Introduction
Polycystic ovary syndrome (PCOS) is the most
prevalent hormonal and metabolic disorder that significantly
impacts women during their reproductive years (1), with a global prevalence estimated to
range from 4-20% (2). Women with
PCOS often experience ovulatory irregularities, leading to
infertility, and face an increased risk of adverse pregnancy
outcomes (3,4). The management of PCOS-related
infertility typically involves first-line therapies such as
lifestyle modifications and pharmacological interventions aimed at
inducing ovulation (5,6). However, for women who are resistant to
these treatments, surgical interventions such as laparoscopic
ovarian drilling, have proven to be effective strategies (7).
The pathophysiology of PCOS is complex,
characterized by hormonal imbalances, neuroendocrine disruptions,
alterations in adipocytes function, changes in gut microbiota,
insulin resistance and hyperandrogenism which impair
folliculogenesis and increase the risk of related comorbidities,
including endometrial cancer and type 2 diabetes (T2D) (8,9). PCOS
is also recognized as a complex polygenic disease influenced by
various factors such as gene variants, epigenetic and ethnicity
(10). A previous genetic study
identified various potential genes associated with single
nucleotide polymorphisms (SNPs) or mutations which are linked to a
range of PCOS symptoms (11).
KCNQ1 are potassium voltage-gated channels that play
pivotal role in various physiological processes, including cell
homeostasis, electrical signalling in cardiac myocytes, gastric
acid secretion, sperm motility, glucose metabolism and insulin
secretion (12-19).
The activity of KCNQ1 is primarily dependent on its interactions
with KCNE (Potassium Voltage-Gated Channel Subfamily E Regulatory)
family proteins, particularly KCNE1, and is further regulated by
cellular factors such as calmodulin, PIP2, PKA and
post-translational modifications (13).
The KCNQ1 gene is located on chromosome
11p15.5 and spanning over 400 kb (17) and mutations in this gene can lead to
channel dysfunction or loss of function, resulting in multiple
diseases (18-22).
SNPs within the KCNQ1 gene have been
identified and linked to T2D risk with evidence across diverse
populations including, Chinese, Singaporean, Indian and
Euro-Caucasian individuals (23-28).
Given the well-established intrinsic connection and
shared pathophysiological mechanisms between PCOS and T2D (29-31),
several genes associated with T2D, have been identified as
potential candidates for the development of PCOS (32-39),
yet the potential role of KCNQ1 in PCOS remains
unexplored.
In the present study, a critical knowledge gap in
the field of PCOS research was addressed by investigating the
potential relationship between three KCNQ1 variants
(rs151290, rs231361 and rs2237895) and PCOS in the Tunisian Arab
population. The study sought to provide valuable insights into the
genetic basis of PCOS, ultimately leading to the development of
improved diagnostic and therapeutic strategies and enhancing the
overall health and well-being of women affected by this
syndrome.
Materials and methods
Study subjects
The present retrospective case-control study
enrolled a total of 460 women, comprising 230 patients diagnosed
with PCOS and 230 ethnically matching control individuals. The age
range of the PCOS participants was 28-37 years (average, 32.5
years), while the control subjects had an age range of 27-33.25
years (average, 30 years). Participants were recruited from the
outpatient obstetrics, gynaecology and endocrinology clinics at
Farhat Hached Hospital in Tunisia between December 2023 and June
2024.
The three diagnostic criteria outlined for PCOS
encompass the identification and quantification of its classical
features, namely hyperandrogenism, ovulatory dysfunction and
polycystic ovarian morphology (40,41).
While these criteria are essential for a definitive diagnosis, it
is crucial to consider that relying on any single criterion may not
provide a conclusive result.
Other endocrine disorders, such as
hyperprolactinemia, thyroid disease and non-classical congenital
adrenal hyperplasia, can present with similar symptoms and
manifestations as PCOS. Therefore, to ensure a homogeneous study
population and accurate diagnosis, the study excluded women with
these potential disorders.
The control group comprised women examined during
the follicular phase (day 2-5) of their menstrual cycle, meeting
the following criteria: i) Regular menstrual cycles; ii) androgen
and hormonal levels within range; iii) and ovaries that were free
of any cysts on an ultrasound examination by the study
gynaecologist.
To ensure the integrity of the study, participants
were carefully screened to exclude those who had been on hormonal
therapy, or any medication known to influence carbohydrate
metabolism or endocrine parameters for at least 6 months prior to
their inclusion in the study. This exclusion criterion was
implemented to minimize the potential confounding effects of these
interventions on the study outcomes.
Data collection
The present study was conducted in adherence with
the principles outlined in the Helsinki Declaration (2014)
(42), and received ethical
approval from the local research and ethics committee of Farhat
Hached Hospital (approval no. 35220228; Sousse, Tunisia). All
patients who met the eligibility criteria were recruited and their
voluntary participation was confirmed through signed informed
consent.
The collected data encompassed demographic
information such as age and body mass index (BMI). Additionally,
lipide profiles, sexual hormone levels were collected from all
participants.
Biochemical and hormonal analysis
During the early follicular phase of the menstrual
cycle (days 2-5), venous blood samples were collected from control
subjects or any day for women with PCOS after an overnight fast.
Blood was collected into two types of tubes: Plain tubes for serum
collection and EDTA-containing tubes for plasma and DNA
preparation.
Serum samples underwent analysis for fasting blood
glucose using the hexokinase method in fluoride oxalate tube, with
the Cobas Integra 800 from Roche Diagnostics GmbH.
Immunofluorometric assay or radioimmunoassay were employed to
measure follicle-stimulating hormone (FSH), luteinizing hormone
(LH) and testosterone with both intra- and inter-assay coefficients
of variation <10%. Serum lipid levels, including total
cholesterol, high-density lipoprotein (HDL), low-density
lipoprotein (LDL) and triglycerides, were assessed using an
enzymatic colorimetric method on the Integra 800 from Roche.
Genomic DNA extraction and
genotyping
Genomic DNA was extracted from peripheral blood
leukocytes using Qiagen QIAamp DNA blood Mini Kit (Qiagen GmbH).
Quantification and purity assessment of the extracted DNA were
conducted using an ND-2000 Nanodrop spectrophotometer (Thermo
Fisher Scientific, Inc.). The purity of the DNA was considered
satisfactory with an A260/A280 ratio of ~1.8.
The potential association between KCNQ1
variants and PCOS susceptibility has not yet been explored and
given the known genetic overlap between PCOS and T2D, the selection
of the three KCNQ1 variants was based on their established
association with T2D on prior research studies as well as their
frequency in Caucasians with a minor allele frequency (MAF) >5%.
The National Center for Biotechnology Information Gene SNP Geneview
data base was utilized for this purpose (www.ncbi.nlm.nih.gov/snp).
Genotyping of the variants was performed using the
allelic discrimination method on StepOne real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Commercially
available primers from the assay-on-demand system were utilized and
well-defined genotype clusters were obtained. Specific TaqMan assay
IDs were used to target each variant: C_3075843_1_ (rs231361),
C_3075727_1_(rs151290) and C_16171034_10 (rs2237895). Replicated
blinded quality control samples were included to assess
reproducibility, which indicated concordance >99%.
