Shi Y, Li W, Liu L, et al: Tumor
PDCD1LG2 (PD-L2) expression and the lymphocytic reaction to
colorectal cancer. Cancer Immunol Res. 5:1046–1055. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Wolf Y, Anderson AC and Kuchroo VK: TIM3
comes of age as an inhibitory receptor. Nat Rev Immunol.
20:173–185. 2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Anderson AC, Joller N and Kuchroo VK:
Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized
functions in immune regulation. Immunity. 44:989–1004.
2016.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Reschke R and Gajewski TF: CXCL9 and
CXCL10 bring the heat to tumors. Sci Immunol.
7(eabq6509)2022.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Duhen R, Fesneau O, Samson KA, Frye AK,
Beymer M, Rajamanickam V, Ross D, Tran E, Bernard B, Weinberg AD
and Duhen T: PD-1 and ICOS coexpression identifies tumor-reactive
CD4+ T cells in human solid tumors. J Clin Invest.
132(e156821)2022.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Yan C and Richmond A: Hiding in the dark:
Pan-cancer characterization of expression and clinical relevance of
CD40 to immune checkpoint blockade therapy. Mol Cancer.
20(146)2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Sugiura D, Maruhashi T, Okazaki IM,
Shimizu K, Maeda TK, Takemoto T and Okazaki T: Restriction of PD-1
function by cis-PD-L1/CD80 interactions is required for optimal T
cell responses. Science. 364:558–566. 2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Chen L, Diao L, Yang Y, Yi X, Rodriguez
BL, Li Y, Villalobos PA, Cascone T, Liu X, Tan L, et al:
CD38-mediated immunosuppression as a mechanism of tumor cell escape
from PD-1/PD-L1 blockade. Cancer Discov. 8:1156–1175.
2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Liu Y, Wang L, Predina J, Han R, Beier UH,
Wang LC, Kapoor V, Bhatti TR, Akimova T, Singhal S, et al:
Inhibition of p300 impairs Foxp3+ T regulatory cell function and
promotes antitumor immunity. Nat Med. 19:1173–1177. 2013.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Grebinoski S, Zhang Q, Cillo AR, Manne S,
Xiao H, Brunazzi EA, Tabib T, Cardello C, Lian CG, Murphy GF, et
al: Autoreactive CD8+ T cells are restrained by an
exhaustion-like program that is maintained by LAG3. Nat Immunol.
23:868–877. 2022.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Klepsch V, Hermann-Kleiter N, Do-Dinh P,
Jakic B, Offermann A, Efremova M, Sopper S, Rieder D, Krogsdam A,
Gamerith G, et al: Nuclear receptor NR2F6 inhibition potentiates
responses to PD-L1/PD-1 cancer immune checkpoint blockade. Nat
Commun. 9(1538)2018.PubMed/NCBI View Article : Google Scholar
|