Statistical analysis
The sample size and power analysis for detecting an
association between KCNQ1 variants and PCOS were calculated
using the G*power calculator, version 3.1.9.7 (Heinrich Heine
University). The parameters considered included the number of study
subjects (230 cases and 230 controls), genotypic odds ratio (OR)
for heterozygotes and mutant homozygotes, MAF for PCOS cases and
controls across the three tested variants, and a 12% prevalence of
PCOS in the general population according to the Rotterdam criteria
(43). Under these assumptions, the
overall power was calculated as the average power of the tested
variants and was found to be 80%.
Statistical analysis for the study was performed
using SPSS Statistics for Windows, Version 28 (IBM Corp.). The
clinical characteristics were presented as median (25-75th
percentile). To compare quantitative variables, Mann-Whitney U
(Wilcoxon) was used.
Allelic and genotypic frequencies were determined
using the gene-counting method implemented through SNPassoc R
package (https://github.com/isglobal-brge/SNPassoc). The
Hardy-Weinberg equilibrium (HWE) for each variant, was assessed
using a chi-squared test, also performed with the SNPassoc R
package.
The differences in PCOS traits levels between the
three genotypes (wild homozygous, heterozygous and mutant
homozygous) of the three tested variants were assessed with the
non-parametric Kruskal Wallis test.
Correlation between covariate traits and the three
genotypes, was estimated by Spearman coefficient. A positive
correlation is considered when the coefficient ranges between 0 to
1. Conversely, a negative correlation corresponds to a coefficient
ranging between 0 to -1.
The differences in allele and genotype frequencies
between cases and controls were calculated for each variant,
utilizing the gene-counting method using the SNPassoc R
package.
Genetic association analysis was performed using
binary logistic regression under four genetic models (additive,
codominant, dominant and recessive). The control group served as
the reference for calculating ORs. Suppose (A) is the ancestral
allele and (a) is the altered allele. The codominant model compared
heterozygous (Aa) and homozygous variant (aa) genotypes to the
ancestral homozygous (AA) genotype. The dominant model compared
carriers of the altered allele (Aa + aa) to non-carriers (AA),
while the recessive model compared homozygous variant (aa)
individuals with those with the (AA + Aa) genotype. The additive
model assumed a proportional increase in risk with each additional
altered allele (a).
SHEsisPlus software (http://shesisplus.bio-x.cn/SHEsis.html) was used for
binary variant interactions and haplotype association analysis. The
multifactor dimensionality reduction (MDR) method was applied to
analyse epistasis (44) which helps
identify genetic variant combinations that influence disease
susceptibility, by reducing the data's dimensionality and
evaluating the predictive power of these combinations.
Corrected P-values for multiple hypothesis testing
for analysis was performed with false discovery rate (FDR) method
by Benjamini-Hochberg. P<0.05 was considered to indicate a
statistically significant difference.
Results
Study subjects
The demographic and biochemical parameters of all
the study participants are presented in Table I. A significant difference in age
was revealed at examination between PCOS and controls subjects
after applying the Benjamini-Hochberg adjustment. However, no
substantial disparities were observed in BMI, fasting glucose,
total testosterone, LDL, triglycerides, total cholesterol, FSH or
LH in women with PCOS compared with controls (P>0.05).
 | Table IClinical and biochemical
characteristics of study subjects. |
Table I
Clinical and biochemical
characteristics of study subjects.
| Characteristic | Polycystic ovary
syndromea (n=230) |
Controlsa (n=230) |
P-valueb | FDR
P-valuec |
|---|
| Age, years | 32.5 (28-37) | 30 (27-33.25) | 0.001 | 0.011 |
| Body mass index
(kg/m2) | 26 (23-29) | 27 (24-31) | 0.018 | 0.099 |
| High-density
lipoprotein (mmol/l) | 1.38
(1.24-1.42) | 1.36
(1.26-1.42) | 0.876 | 0.938 |
| Low-density
lipoprotein (mmol/l) | 2.97
(2.58-3.03) | 2.92
(2.38-3.08) | 0.402 | 0.816 |
| Triglycerides
(mmol/l) | 1.51
(1.03-1.62) | 1.58
(1.42-2.09) | 0.055 | 0.201 |
| Cholesterol
(mmol/l) | 5.80
(5.25-5.94) | 5.91
(5.20-6.32) | 0.478 | 0.816 |
| Luteinizing hormone
levels (mlU/mol) | 2.99
(0.49-5.86) | 2.94
(0.48-4.51) | 0.633 | 0.816 |
|
Follicle-stimulating hormone levels
(mlU/mol) | 4.67
(3.82-6.52) | 4.45
(3.34-5.01) | 0.215 | 0.591 |
| Progesterone
(mlU/mol) | 0.97
(0.92-1.02) | 0.94 (0.92-1) | 0.619 | 0.816 |
| Testosterone
(nmol/l) | 2.75
(1.59-3.91) | 2.97
(1.89-3.84) | 0.668 | 0.816 |
| Prolactin
(ng/ml) | 114.87
(84.74-135.41) | 99.95
(77.38-130.37) | 0.938 | 0.938 |
Correlation between clinical,
biochemical and hormonal parameters
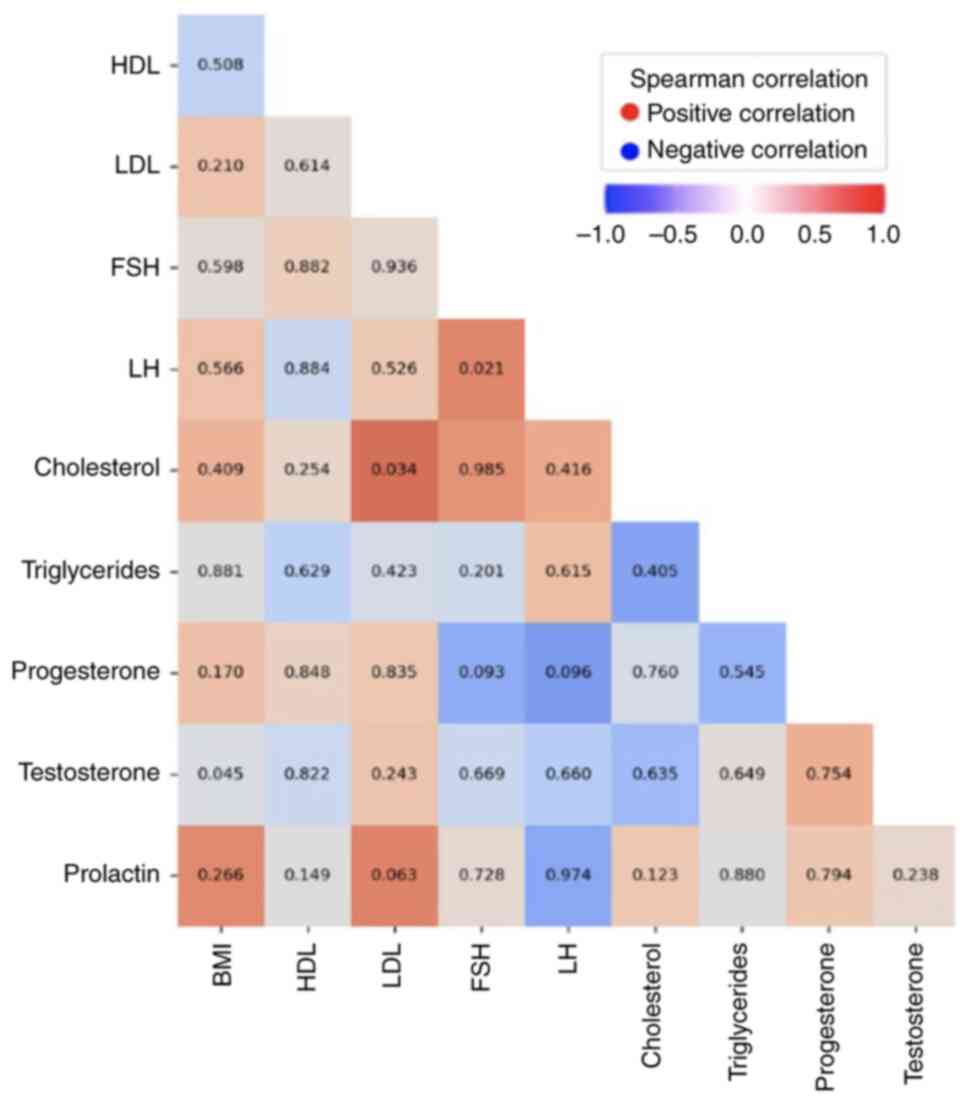
The correlation between the different quantitative
parameters including BMI, cholesterol, triglycerides, HDL, LDL,
FSH, LH, progesterone, testosterone and prolactin was assessed by
Spearman's coefficient. The results showed a positive correlation
between LDL and cholesterol (P=0.034) consistent with the
well-established relationship between these lipids, as well as
between FSH and LH (P=0.021) (Fig.
1). Additionally, a weak negative correlation was detected
between testosterone and BMI (P=0.045). No other statistically
significant differences were observed between the other parameters
(Fig. 1).
Variants association analysis
The HWE and MAF for the three KCNQ1 variants
(rs231361, rs151290 and rs2237895) in both case and control
subjects are summarized in Table
II. All variants were in HWE (P>0.05), indicating that the
genotype distributions follow the expected pattern, and no
population stratification was detected.
 | Table IIGenotype and risk allele frequencies
of KCNQ1 gene variants in PCOS cases and controls. |
Table II
Genotype and risk allele frequencies
of KCNQ1 gene variants in PCOS cases and controls.
| Variant | PCOS, n (%) | Controls, n
(%) | HWE-
P-valuea |
|---|
| rs231361 | | | 0.063 |
|
G/G | 130 (56.5) | 148 (64.3) | |
|
G/A | 74 (32.2) | 76 (33.0) | |
|
A/A | 26 (11.3) | 6 (2.6) | |
|
RA (A) | 126 (27.4) | 88 (19.1) | |
| rs151290 | | | 0.297 |
|
C/C | 112 (48.7) | 132 (57.4) | |
|
C/A | 108 (47.0) | 80 (34.8) | |
|
A/A | 10 (4.3) | 18 (7.8) | |
|
RA (A) | 128 (27.8) | 116 (25.2) | |
| rs2237895 | | | 0.132 |
|
A/A | 70 (30.4) | 94 (40.9) | |
|
A/C | 106 (46.1) | 102 (44.3) | |
|
C/C | 54 (23.5) | 34 (14.8) | |
|
RA (C) | 214 (46.5) | 170 (36.9) | |
The subsequent genetic analysis examined the
relationship between PCOS susceptibility and these variants using
various genetic models, including additive, codominant, dominant
and recessive models. The results of the data analysis, presented
in Table III, revealed that only
the rs231361 variant was significantly associated with an increased
risk of PCOS across multiple genetic models.
 | Table IIIPCOS association for KCNQ1
gene variants in the Tunisian study samples. |
Table III
PCOS association for KCNQ1
gene variants in the Tunisian study samples.
| Variant | Model |
P-valuea | FDR
P-valueb | FDR
P-valuec | OR (95% CI) | Power |
|---|
| rs231361 | Additive: (A vs.
G) | 0.003 | 0.004 | 0.011 | 1.59
(1.17-2.17) | 0.847 |
| | Codominant: (G/A
vs. G/G) | 0.611 | 0.611 | 0.611 | 1.11
(0.73-1.64) | |
| | Codominant: (A/A
vs. G/G) |
2.2x10-4 |
6.6x10-4 | 0.002 | 4.93
(1.35-18.8) | 0.959 |
| | Dominant: (G/A+A/A
vs. G/G) | 0.087 | 0.087 | 0.130 | 1.39
(0.95-2.02) | |
| | Recessive: (A/A vs.
G/A+G/G) |
2.47x10-4 |
7.4x10-4 | 0.002 | 4.76
(1.92-11.79) | 0.958 |
| rs151290 | Additive: (A vs.
C) | 0.526 | 0.526 | 0.564 | 1.14
(0.85-1.53) | |
| | Codominant: (C/A
vs. C/C) | 0.017 | 0.051 | 0.036 | 1.59
(1.08-2.34) | 0.760 |
| | Codominant: (A/A
vs. C/C) | 0.305 | 0.305 | 0.352 | 0.65
(0.29-1.48) | |
| | Dominant: (C/A+A/A
vs. C/C) | 0.061 | 0.087 | 0.102 | 1.42
(0.98-2.05) | |
| | Recessive: (A/A vs.
C/A+C/C) | 0.119 | 0.119 | 0.149 | 0.54
(0.24-1.19) | |
| rs2237895 | Additive: (C vs.
A) | 0.003 | 0.004 | 0.011 | 1.48
(1.14-1.93) | 0.551 |
| | Codominant: (A/C
vs. A/A) | 0.112 | 0.168 | 0.149 | 1.40
(0.92-2.11) | |
| | Codominant: (C/C
vs. A/A) | 0.005 | 0.007 | 0.015 | 2.13
(1.26-3.62) | 0.660 |
| | Dominant: (A/C+C/C
vs. A/A) | 0.019 | 0.057 | 0.036 | 1.58
(1.08-2.32) | 0.662 |
| | Recessive: (C/C vs.
A/C+A/A) | 0.018 | 0.027 | 0.036 | 1.77
(1.10-2.84) | 0.551 |
The additive model showed an OR of 1.59, with an FDR
P-value of 0.011 and a power of 84.7%. Similarly, the codominant
and recessive models also demonstrated significant associations,
with ORs of 4.93 and 4.76, respectively, and FDR P-values of 0.002,
along with high statistical power (>95%). However, while the
rs151290 and rs2237895 variants showed association with an
increased risk of PCOS, these associations were not considered
statistically significant as the statistical power did not exceed
the threshold of 80%.
Correlation between KCNQ1 variants and
PCOS-associated features
The correlation between KCNQ1 tested variants
and PCOS-associated metabolic and endocrine traits was assessed.
One of the key findings was a positive correlation between the
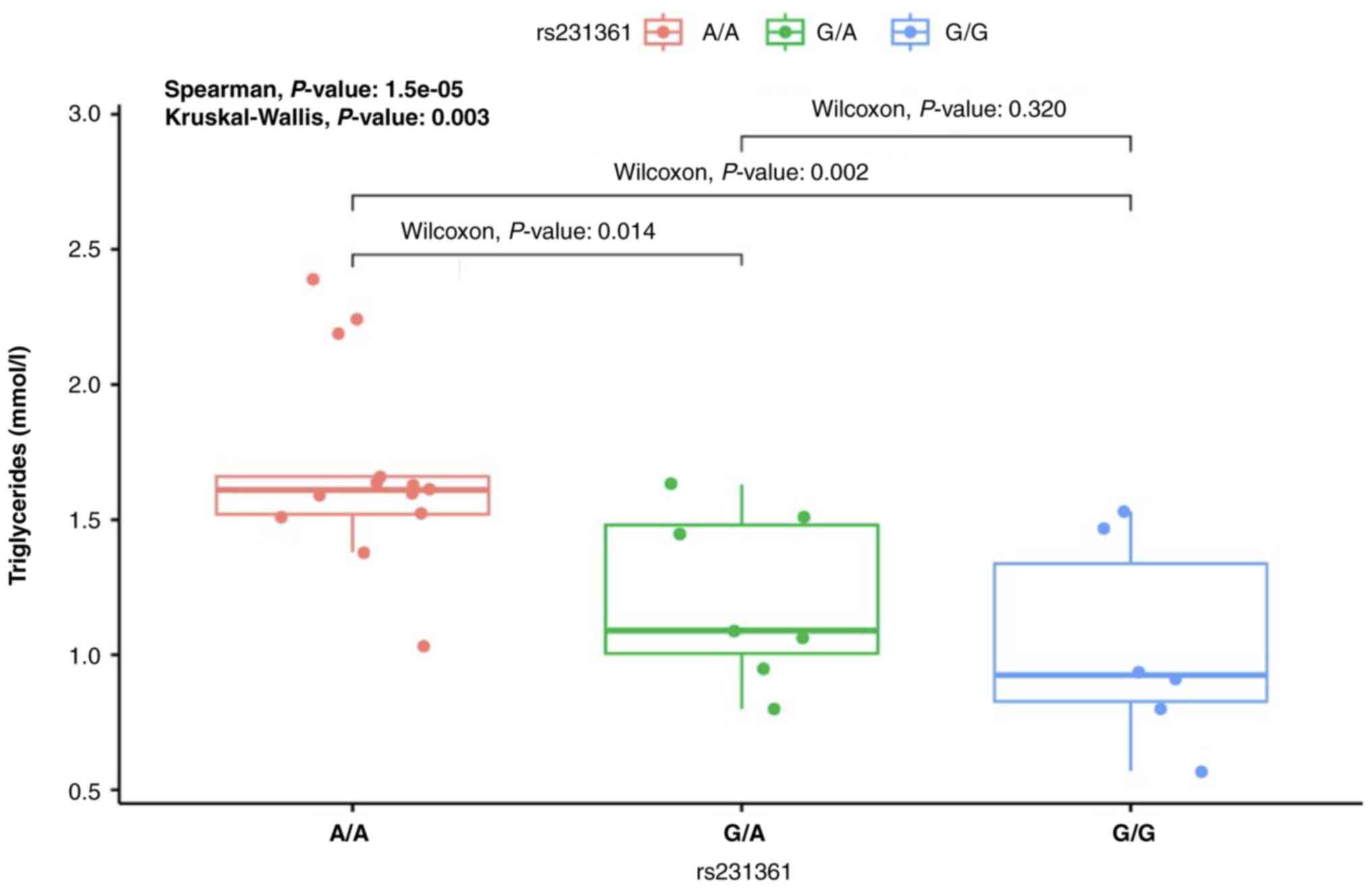
rs231361 variant and increased triglyceride levels (Table IV). Furthermore, when comparing
different genotypes of the rs231361 variant, individuals carrying
the A/A genotype displayed a statistically significant correlation
with higher triglyceride levels compared with those with the
wild-type G/G genotype (P=0.002) and the heterozygous G/A genotype
(P=0.014) (Fig. 2). This indicates
that the individuals carrying A/A genotype may be more prone to
elevated triglyceride levels, which is a metabolic parameter often
associated with PCOS. This genotype-specific association highlights
the importance of considering genetic variations when studying the
metabolic disturbances in PCOS and the importance of personalized
medicine in PCOS management. No statistically significant
differences were observed between the other KCNQ1 variants
(rs151290 and rs2237895) and any of the metabolic or endocrine
parameters tested (Table IV),
indicating that these variants may not have a major impact on
PCOS-related traits.
 | Table IVCorrelation between KCNQ1 gene
variants with PCOS features. |
Table IV
Correlation between KCNQ1 gene
variants with PCOS features.
| Trait | Variant | Risk allele |
P-valuea |
rb |
|---|
| Body mass
index | rs231361 | A | 0.942 | 0.008 |
| | rs151290 | A | 0.464 | 0.078 |
| | rs2237895 | C | 0.167 | 0.146 |
| Cholesterol | rs231361 | A | 0.173 | 0.467 |
| | rs151290 | A | 0.314 | -0.355 |
| | rs2237895 | C | 0.155 | 0.485 |
| Triglycerides | rs231361 | A |
1.5x10-5 | 0.676 |
| | rs151290 | A | 0.600 | 0.108 |
| | rs2237895 | C | 0.929 | 0.018 |
| High-density
lipoprotein | rs231361 | A | 0.639 | 0.074 |
| | rs151290 | A | 0.133 | 0.233 |
| | rs2237895 | C | 0.182 | -0.207 |
| Low-density
lipoprotein | rs231361 | A | 0.539 | -0.096 |
| | rs151290 | A | 0.983 | 0.003 |
| | rs2237895 | C | 0.336 | 0.150 |
|
Follicle-stimulating hormone | rs231361 | A | 0.302 | 0.250 |
| | rs151290 | A | 0.638 | -0.115 |
| | rs2237895 | C | 0.709 | 0.092 |
| Luteinizing
hormone | rs231361 | A | 0.761 | 0.083 |
| | rs151290 | A | 0.548 | -0.150 |
| | rs2237895 | C | 0.107 | -0.418 |
| Progesterone | rs231361 | A | 0.704 | 0.107 |
| | rs151290 | A | 0.491 | 0.193 |
| | rs2237895 | C | 0.975 | -0.009 |
| Testosterone | rs231361 | A | 0.554 | -0.077 |
| | rs151290 | A | 0.582 | -0.072 |
| | rs2237895 | C | 0.826 | -0.029 |
| Prolactin | rs231361 | A | 0.150 | 0.405 |
| | rs151290 | A | 0.659 | 0.129 |
| | rs2237895 | C | 0.062 | 0.590 |
Variants interaction analysis
The binary variant interaction analysis aimed to
investigate whether specific combinations of KCNQ1 variants
(rs231361, rs151290 and rs2237895) were associated with PCOS
susceptibility. However, the results in Table V indicate that none of the variant
interactions showed statistically significant differences between
individuals with PCOS and controls. Both the nominal P-values and
the more stringent FDR P-values were above the 0.05 threshold
(Table V), suggesting that variant
interactions may not contribute to PCOS susceptibility in this
study population.
 | Table VBinary variant interactions
analysis. |
Table V
Binary variant interactions
analysis.
| KCNQ1
variants | PCOSa |
Controlsb | Difference |
P-valuec | FDR
P-valueb |
|---|
|
rs231361-rs151290 | -0.009 | -0.014 | 0.005 | 0.309 | 0.309 |
|
rs231361-rs2237895 | -0.011 | -0.024 | 0.012 | 0.189 | 0.309 |
|
rs151290-rs2237895 | -0.005 | -0.016 | 0.011 | 0.249 | 0.309 |
MDR simulation for the effect of KCNQ1
interaction on PCOS risk
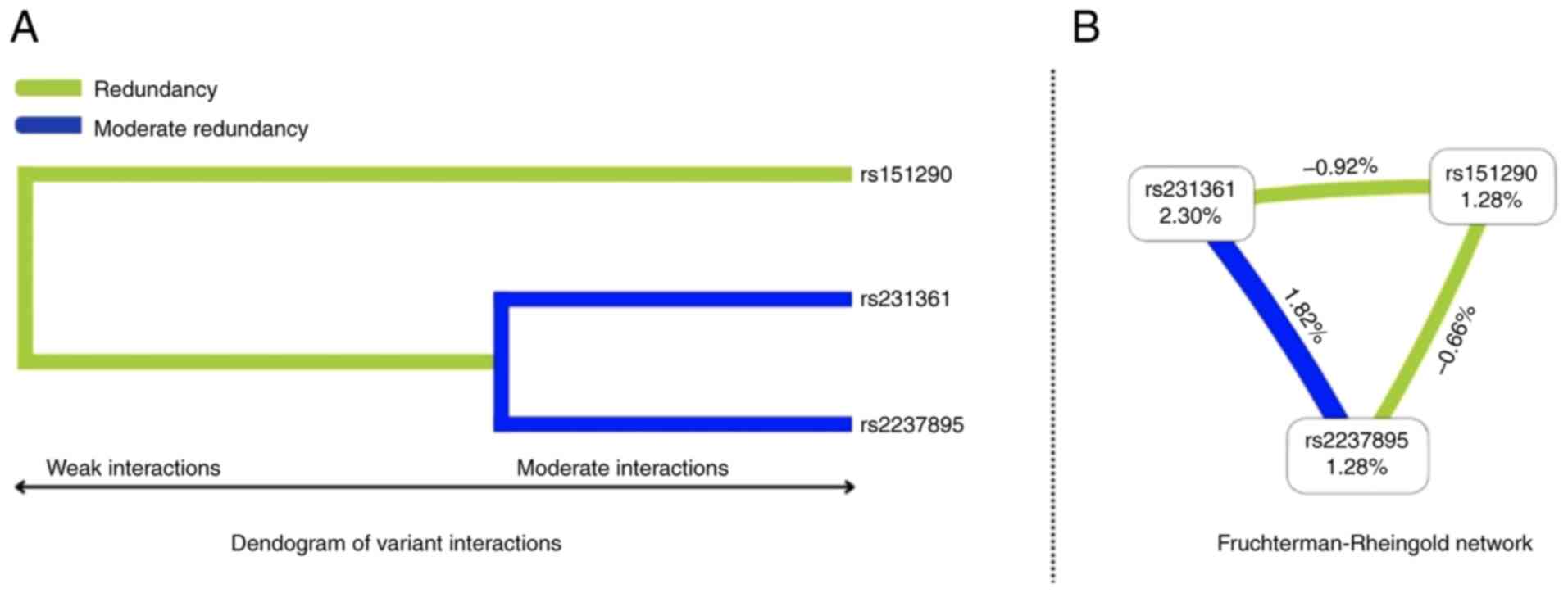
To further explore the epistatic interactions of
KCNQ1 variants, a MDR simulation was conducted. A total of
two visualization methods were employed: A dendrogram illustrating
variant interaction patterns and a Fruchterman-Reingold
network.
The visualizations in Fig. 3A (dendogram) and Fig. 3B (Fruchterman-Rheingold) suggest a
moderate synergy between rs231361 and rs2237895 variants, forming a
cluster indicative of a combined, amplified effect. This indicates
that these two variants may interact in a positively synergistic
manner, influencing PCOS risk. By contrast, the rs151290 variant
showed weak and antagonistic interactions with both rs231361 and
rs2237895 indicating that its presence may counteract the effects
of the aforementioned variants on PCOS risk.
The best models of KCNQ1 variant interactions
and their potential association with PCOS susceptibility are
reported in Table VI. Although
multiple models were identified, none achieved statistical
significance. Future studies using larger sample sizes are
warranted to validate potential interactions.
 | Table VIBest models for variant interactions
of the KCNQ1 gene and the predisposition to PCOS. |
Table VI
Best models for variant interactions
of the KCNQ1 gene and the predisposition to PCOS.
| Models | Testing balance
accuracy | Cross validation
consistency | OR (95% CI) |
P-valuea | FDR
P-valueb |
|---|
| rs231361 | 0.448 | 6/10 | 1.68
(0.69-4.07) | 0.250 | 0.337 |
|
rs231361-rs2237895 | 0.583 | 10/10 | 2.02
(0.37-11.17) | 0.189 | 0.337 |
|
rs231361-rs151290-rs2237895 | 0.483 | 10/10 | 2.55
(0.35-18.47) | 0.337 | 0.337 |
Haplotype analysis
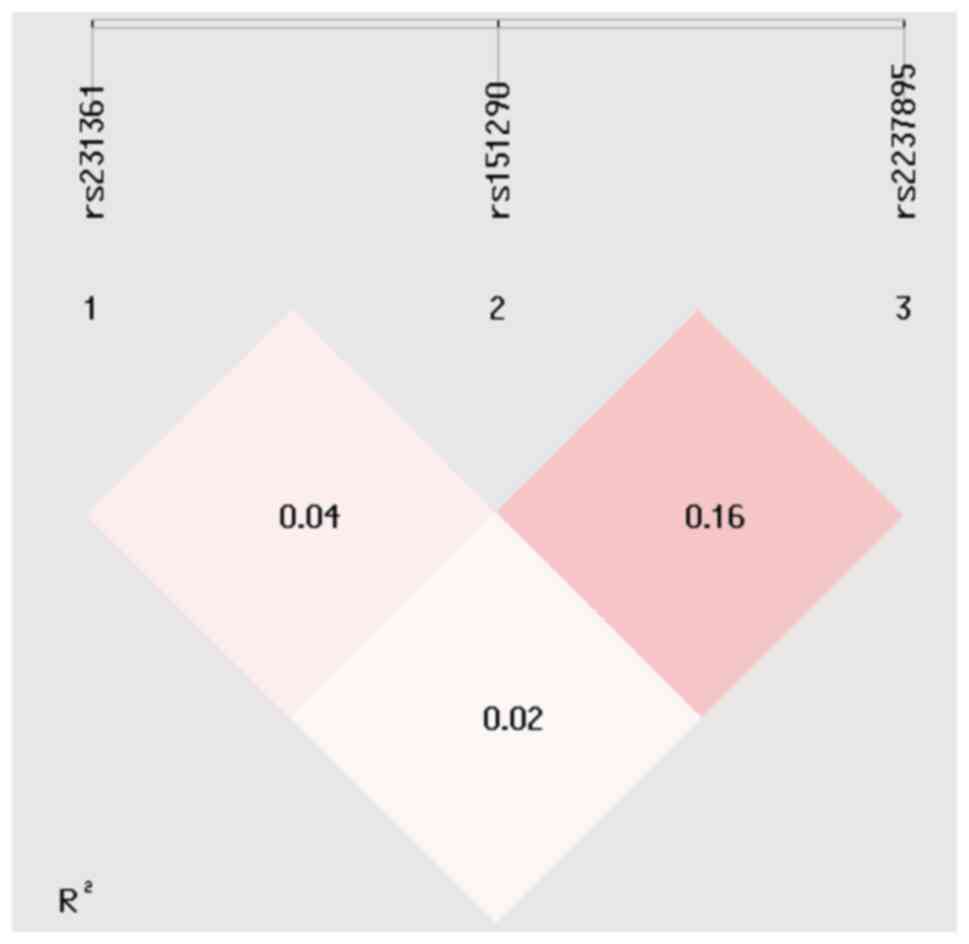
The haplotype analysis provides further insights
into the potential associations between KCNQ1 gene variants
and PCOS susceptibility. Linkage disequilibrium (LD) analysis,
which refers to the random association of three alleles, revealed
limited LD between the three tested KCNQ1 variants (Fig. 4), suggesting that these variants act
independently rather than as a tightly linked block. Several
haplotypes were identified in the study population. The frequencies
of these haplotypes differed between cases and control subjects as
shown in Table VII.
 | Table VIIHaplotype association analysis of
KCNQ1 gene variants with PCOS. |
Table VII
Haplotype association analysis of
KCNQ1 gene variants with PCOS.
|
Haplotypea | PCOS, n
(%)b | Controls, n
(%)b |
P-valuec | FDR
P-valued | OR (95% CI) | Power |
|---|
| GCA | 112 (24.3) | 158 (34.3) | - | - | Reference | |
| GAA | 82 (17.8) | 92 (20.0) | 0.242 | 0.242 | 1.26
(0.86-1.85) | |
| GCC | 108 (23.4) | 104 (22.6) | 0.038 | 0.057 | 1.46
(1.02-2.10) | |
| GAC | 32 (6.9) | 18 (3.9) | 0.003 | 0.009 | 2.51
(1.34-4.69) | 0.521 |
| ACA | 40 (8.6) | 34 (7.3) | 0.054 | 0.065 | 1.66
(0.99-2.78) | |
|
ACC | 72 (15.6) | 48 (10.4) |
7.22x10-4 | 0.004 | 2.12
(1.37-3.29) | 0.650 |
| AAA | 12 (12.6) | 6 (1.3) | 0.037 | 0.057 | 2.82
(1.03-7.74) | |
Taking the GCA haplotype as a common (reference),
two haplotypes showed positive associations with PCOS risk. The GAC
haplotype had an OR of 2.51, indicating that individuals carrying
this haplotype were ~2.5 times more likely to develop PCOS compared
with those with the reference haplotype. Similarly, the ACC
haplotype had an OR of 2.12, suggesting a slightly lower but still
significant association with PCOS risk (Table VII). However, despite these
observed associations, the statistical power did not reach the
threshold of >80%, thus none of these haplotypes are considered
associated with PCOS susceptibility.
Discussion
The present study provides new insights into the
potential role of the KCNQ1 gene in the genetic
predisposition to PCOS. Although the KCNQ1 gene is
well-recognized as a risk factor for T2D in multiple ethnic
populations (24-28),
its association with PCOS has remained unexplored. To the best of
our knowledge, the present study is the first to investigate the
potential relationship between three KCNQ1 variants
(rs231361, rs151290 and rs2237895) and the risk of developing PCOS,
offering a new perspective on the shared genetic mechanisms
underlying these complex disorders. Nevertheless, given the absence
of prior research on the association between PCOS and the
KCNQ1 gene, direct comparisons are currently
challenging.
One of the key outcomes of the present study is the
identification of the KCNQ1 rs21361 variant as a novel risk
factor for PCOS, with statistical analyses demonstrating its strong
association across multiple genetic models. Interestingly, this
variant was also found to be significantly correlated with
increased triglycerides levels in affected women, suggesting a
potential mechanistic link between KCNQ1 genetic variation
and metabolic dysfunction in PCOS.
Additionally, previous research has indicated that
the rs231361 variant does not contribute to T2D susceptibility in
Tunisian Arab populations (45),
suggesting a unique role in PCOS development, independent of its
relationship with T2D in this ethnic group. This specificity
suggests that the rs231361 variant may act through distinct
molecular pathways independent of those involved in T2D,
highlighting the need for further functional studies to elucidate
the precise mechanisms through which this variant contributes to
PCOS pathogenesis.
The analysis of the remaining KCNQ1 variants,
including rs151290 and rs2237895, did not reveal statistically
significant associations with PCOS in the studied population,
indicating their unlikely involvement in the pathogenesis of PCOS
in Tunisian Arabs. Interestingly, previous studies have observed
the absence of association between these variants and T2D in the
Tunisian Arab population (45),
contrasting with findings in other ethnic groups (25,46).
This highlights the potential influence of ethnic-specific genetic
factors on disease susceptibility. It also indicates that these
KCNQ1 variants may have a limited role in the genetic
susceptibility to both PCOS and T2D within the Tunisian population.
This emphasizes the significance of investigating diverse
populations to gain a more comprehensive understanding of the
genetic complexities underlying these disorders.
Beyond identifying specific genetic associations,
the present study also explored the phenotypic correlations between
KCNQ1 variants with PCOS-related metabolic features. A
significant association was observed between the risk allele (A) of
the rs231361 variant and elevated triglyceride levels in women with
PCOS, underscoring the potential role of KCNQ1 variants in
lipid metabolism and their potential contribution to the metabolic
disturbances commonly observed in PCOS. The aforementioned results
align with previous research on other KCNQ1 variants such as
r2283228 and rs2237892, which have been linked to higher
triglycerides levels, and lower HDL levels (47). The clinical implications of the
current findings are particularly relevant for the development of
personalized treatment strategies for individuals with PCOS. Given
the association between KCNQ1 variants and lipid metabolism,
targeted interventions aimed at modulating triglyceride levels may
prove beneficial for patients with PCOS carrying the rs231361
variant. Furthermore, pharmacogenomic studies could further explore
how genetic variants influence individual responses to
lipid-lowering agents, such as statins or fibrates, in PCOS
management.
The MDR analysis revealed epistatic interactions
between KCNQ1 variants and their collective impact on PCOS
risk. Specifically, the rs231361 and rs2237895 variants exhibited a
moderate synergistic effect, increasing susceptibility to PCOS. By
contrast, the rs151290 variant displayed weaker, antagonistic
interactions with both rs231361 and rs2237895 variants. These
findings suggest a complex interplay between KCNQ1 genetic
variants in modulating PCOS risk, emphasizing the importance of
considering epistatic effects in pharmacogenomic strategies, rather
than solely focusing on the independent contributions of individual
variants. Furthermore, given the association of rs231361 with
elevated triglyceride levels, and its synergistic effect with
rs2237895, patients carrying these variants may benefit from
personalized lipid-modulating interventions, such as statins,
fibrates, or novel lipid-lowering agents. This opens avenues for
gene-based therapeutic strategies, including the potential for
selective potassium channel modulators to regulate metabolic
imbalances in PCOS. Furthermore, the antagonistic effect of
rs151290 suggests that its presence may influence drug response in
patients with PCOS, warranting further investigation. These
findings highlight the necessity of integrating pharmacogenomic
screening into PCOS treatment protocols, to ensure that genetic
variations are accounted for when prescribing metabolic and
hormonal therapies.
The haplotype analysis identified two haplotypes
(GAC and AAC) which exhibited positive associations with PCOS risk.
However, despite the observed associations, statistical power
limitations of the study prevent us from drawing definitive
conclusions regarding their contribution to disease susceptibility.
Larger-scale studies are warranted to validate these findings and
determine whether specific KCNQ1 haplotypes indeed increase
the risk of developing PCOS.
While the present study possesses several strengths
that contribute to its validity, it is essential to acknowledge its
limitations. The homogenous ethnic origin (Tunisian Arabs) of the
present study population reduces confounding effects from genetic
admixture but may limit the generalizability of our findings to
other populations. Additionally, the present study focused on only
three KCNQ1 variants, potentially overlooking other variants
implicated in PCOS pathogenesis. The retrospective nature of our
case-control study design, further limits causal inference. Future
research should encompass these limitations by incorporating
larger, multi-ethnic cohorts, performing comprehensive genomic
analyses, and utilizing longitudinal study designs to establish
causal relationships.
In conclusion, the current findings pave the way for
a deeper understanding of the complex genetic architecture of PCOS,
identifying KCNQ1 rs21361 variant as an important factor in
disease susceptibility. The observed associations between this
variant and triglyceride levels underscore the potential metabolic
implications of KCNQ1 variation, opening avenues for
targeted pharmaceutical interventions. Future studies, integrating
genomics, pharmacogenomics and larger-scale genetic analyses will
be crucial for elucidating the biological mechanisms linking
KCNQ1 to PCOS and optimizing treatment strategies for
affected individuals.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ABS designed the study, collected samples and
selected data. IE collected samples and selected data. HBA
contributed to data acquisition and critically reviewed the
manuscript. NM analysed and interpreted data and critically
reviewed the manuscript. SS was the project leader, interpretated
data, and wrote and reviewed the manuscript. All authors read and
approved the final version of the manuscript. ABS and NM confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was conducted in adherence with
the principles outlined in the Helsinki Declaration (2014) and
received ethical approval from the local research and ethics
committee of Farhat Hached Hospital (approval no. 35220228; Sousse,
Tunisia). Written informed consent was provided by all participants
before the start of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Escobar-Morreale HF: Polycystic ovary
syndrome: Definition, aetiology, diagnosis and treatment. Nat Rev
Endocrinol. 14:270–284. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Deswal R, Narwal V, Dang A and Pundir CS:
The prevalence of polycystic ovary syndrome: A brief systematic
review. J Hum Reprod Sci. 13:261–271. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li M, Ruan X and Mueck AO: Management
strategy of infertility in polycystic ovary syndrome. Glob Health
J. 6:70–74. 2022.
|
|
4
|
Goodarzi MO, Dumesic DA, Chazenbalk G and
Azziz R: Polycystic ovary syndrome: etiology, pathogenesis and
diagnosis. Nat Rev Endocrinol. 7:219–231. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Costello M, Garad R, Hart R, Homer H,
Johnson L, Jordan C, Mocanu E, Qiao J, Rombauts L, Teede HJ, et al:
A review of first line infertility treatments and supporting
evidence in women with polycystic ovary syndrome. Med Sci (Basel).
7(95)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Singh S, Pal N, Shubham S, Sarma DK, Verma
V, Marotta F and Kumar M: Polycystic ovary syndrome: Etiology,
current management, and future therapeutics. J Clin Med.
12(1454)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bordewijk EM, Ng KYB, Rakic L, Mol BWJ,
Brown J, Crawford TJ and van Wely M: Laparoscopic ovarian drilling
for ovulation induction in women with anovulatory polycystic ovary
syndrome. Cochrane Database Syst Rev. 2(CD001122)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dong J and Rees DA: Polycystic ovary
syndrome: Pathophysiology and therapeutic opportunities. BMJ Med.
2(e000548)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Barry JA, Azizia MM and Hardiman PJ: Risk
of endometrial, ovarian and breast cancer in women with polycystic
ovary syndrome: A systematic review and meta-analysis. Hum Reprod
Update. 20:748–758. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen Q, Zheng B, Du S and Lin Y: Explore
the potential molecular mechanism of polycystic ovarian syndrome by
protein-protein interaction network analysis. Taiwan J Obstet
Gynecol. 60:807–815. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hong SH, Hong YS, Jeong K, Chung H, Lee H
and Sung YA: Relationship between the characteristic traits of
polycystic ovary syndrome and susceptibility genes. Sci Rep.
10(10479)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dixit G, Dabney-Smith C and Lorigan GA:
The membrane protein KCNQ1 potassium ion channel: Functional
diversity and current structural insights. Biochim Biophys Acta
Biomembr. 1862(183148)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kuenze G, Duran AM, Woods H, Brewer KR,
McDonald EF, Vanoye CG, George AL Jr, Sanders CR and Meiler J:
Upgraded molecular models of the human KCNQ1 potassium channel.
PLoS One. 14(e0220415)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jespersen T, Grunnet M and Olesen SP: The
KCNQ1 potassium channel: From gene to physiological function.
Physiology (Bethesda). 20:408–416. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gao T, Li K, Liang F, Yu J, Liu A, Ni Y
and Sun P: KCNQ1 potassium channel expressed in human sperm is
involved in sperm motility, acrosome reaction, protein tyrosine
phosphorylation, and ion homeostasis during capacitation. Front
Physiol. 12(761910)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Grahammer F, Herling AW, Lang HJ,
Schmitt-Gräff A, Wittekindt OH, Nitschke R, Bleich M, Barhanin J
and Warth R: The cardiac K+ channel KCNQ1 is essential for gastric
acid secretion. Gastroenterology. 120:1363–1371. 2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Neyroud N, Richard P, Vignier N, Donger C,
Denjoy I, Demay L, Shkolnikova M, Pesce R, Chevalier P, Hainque B,
et al: Genomic organization of the KCNQ1 K+ channel gene and
identification of C-terminal mutations in the long-QT syndrome.
Circ Res. 84:290–297. 1999.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Neyroud N, Tesson F, Denjoy I, Leibovici
M, Donger C, Barhanin J, Fauré S, Gary F, Coumel P, Petit C, et al:
A novel mutation in the potassium channel gene KVLQT1 causes the
Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet.
15:186–189. 1997.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Rosengren AH, Braun M, Mahdi T, Andersson
SA, Travers ME, Shigeto M, Zhang E, Almgren P, Ladenvall C,
Axelsson AS, et al: Reduced insulin exocytosis in human pancreatic
β-cells with gene variants linked to type 2 diabetes. Diabetes.
61:1726–1733. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tiron C, Campuzano O, Pérez-Serra A,
Mademont I, Coll M, Allegue C, Iglesias A, Partemi S, Striano P,
Oliva A and Brugada R: Further evidence of the association between
LQT syndrome and epilepsy in a family with KCNQ1 pathogenic
variant. Seizure. 25:65–67. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Goldman AM, Glasscock E, Yoo J, Chen TT,
Klassen TL and Noebels JL: Arrhythmia in heart and brain: KCNQ1
mutations link epilepsy and sudden unexplained death. Sci Transl
Med. 1(2ra6)2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lieve KVV and Wilde AAM: Inherited ion
channel diseases: A brief review. Europace. 17 (Suppl 2):ii1–ii6.
2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Morris AP: Fine mapping of type 2 diabetes
susceptibility loci. Curr Diab Rep. 14(549)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ma Q, Wang L, Yao H, Wang TT, Ma Y, Su YX,
Wang ZQ, Zhu J, Wang SX, Zhang ZX, et al: Association between KCNQ1
genetic variants and type 2 diabetes in the uyghur population.
Genet Test Mol Biomarkers. 19:698–702. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Been LF, Ralhan S, Wander GS, Mehra NK,
Singh J, Mulvihill JJ, Aston CE and Sanghera DK: Variants in KCNQ1
increase type II diabetes susceptibility in South Asians: A study
of 3,310 subjects from India and the US. BMC Med Genet.
12(18)2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Unoki H, Takahashi A, Kawaguchi T, Hara K,
Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K,
Jørgensen T, et al: SNPs in KCNQ1 are associated with
susceptibility to type 2 diabetes in East Asian and European
populations. Nat Genet. 40:1098–1102. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Yasuda K, Miyake K, Horikawa Y, Hara K,
Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Sun Q, Song K, Shen X and Cai Y: The
association between KCNQ1 gene polymorphism and type 2 diabetes
risk: A meta-analysis. PLoS One. 7(e48578)2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Thong EP, Codner E, Laven JSE and Teede H:
Diabetes: A metabolic and reproductive disorder in women. Lancet
Diabetes Endocrinol. 8:134–149. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Glintborg D, Ollila MM, Møller JK, Pesonen
P, Persson S, Elenis E, Rubin KH, Gissler M, Andersen MS,
Sundström-Poromaa I and Piltonen T: Prospective risk of type 2
diabetes in 99 892 Nordic women with polycystic ovary syndrome and
446 055 controls: National cohort study from Denmark, Finland, and
Sweden. Hum Reprod. 39:1823–1834. 2024.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hossain MA, Al Ashik SA, Mahin MR, Al Amin
M, Rahman MH, Khan MA and Emran AA: Systems biology and in
silico-based analysis of PCOS revealed the risk of metabolic
disorders. Heliyon. 8(e12480)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Márquez JL, Pacheco A, Valdés P and
Salazar LA: Association between CAPN10 UCSNP-43 gene polymorphism
and polycystic ovary syndrome in Chilean women. Clin Chim Acta.
398:5–9. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gonzalez A, Abril E, Roca A, Aragón MJ,
Figueroa MJ, Velarde P, Royo JL, Real LM and Ruiz A: Comment:
CAPN10 alleles are associated with polycystic ovary syndrome. J
Clin Endocrinol Metab. 87:3971–3976. 2002.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tang ST, Wang CJ, Tang HQ, Peng WJ, Wang
YM and Zhang Q: Association of Pro12Ala polymorphism in peroxisome
proliferator-activated receptor gamma with polycystic ovary
syndrome: A meta-analysis. Mol Biol Rep. 39:9649–9660.
2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xu X, Zhao H, Shi Y, You L, Bian Y, Zhao Y
and Chen ZJ: Family association study between INSR gene
polymorphisms and PCOS in Han Chinese. Reprod Biol Endocrinol.
9(76)2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Christopoulos P, Mastorakos G, Gazouli M,
Panidis D, Deligeoroglou E, Katsikis I, Papadias K,
Diamandi-Kandarakis E and Creatsas G: Genetic variants in TCF7L2
and KCNJ11 genes in a Greek population with polycystic ovary
syndrome. Gynecol Endocrinol. 24:486–490. 2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wojciechowski P, Lipowska A, Rys P, Ewens
KG, Franks S, Tan S, Lerchbaum E, Vcelak J, Attaoua R, Straczkowski
M, et al: Impact of FTO genotypes on BMI and weight in polycystic
ovary syndrome: A systematic review and meta-analysis.
Diabetologia. 55:2636–2645. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Waterworth DM, Bennett ST, Gharani N,
McCarthy MI, Hague S, Batty S, Conway GS, White D, Todd JA, Franks
S and Williamson R: Linkage and association of insulin gene VNTR
regulatory polymorphism with polycystic ovary syndrome. Lancet.
349:986–990. 1997.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li X, Xiao H, Ma Y, Zhou Z and Chen D:
Identifying novel genetic loci associated with polycystic ovary
syndrome based on its shared genetic architecture with type 2
diabetes. Front Genet. 13(905716)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Rotterdam ESHRE/ASRM-Sponsored PCOS
consensus workshop group. Revised 2003 consensus on diagnostic
criteria and long-term health risks related to polycystic ovary
syndrome (PCOS). Hum Reprod. 19:41–47. 2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Christ JP and Cedars MI: Current
guidelines for diagnosing PCOS. Diagnostics (Basel).
13(1113)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
General Assembly of the World Medical
Association. World medical association declaration of Helsinki:
Ethical principles for medical research involving human subjects. J
Am Coll Dent. 81:14–18. 2014.PubMed/NCBI
|
|
43
|
Mousa M, Al-Jefout M, Alsafar H, Kirtley
S, Lindgren CM, Missmer SA, Becker CM, Zondervan KT and Rahmioglu
N: Prevalence of common gynecological conditions in the middle
east: Systematic review and meta-analysis. Front Reprod Health.
3(661360)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Leem S and Park T: An empirical fuzzy
multifactor dimensionality reduction method for detecting gene-gene
interactions. BMC Genomics. 18 (Suppl 2)(S115)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Turki A, Mtiraoui N, Al-Busaidi AS,
Khirallah M, Mahjoub T and Almawi WY: Lack of association between
genetic polymorphisms within KCNQ1 locus and type 2 diabetes in
Tunisian Arabs. Diabetes Res Clin Pract. 98:452–458.
2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Holmkvist J, Banasik K, Andersen G, Unoki
H, Jensen TS, Pisinger C, Borch-Johnsen K, Sandbaek A, Lauritzen T,
Brunak S, et al: The type 2 diabetes associated minor allele of
rs2237895 KCNQ1 associates with reduced insulin release following
an oral glucose load. PLoS One. 4(e5872)2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chen Z, Yin Q, Ma G and Qian Q: KCNQ1 gene
polymorphisms are associated with lipid parameters in a Chinese Han
population. Cardiovasc Diabetol. 9(35)2010.PubMed/NCBI View Article : Google Scholar
|